Abstract
The suprachiasmatic nucleus (SCN) of the hypothalamus is the master mammalian circadian clock. The SCN is highly specialized because it is responsible for generating a near 24 h rhythm, integrating external cues, and translating the rhythm throughout the body. Currently, our understanding of the developmental origin and genetic program involved in the proper specification and maturation of the SCN is limited. Herein, we provide a detailed analysis of transcription factor (TF) and developmental-gene expression in the SCN from neurogenesis to adulthood in mice (Mus musculus). TF expression within the postmitotic SCN was not static but rather showed specific temporal and spatial changes during prenatal and postnatal development. In addition, we found both global and regional patterns of TF expression extending into the adult. We found that the SCN is derived from a distinct region of the neuroepithelium expressing a combination of developmental genes: Six3, Six6, Fzd5, and transient Rx, allowing us to pinpoint the origin of this region within the broader developing telencephalon/diencephalon. We tested the necessity of two TFs in SCN development, RORα and Six3, which were expressed during SCN development, persisted into adulthood, and showed diurnal rhythmicity. Loss of RORα function had no effect on SCN peptide expression or localization. In marked contrast, the conditional deletion of Six3 from early neural progenitors completely eliminated the formation of the SCN. Our results provide the first description of the involvement of TFs in the specification and maturation of a neural population necessary for circadian behavior.
Introduction
The suprachiasmatic nucleus (SCN) of the hypothalamus acts as the master mammalian circadian clock generating a near 24 h rhythm through the cyclic expression of key genes, including crucial transcription factors (TFs). The ability of the SCN to integrate external cues and translate rhythms throughout the body reflects a unique specialization of the nucleus and neurons within (Herzog and Schwartz, 2002). Properties underlying the production of 24 h rhythms have been well characterized; however, the genes involved in specification and development of the SCN are unknown.
The SCN is a heterogeneous group of neurons able to generate unified rhythmic output from individual neuronal oscillators (Webb et al., 2009). Expression of arginine vasopressin (AVP) and vasoactive intestinal peptide (VIP) divide the nucleus into two subdivisions, termed dorsomedial/shell and ventrolateral/core, respectively. Although these gross divisions do not necessarily reflect differences in cellular properties, these areas have been shown to differ in their projection patterns (Leak and Moore, 2001), innervation (Moga and Moore, 1997; Abrahamson and Moore, 2001), and rhythmicity (Hamada et al., 2001; Yan and Okamura, 2002; Bendová et al., 2004; Karatsoreos et al., 2004).
There is an extensive literature describing the birth dating, onset of neurogenesis, and migration of neurons in the SCN of mammals. Initial experiments in rat estimated that the SCN is derived from neuroepithelium caudal to the optic recess during the third wave of hypothalamic development (Altman and Bayer, 1986). In addition, studies in mice have shown that SCN neurogenesis occurs in a ventrolateral to dorsomedial progression approximately 5 d before birth (Shimada and Nakamura, 1973; Okamura et al., 1983; Kabrita and Davis, 2008), consistent with previous findings in rat and hamster (Altman and Bayer, 1978, 1986; Davis et al., 1990; Antle et al., 2005). Additional information regarding the prenatal SCN remains limited, in part because of the difficulty in identifying SCN boundaries beyond cell density estimates before peptide expression. In addition, no single gene has yet been identified that unambiguously distinguishes all SCN neurons from adjacent neural populations.
Individual TFs can distinguish subsets of cells within heterogeneous populations and provide insight into both development and function (Dasen et al., 2005; Gray, 2008). To date, the study of TFs within the SCN has been either in their relation to clock function or as a consequence of focus on neighboring hypothalamic regions. Investigation of TFs specifically within the SCN may provide insight into SCN development, the presence of genetically distinct areas, and relationships between the SCN and other brain regions.
We hypothesized that SCN function is influenced by TF expression and that these genes play a role in both the formation and maturation of SCN rhythmicity. Thus, we provide a detailed spatiotemporal description of TF and developmental-gene expression within the mouse SCN. We find that the SCN undergoes prenatal and postnatal maturation of gene expression. Furthermore, we find that patterns of TF expression reveal a specific gene cascade from which SCN neurons are derived and can be used to identify the developing SCN before published markers. In addition, we demonstrate a crucial role for an evolutionarily conserved TF in SCN formation.
