Abstract
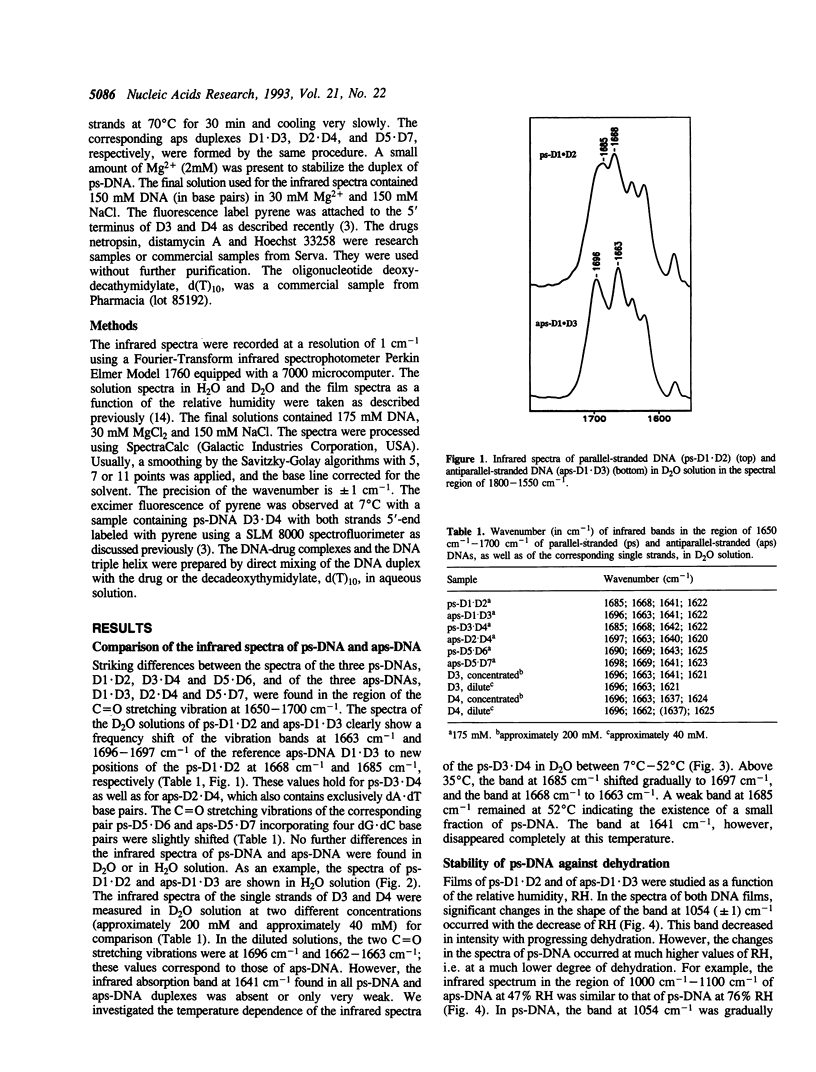
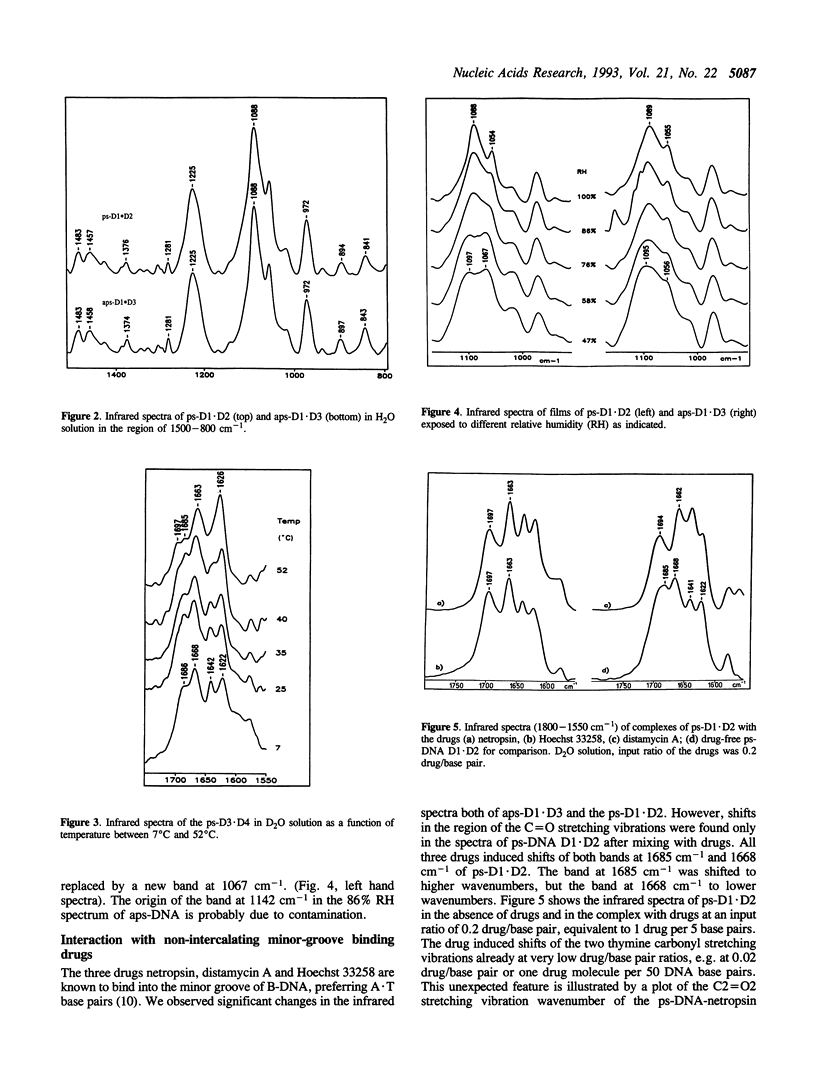
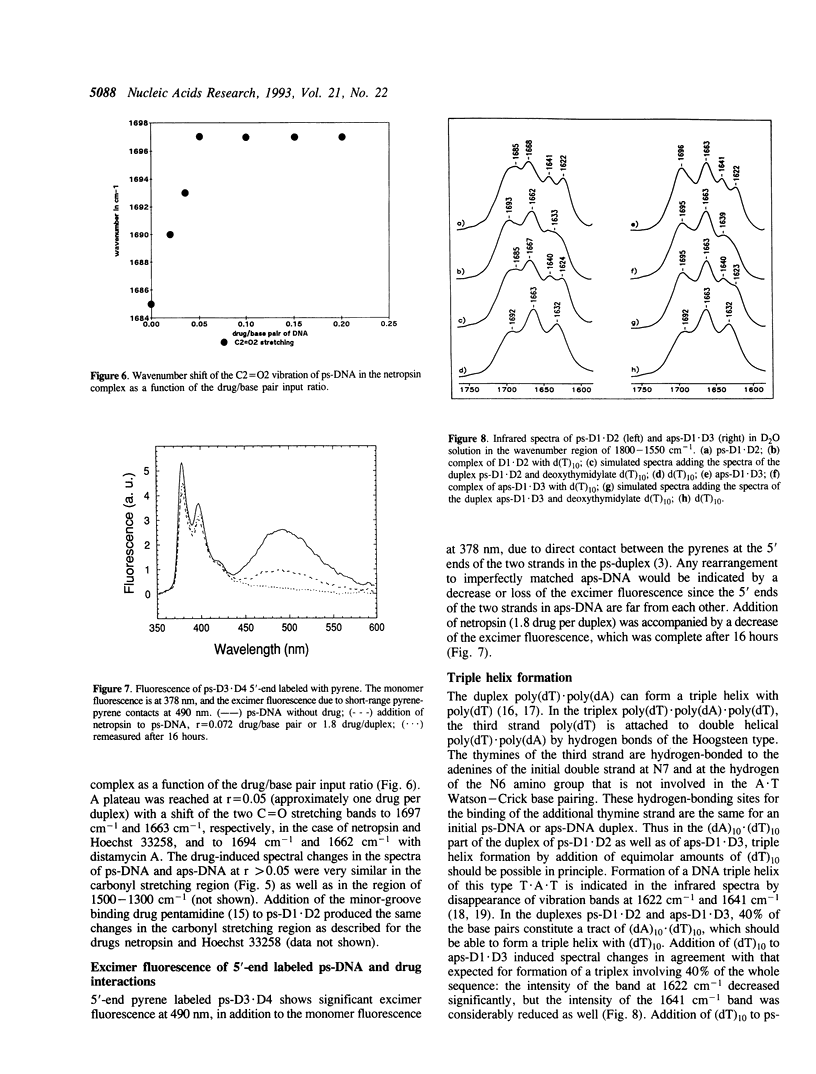
The infrared spectra of three different 25-mer parallel-stranded DNAs (ps-DNA) have been studied. We have used ps-DNAs containing either exclusively dA x dT base pairs or substitution with four dG x dC base pairs and have them compared with their antiparallel-stranded (aps) reference duplexes in a conventional B-DNA conformation. Significant differences have been found in the region of the thymine C = O stretching vibrations. The parallel-stranded duplexes showed characteristic marker bands for the C2 = O2 and C4 = O4 carbonyl stretching vibrations of thymine at 1685 cm-1 and 1668 cm-1, respectively, as compared to values of 1696 cm-1 and 1663 cm-1 for the antiparallel-stranded reference duplexes. The results confirm previous studies indicating that the secondary structure in parallel-stranded DNA is established by reversed Watson--Crick base pairing of dA x dT with hydrogen bonds between N6H...O2 and N1...HN3. The duplex structure of the ps-DNA is much more sensitive to dehydration than that of the aps-DNA. Interaction with three drugs known to bind in the minor groove of aps-DNA--netropsin, distamycin A and Hoechst 33258--induces shifts of the C = O stretching vibrations of ps-DNA even at low ratio of drug per DNA base pair. These results suggest a conformational change of the ps-DNA to optimize the DNA-drug interaction. As demonstrated by excimer fluorescence of strands labeled with pyrene at the 5'-end, the drugs induce dissociation of the ps-DNA duplex with subsequent formation of imperfectly matched aps-DNA to allow the more favorable drug binding to aps-DNA. Similarly, attempts to form a triple helix of the type d(T)n.d(A)n.d(T)n with ps-DNA failed and resulted in the dissociation of the ps-DNA duplex and reformation of a triple helix based upon an aps-DNA duplex core d(T)10.d(A)10.