Materials and Methods
Animals and housing.
Male and female wild-type CD1 (Charles River), Six3flox (Guillermo Oliver, St. Jude Children's Research Hospital, Memphis, TN), RORαsg (002651; The Jackson Laboratory), and Nestin-cre (003771; The Jackson Laboratory) mice were used. Mice were maintained in a normal 12 h light/dark schedule under standard care conditions. Mutant animals were maintained on a C57BL/6 background (Charles River). All experiments were approved by the Animal Studies Committee at Washington University School of Medicine.
Genotyping.
Mice were genotyped by PCR for disrupted RORα gene, Six3 floxed, and/or wild-type alleles as described previously (Liu et al., 2006) or as outlined by The Jackson Laboratory. The presence of Cre was determined by the presence or absence of an amplified band using primers 5′ GAGTGAACGAACCTGGTCGAAATCAGTGCG 3′ and 5′ GCATTACCGGTCGATGCAACGAGTGATGAG 3′.
Tissue acquisition.
Unless otherwise stated, all collections were done at approximately Zeitgeber time 8 (ZT8). Neonatal pups [postnatal day 0 (P0) to P2] or embryos [embryonic day 10.5 (E10.5) to E18.5] from timed pregnant females (morning of plug was E0.5) were anesthetized on ice and either transcardially perfused (≥E16.5) or immersion fixed in 4% paraformaldehyde (PFA) in 0.1 m PBS, pH 7.4. Older mice were anesthetized with a ketamine/xylazine mixture before perfusion with 4% PFA. All tissues were postfixed in 4% PFA overnight at 4°C, cryoprotected in 20% sucrose in PBS, frozen in O.C.T. Compound Embedding Medium (Tissue-Tek), and stored at −75°C. Serial sections (14 or 20 μm) were cut on a Hacker cryostat and thaw mounted on Superfrost Plus slides (Thermo Fisher Scientific). Five adjacent sets of sections were prepared from each embryonic and postnatal age and stored at −20°C.
ZT tissue collection.
Tissue was collected as outlined above every 4 h for 24 h from animals (P21–P23) kept in a 12 h light/dark schedule starting at ZT2, 2 h after lights on (ZT0). For dark time points, animals were enucleated under red light before perfusion.
Probe synthesis.
Plasmids for in situ hybridization (ISH) probes were acquired from published sources (Gray et al., 2004; Burns et al., 2008; Zhang et al., 2008) or purchased from Open Biosystems. Gene fragments from verified plasmids were linearized by direct amplification by sequence or vector-specific PCR (gene, GenBank accession number, region, forward primer, reverse primer); AVP, BC051997, GTAATACGACTCACTATAGG (T7), AATTAACCCTCACTAAAGGG (T3); BMP7, BC010771, 281–993, GCCTCTGTTCTTGCTGCGCTCC, CCTGGATGGGCAGAGCATCA; Dlx2, BQ443330, 75–797, CGGGCTCCGCTTCACACCTG, ACGGAGTGTCCTGGGAAAGTGGA; Hmx2, BC023402, T7, ATTTAGGTGACACTATAG (SP6); Per1, BC039768, 532–1750, CCCAGAAAGAACTCATGACTG, GCATCTGGTAAAGCACCAGGG; Ptch1, CB723783, T7, T3; RORα, BC003757, 273–1054, GCCACCTACTCCTGTCCTCG, GCAGGCTCGCTAGAGGTGGTGT; RORβ, BC024842, 331–987, CAGGAACCGTTGCCAACACTGC, CCAGTACGTGGTGGAGTTCGC; Rx, BC024731, 539–1261, CCCGGGATTCGTCCCGGAGT, TTGGCCTTCAGGCGCAGAGC; sonic hedgehog (SHH), BC063087, 51–998, GAGACCCAACTCCGATGTG, GAAAGCAGAGAACTCCGTGGC. Digoxigenin (DIG)-labeled antisense and sense RNA probes were made using PCR products as template and T7, T3, or SP6 RNA polymerases (Roche). cRNA probes were purified using Quick Spin columns (Roche) and quantified by spectrophotometry. Probes were used at a concentration of 1–2 μg/ml. Sense counterparts of all probes were tested to ensure probe specificity.
In situ hybridization.