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam S., Bourtayre P., Liquier J., Taillandier E. Interaction of transition metal ions with Z form poly d(A-C).poly d(G-T) and poly d(A-T) studied by I.R. spectroscopy. Nucleic Acids Res. 1986 Apr 25;14(8):3501–3513. doi: 10.1093/nar/14.8.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adnet F., Liquier J., Taillandier E., Singh M. P., Rao K. E., Lown J. W. FTIR study of specific binding interactions between DNA minor groove binding ligands and polynucleotides. J Biomol Struct Dyn. 1992 Dec;10(3):565–575. doi: 10.1080/07391102.1992.10508668. [DOI] [PubMed] [Google Scholar]
- Arnott S., Selsing E. Structures for the polynucleotide complexes poly(dA) with poly (dT) and poly(dT) with poly(dA) with poly (dT). J Mol Biol. 1974 Sep 15;88(2):509–521. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- Cheng Y. K., Pettitt B. M. Stabilities of double- and triple-strand helical nucleic acids. Prog Biophys Mol Biol. 1992;58(3):225–257. doi: 10.1016/0079-6107(92)90007-s. [DOI] [PubMed] [Google Scholar]
- Edwards K. J., Jenkins T. C., Neidle S. Crystal structure of a pentamidine-oligonucleotide complex: implications for DNA-binding properties. Biochemistry. 1992 Aug 11;31(31):7104–7109. doi: 10.1021/bi00146a011. [DOI] [PubMed] [Google Scholar]
- Ghomi M., Letellier R., Liquier J., Taillandier E. Interpretation of DNA vibrational spectra by normal coordinate analysis. Int J Biochem. 1990;22(7):691–699. doi: 10.1016/0020-711x(90)90003-l. [DOI] [PubMed] [Google Scholar]