Slides were immersed in 4% PFA, permeabilized with proteinase K, and returned to 4% PFA before being washed in 0.1 m triethanolamine-HCl with 0.25% acetic anhydride. Once blocked in hybridization buffer at 65°C slides were incubated in hybridization buffer containing 1–2 μg/ml DIG-labeled antisense cRNA overnight at 65°C. Slides were then washed in 2× SSC at 62°C, washed in 0.2× SSC at 65°C, blocked with 10% normal horse serum (NHS) in 0.1 m PBS, and incubated in alkaline phosphatase-labeled anti-DIG antibody (1:2000 in 10% NHS; Roche) overnight. Sections were washed and color was visualized using nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Roche). Staining was stopped after visual inspection. Sections were washed, fixed in 4% PFA, and coverslipped in 90% glycerol, Vectashield Mounting Medium (Vector Laboratories), or UltraCruz Mounting Media with 4′,6′-diamidino-2-phenylindole (DAPI) (Santa Cruz Biotechnology).
Immunohistochemistry.
Sections on slides were dried at room temperature and rinsed in 0.5% Triton X-100 in PBS (PBS-T) before incubating in 10% NHS in PBS-T. Slides were then placed into primary antibody overnight at 4°C. Primary antibodies used include the following: rabbit (Rb) AVP (1:800; Immunostar), Rb Ki67 (1:500; Thermo Fisher Scientific), goat (Gt) Lhx2 (1:500; Santa Cruz Biotechnology), Gt Lhx9 (1:750; Santa Cruz Biotechnology), mouse TuJ1 (1:1000; Covance), and Rb VIP (1:8000; Immunostar). Primary antibodies were removed, and sections were rinsed in PBS-T before incubation in secondary antibodies for 2 h at room temperature. Secondary antibodies used were either the appropriate Alexa Fluor 488 (1:1000; Invitrogen) or cyanine 3 (1:500; Jackson ImmunoResearch). Sections were washed and coverslipped with UltraCruz Mounting Media with DAPI.
Image acquisition.
Images were acquired using a Nikon Eclipse 90i microscope, Photometrics Coolsnap HQ2 camera with a Prior Scientific ProScan II motorized translation stage, and acquired in Volocity (PerkinElmer Life and Analytical Sciences) or a Nikon Coolscan V Ed slide scanner with SilverFast SE software (LaserSoft Imaging). Images were exported as eight-bit JPEG or TIFF files. All images were adjusted for clarity by filtering and/or modifying levels, as necessary, in Photoshop (Adobe Systems).
mRNA quantification and analysis.
Percentage of SCN area containing either Lhx1 or RORα expression was calculated by dividing area expressing mRNA by the total estimated SCN area marked with DAPI, in which area was calculated by outlining regions of interest within NIH ImageJ. Whole SCN quantifications of mRNA expression were performed by quantifying intensity of staining relative to background from 10× ISH images using NIH ImageJ. Margins of the SCN in E17.5 to adult animals were determined by cell density marked with DAPI nuclear stain. For statistical analysis of diurnal variation, one-way ANOVA was applied to relative intensity measurements obtained from ZT2, ZT6, ZT10, ZT14, and ZT18 for Six3, RORα, and Per1. The differences of mRNA relative intensity between ZT6 and ZT18 were analyzed using Student's t test.
Results
Identification of SCN candidate transcription factors
The SCN is a specialized neural structure with an essential behavioral function. We hypothesized that aspects of SCN neurons and SCN function are a direct consequence of the TFs they express and that underlie specification. We set out to identify TFs that might be important for both the development and function of the adult SCN by reexamining a published analysis of nearly 1000 TF and TF-related gene expression patterns in mice at E13.5 and P0 (Gray et al., 2004). Two hundred twenty TFs were previously identified as restricted to populations within the hypothalamus of E13.5 and/or P0 mice. From these data as well as other published work (Rivkees et al., 1992; Schaeren-Wiemers et al., 1997; Jean et al., 1999; Wang and Lufkin, 2000), we identified 72 TFs as possibly expressed in discrete patterns of the postnatal (P0; supplemental Table 1, available at www.jneurosci.org as supplemental material) and/or prenatal (E13.5; supplemental Table 2, available at www.jneurosci.org as supplemental material) anterior hypothalamus that includes the SCN. Of these 72 TFs, 28 appeared potentially expressed within the postnatal SCN itself (supplemental Table 1, available at www.jneurosci.org as supplemental material). We confirmed the perinatal or adult hypothalamic expression of at least 68% of these 28 TFs using data from the Allen Brain Atlas (Lein et al., 2007).
SCN transcription factor expression is diverse and dynamic
Previous large screens of gene expression within the brain, although extremely informative, have generally lacked the spatial and/or temporal precision to determine the detailed boundaries of expression within the SCN (Gong et al., 2003; Gray et al., 2004; Lein et al., 2007; Shimogori et al., 2010). To directly address the range of TF expression patterns within the early SCN, we selected a subset of nine postnatal TFs [Lhx1, Lhx2, Lhx9, Otp, Pou2f2 (Oct-2), RORα (Rora), RORβ (Rorb), Six3, and Six6] for detailed ISH or immunohistochemical (IHC) analysis within the P0 mouse SCN. These genes were selected based on their relative levels of expression and specificity within the SCN region and/or previous work, suggesting a possible role in aspects of SCN function (Rivkees et al., 1992; Schaeren-Wiemers et al., 1997; Sato et al., 2004).
We found that TFs showed a range of expression patterns suggesting a diverse transcriptional environment (Fig. 1). Of the nine postnatal TFs analyzed, six were found specifically expressed in the SCN but not in adjacent hypothalamic nuclei. Compared with a cell density estimate of SCN margins, Six3, Six6, RORα, and Pou2f2 were expressed throughout the entire SCN, whereas Lhx1 and RORβ both displayed regional specificity within the SCN. Conversely, we found two TFs (Lhx2 and Otp) expressed at lateral and dorsal boundaries but not within the SCN. Lhx9 was neither expressed within the SCN nor served as a boundary marker (data not shown).
Figure 1.
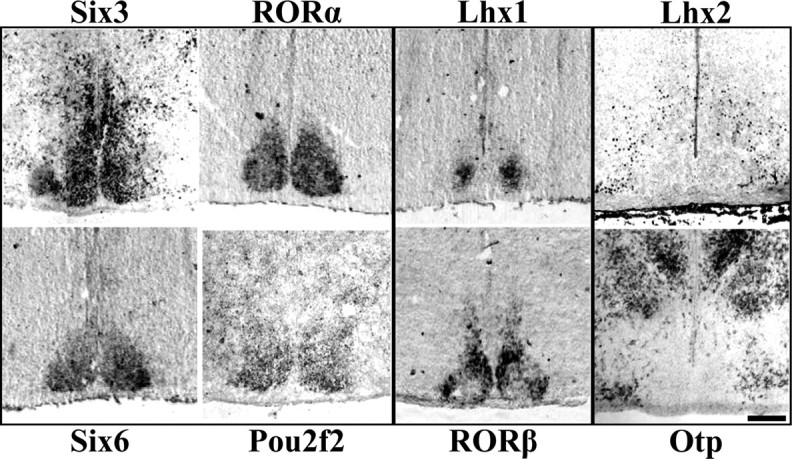
Diverse patterns of transcription factor expression in P0 mouse SCN. Representative ISH and IHC images of mid-SCN coronal sections showing specific expression throughout the entire SCN but not adjacent hypothalamus for Six3, RORα, Six6, and Pou2f2, but restricted localization within the SCN for Lhx1 and RORβ. Lhx2 and Otp are expressed outside the SCN, indicating a transcription factor boundary. Scale bar, 250 μm.
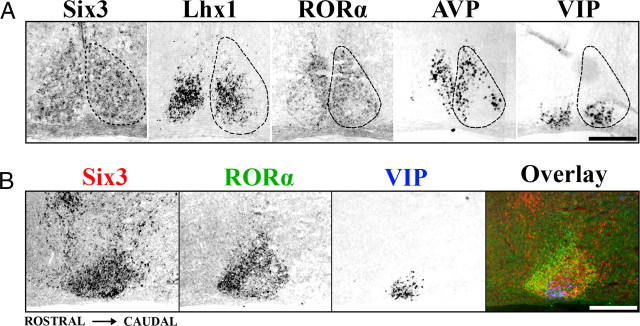
To determine whether representative patterns of early postnatal TF expression are modified by postnatal maturation, we examined Six3, Lhx1, and RORα further for expression at P15, just after eyes open. Six3 retained expression throughout the P15 SCN (Fig. 2A). Similar to expression at P0, Lhx1 displayed regionalized specificity within the SCN at P15. Expression of Lhx1 covered 52 ± 2.6% of the central SCN and partially overlapped both SCN ventrolateral/core and dorsomedial/shell (Fig. 2A). In contrast to expression at P0, RORα was no longer present throughout the entire SCN; instead, expression was restricted to only the outer 71 ± 2.2% of the SCN, similar in distribution to AVP and RORβ. RORα expression did not overlap the VIP-expressing region, seen clearly in both transverse (Fig. 2A) and sagittal (Fig. 2B) sections through the SCN. We found that expression patterns of Six3, Lhx1, and RORα at P15 were identical to those at P21 and adult (data not shown).