- Letellier R., Ghomi M., Taillandier E. Interpretation of DNA vibration modes. II--The adenosine and thymidine residues involved in oligonucleotides and polynucleotides. J Biomol Struct Dyn. 1987 Feb;4(4):663–683. doi: 10.1080/07391102.1987.10507667. [DOI] [PubMed] [Google Scholar]
- Liquier J., Mchami A., Taillandier E. FTIR study of netropsin binding to poly d(A-T) and poly dA.poly dT. J Biomol Struct Dyn. 1989 Aug;7(1):119–126. doi: 10.1080/07391102.1989.10507755. [DOI] [PubMed] [Google Scholar]
- Otto C., Thomas G. A., Rippe K., Jovin T. M., Peticolas W. L. The hydrogen-bonding structure in parallel-stranded duplex DNA is reverse Watson-Crick. Biochemistry. 1991 Mar 26;30(12):3062–3069. doi: 10.1021/bi00226a012. [DOI] [PubMed] [Google Scholar]
- Pattabiraman N. Can the double helix be parallel? Biopolymers. 1986 Sep;25(9):1603–1606. doi: 10.1002/bip.360250903. [DOI] [PubMed] [Google Scholar]
- Pilet J., Brahms J. Dependence of B-A conformational change in DNA on base composition. Nat New Biol. 1972 Mar 29;236(65):99–100. doi: 10.1038/newbio236099a0. [DOI] [PubMed] [Google Scholar]
- Rippe K., Fritsch V., Westhof E., Jovin T. M. Alternating d(G-A) sequences form a parallel-stranded DNA homoduplex. EMBO J. 1992 Oct;11(10):3777–3786. doi: 10.1002/j.1460-2075.1992.tb05463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K., Jovin T. M. Parallel-stranded duplex DNA. Methods Enzymol. 1992;211:199–220. doi: 10.1016/0076-6879(92)11013-9. [DOI] [PubMed] [Google Scholar]
- Rippe K., Ramsing N. B., Jovin T. M. Spectroscopic properties and helical stabilities of 25-nt parallel-stranded linear DNA duplexes. Biochemistry. 1989 Nov 28;28(24):9536–9541. doi: 10.1021/bi00450a043. [DOI] [PubMed] [Google Scholar]