Figure 2.
The adolescent mouse SCN shows discrete patterns of transcription factor expression. A, Representative ISH images showing localization of Six3, Lhx1, and RORα expression within coronal sections of mid-SCN at P15. Note the discrete localization of RORα to a region overlapping AVP but not VIP expression, whereas Lhx1 overlaps both. Dotted lines indicate the margins of the SCN. B, ISH images from adjacent sagittal sections of Six3 (red), RORα (green), and VIP (blue) are overlaid in pseudocolor, showing the full rostral to caudal expression of Six3 throughout the SCN and the localized absence of RORα from the VIP-expressing region. Scale bars, 250 μm.
Six3 and RORα show diurnal variation
The SCN can entrain to photoperiods (Sumova et al., 2006) and can be considered mature by P21. Also, by this time, the amplitude of circadian changes in expression level of clock genes has reached adult levels (Kováciková et al., 2006). For clock genes, the differences in phase of expression are a consequence of the interactions in the transcription–translation negative feedback loop that underlie circadian rhythmicity. Much of our understanding of the phasic expression of genes within the SCN has relied on quantitative PCR from whole SCN lysates or radioactive ISH, both of which have limited spatial sensitivity (Sunkin, 2006).
Under standard 12 h light/dark conditions, the mRNA levels of central clock genes Per1, Bmal1, and Cry1 peak and trough at mid-day (ZT6) and mid-night (ZT18) (Oishi et al., 2000; Shearman et al., 2000), with Bmal1 and Cry1 in anti-phase of the mid-day peak of Per1. We tested with nonradioactive ISH whether Per1, Lhx1, RORα, and Six3 mRNA displayed variations at these time points. We found that changes in Six3 and Per1 were significantly different between mid-day and mid-night (p = 0.00064 and 0.028, respectively, Student's t test), whereas RORα and Lhx1 levels were not (p = 0.055 and 0.1065, respectively).
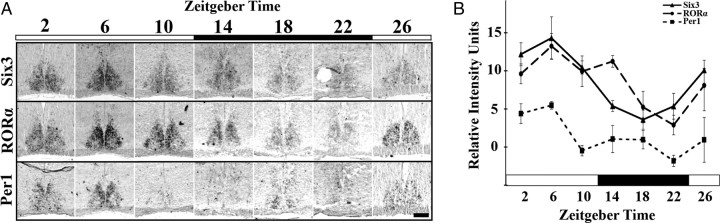
The spatial localization of RORα at ZT8 in the mature SCN suggested that any change in phase of expression might correspond to changes in spatial localization as well as abundance. We tested the expression of Six3 and RORα for changes in mRNA intensity and localization of mRNA over a diurnal period (n = 3 for each time point). We found that RORα displayed temporal variation within the SCN (one-way ANOVA, F(5,47) = 6.64, p = 0.0001), with peak at ZT6 and trough at ZT22, in phase with Per1 (one-way ANOVA, F(5,21) = 4.56, p = 0.0089) (Fig. 3A,B), consistent with previous PCR-based analyses (Sato et al., 2004). RORα was never expressed within the VIP-expressing region, which indicated a previously unrecognized spatial localization. Six3 also showed diurnal variations in expression, with peak at ZT6 and trough at ZT18 (one-way ANOVA, F(5,47) = 10.55, p < 0.0001) (Fig. 3A,B).
Figure 3.
Six3 and RORα show diurnal variation in expression. A, Representative ISH images showing expression of Six3, RORα, and Per1 mRNA in the coronal mid-SCN in P21–P23 mice raised under a 12 h light/dark cycle and sampled every 4 h (ZT0 indicates lights on). In contrast to Per1 and Six3, RORα is expressed only in the outer shell region. B, Quantification of averaged relative mRNA intensity in arbitrary units for Six3, RORα, and Per1 over the same 24 h time period, showing diurnal variation for Six3 (one-way ANOVA, F(5,47) = 10.55, p < 0.0001), RORα (one-way ANOVA, F(5,47) = 6.64, p = 0.0001), and Per1 (one-way ANOVA, F(5,21) = 4.56, p = 0.0089), with peak at ZT6 and trough at ZT22 for RORα and Per1 and ZT18 for Six3 (n = 3). Black bars indicate 12 h of dark. Error bars show SEM. Scale bar, 250 μm.
Transcription factors identify the prenatal SCN
Although the SCN is not functionally mature until postnatal stages, early components of rhythmicity are present in late embryonic life. Some of the earliest rhythms were seen in day/night variations in metabolic activity as early as E19 in the rat (Reppert and Schwartz, 1984) as well as individual rhythmically firing neurons at E22 in the rat (Shibata and Moore, 1987), which corresponds to approximately E16.5 and E19.5 in mouse, respectively. The expression of Six3, Lhx1, and RORα in both the early postnatal and mature SCN suggested that these same genes might provide insight into the development of the SCN during the prenatal stage in which cellular rhythmicity emerges.
To clearly identify the perinatal SCN, we determined the onset and localization of AVP and VIP by ISH and IHC. Consistent with previous work in other rodents (Romero and Silver, 1990; Isobe et al., 1995; Ban et al., 1997), we found both AVP and VIP mRNA expression prenatally but protein-labeled cell bodies only at postnatal stages (data not shown). AVP mRNA expression was present in the SCN by E17.5 (two of three animals), while already strongly expressed in the supraoptic nuclei (SON) at this time (data not shown). VIP mRNA expression was present in the SCN by E18.5 (three of five animals) (data not shown). Within the SCN, both peptides were expressed in spatially restricted regions similar to that seen in the adult from their onset, suggesting that specification of peptidergic identity occurs before E17.5. The global expression of Six3 and RORα in the SCN at P0 led us to theorize that their expression would identify the boundaries of the SCN during prenatal maturation. Thus, we analyzed the expression of Six3, Lhx1, and RORα between the end of neurogenesis at E14.5 and birth (P0).
At E14.5, Six3 was expressed within a portion of the early hypothalamus in both the presumptive SCN region as well as dorsal areas. In contrast, Lhx1 was present in discrete bilateral regions distal to the third ventricle within the broader Six3 domain (Fig. 4). A small subset of the ventral most Lhx1/Six3 region was found to express RORα in two of five animals examined, suggesting that the onset of RORα (Fig. 4) occurs at this time point. At E15.5, before the onset of VIP or AVP mRNA expression, there was an increase in the area and approximate number of cells expressing Lhx1 and RORα. Additionally, nuclear staining with DAPI revealed the first signs of aggregation into a cell-dense nucleus at this stage (Fig. 4, white arrow). However, both Lhx1 and RORα expression extended beyond the newly forming, cell-dense area, which would be consistent with the presence of neurons that will become the SCN before the onset of clustering. We hypothesized that these genes could be used as prenatal genetic markers to define cells of the SCN more precisely than cell density estimates. By E17.5, RORα expression encompassed the entire SCN and completely overlapped with Six3, whereas Lhx1 expression was reduced to a central portion of the SCN (Fig. 4). Six3 expression, however, also marked cells more dorsal and lateral to the SCN (data not shown). Postnatally, these dorsal populations retained their Six3 expression and comprised the anterior paraventricular nucleus (aPV) and subparaventricular regions of the hypothalamus (data not shown). By P0, Six3 and RORα expression completely overlapped cell density estimates of SCN margins, whereas Lhx1 continued to remain centralized (Fig. 4).
Figure 4.