- Rippe K., Ramsing N. B., Klement R., Jovin T. M. A parallel stranded linear DNA duplex incorporating dG.dC base pairs. J Biomol Struct Dyn. 1990 Jun;7(6):1199–1209. doi: 10.1080/07391102.1990.10508559. [DOI] [PubMed] [Google Scholar]
- TSUBOI M., KYOGOKU Y., SHIMANOUCHI T. Infrared absorption spectra of protonated and deprotonated nucleosides. Biochim Biophys Acta. 1962 Jan 22;55:1–12. doi: 10.1016/0006-3002(62)90925-3. [DOI] [PubMed] [Google Scholar]
- Taillandier E., Liquier J. Infrared spectroscopy of DNA. Methods Enzymol. 1992;211:307–335. doi: 10.1016/0076-6879(92)11018-e. [DOI] [PubMed] [Google Scholar]
- Taillandier E., Ridoux J. P., Liquier J., Leupin W., Denny W. A., Wang Y., Thomas G. A., Peticolas W. L. Infrared and Raman studies show that poly(dA).poly(dT) and d(AAAAATTTTT)2 exhibit a heteronomous conformation in films at 75% relative humidity and a B-type conformation at high humidities and in solution. Biochemistry. 1987 Jun 16;26(12):3361–3368. doi: 10.1021/bi00386a017. [DOI] [PubMed] [Google Scholar]
- Tchurikov N. A., Chernov B. K., Golova Y. B., Nechipurenko Y. D. Parallel DNA: generation of a duplex between two Drosophila sequences in vitro. FEBS Lett. 1989 Nov 6;257(2):415–418. doi: 10.1016/0014-5793(89)81585-6. [DOI] [PubMed] [Google Scholar]
- Zhou N., Germann M. W., van de Sande J. H., Pattabiraman N., Vogel H. J. Solution structure of the parallel-stranded hairpin d(T8<text text>C4A8) as determined by two-dimensional NMR. Biochemistry. 1993 Jan 19;32(2):646–656. doi: 10.1021/bi00053a033. [DOI] [PubMed] [Google Scholar]
- Zimmer C., Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47(1):31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]