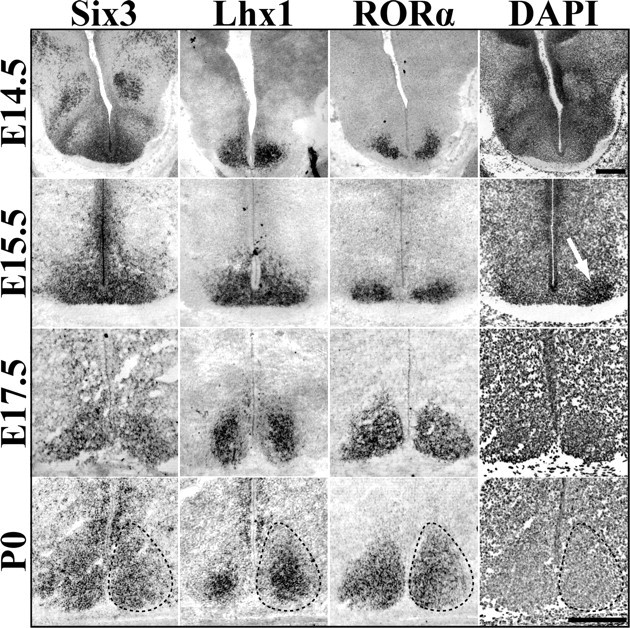
Dynamic transcription factor expression during SCN prenatal maturation. Representative ISH images of Six3, Lhx1, and RORα mRNA expression in the embryonic mouse SCN (E14.5 to P0), showing spatial specificity and prenatal maturation of transcription factor expression. Dotted lines indicate margins of SCN estimated from the cell density marker DAPI (example shown in rightmost panels). White arrow at E15.5 indicates onset of visible clustering of cells. Note change in Lhx1 pattern from global to centrally restricted between E15.5 and P0. E17.5 Lhx1 image was obtained from tissue cut at a different angle and thus the appearance of larger SCN compared with Six3 and RORα. Scale bar, 250 μm.
These data suggest that TFs can be used to identify the prenatal SCN before the onset of peptide markers, dense clustering of cells, and onset of rhythmic activity. The data further suggest that the onset of RORα expression occurs after SCN neurogenesis is complete, consistent with literature describing the delayed onset of RORα expression within other brain regions (Nakagawa and O'Leary, 2003). In addition, these data indicate that the SCN is not fully specified by the end of neurogenesis and that SCN maturation occurs in stages both prenatally and postnatally.
RORα does not specify postnatal SCN structure
In a molecular model of the circadian clock, RORα positively regulates Bmal1, thereby aiding in the stability of the core rhythm (Sato et al., 2004). Knock-outs of RORα show slight behavioral abnormalities in circadian rhythms, leading to a shortened circadian period (Sato et al., 2004). To date, it is unknown whether the change in circadian period is attributable solely to the interaction of RORα with Bmal1 or whether there is an anatomical change within the SCN. Therefore, we investigated the role of RORα in the development of the SCN by comparing the localization of SCN markers in wild-type mice with staggerer mice (RORαsg/sg) containing a deletion of the fifth exon in the RORα gene that causes formation of a premature stop codon and a functionally null protein (Hamilton et al., 1996).
We found that the expression of SCN markers in RORαsg/sg mice were normal. The spatial mRNA expression patterns of AVP and VIP within the SCN as well as that of SCN boundary marker Otp in the hypothalamus (data not shown) remained unaltered in all P15 sg/sg mutants (four of four) compared with wild-type littermates (Fig. 5). Thus, although we found that the postnatal expression of RORα anatomically distinguished the core of the SCN from its shell, RORα expression was not necessary for the differentiation, presence, or segregation of peptidergic cells within the SCN.
Figure 5.
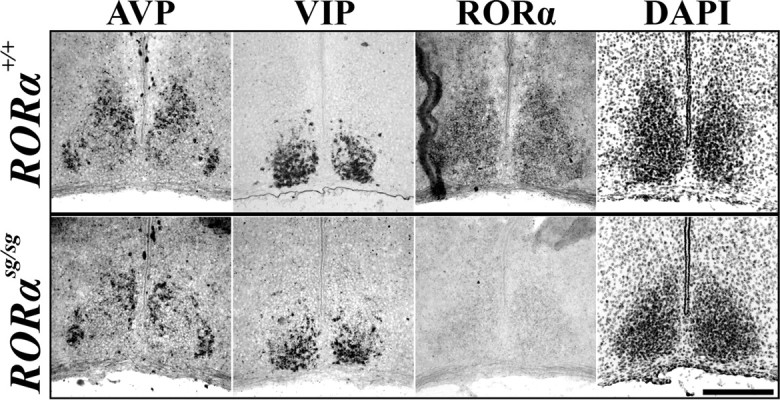
RORα is not necessary for normal SCN peptide localization. Representative ISH images of AVP and VIP mRNA expression, markers of SCN subdivisions, as well as transcription factor RORα and inverted DAPI nuclear stain in P15 RORα mutant (sg/sg, bottom) and wild-type littermate (+/+, top) mice. Note that segregation of peptidergic expression patterns was similar to littermate control. Scale bar, 250 μm.
Identification of the SCN neuroepithelium
The ability of Six3 and Lhx1 to delineate the prenatal, postmitotic SCN suggested that they might also precisely define the suprachiasmatic neuroepithelium within the early telencephalon/diencephalon boundary zone beyond previous anatomical estimates (Altman and Bayer, 1986). Therefore, we assessed the expression of these genes just before neurogenesis, at E10.5 and during neurogenesis (E11.5–E13.5) in mice. Specifically, we examined the region from the rostral eye fields extending caudally to the developing pituitary to identify the exact spatial origin of the SCN.
At E10.5, Six3 marked a broad dorsoventral and rostrocaudal region of the developing floor plate (Fig. 6A and data not shown), consistent with previous reports (Oliver et al., 1995). At E11.5, Six3 was restricted to more ventral regions of the early telencephalon/diencephalon but remained expressed within the developing hypothalamus and pituitary (Fig. 6A,D). Lhx1 expression began at E11.5 in a very small band of cells at the ventral edge of the Six3-expressing zone (Fig. 6A, arrow), allowing for the identification of the early SCN within the telencephalon/diencephalon boundary region (Fig. 6D). Both a dorsal and rostrocaudal expansion in the number of cells expressing Lhx1 occurred from E12.5 to E13.5, whereas Six3 expression became more restricted but still surrounded the base of the third ventricle (Fig. 6A). The coincident onset of and increase in Lhx1 expression during the period of SCN neurogenesis suggests that Lhx1 is one of the first TFs to be expressed in fully postmitotic SCN neurons.
Figure 6.
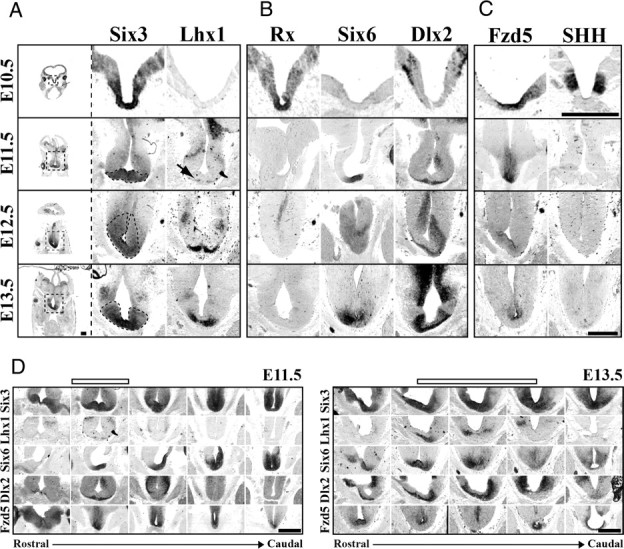
SCN is derived from a distinct progenitor domain. Representative images of Six3 and Lhx1 (A), Rx, Six6, and Dlx2 (B), and Fzd5 and SHH (C) mRNA expression (columns) in SCN progenitor domain from E10.5 to E13.5 (rows). Leftmost column shows entire coronal section, with dotted box indicating approximate region of enlarged images in corresponding row to the right. Dotted black lines in enlarged Six3 images indicate hypothesized early SCN margins outlined by Six3 expression from E11.5 to E13.5. Note at E10.5 that the region that will give rise to SCN neurons expresses a combination of Six3, Rx, and Fzd5. By E11.5, Rx expression is lost and Six6 expression begins. Also note the E11.5 onset of Lhx1 (black arrow) and Dlx2 on the most ventral aspects of tissue beneath the third ventricle. Scale bar, 500 μm. D, Rostrocaudal location of SCN progenitor domain. ISH images depict patterns of Six3, Lhx1, Six6, Dlx2, and Fzd5 mRNA expression (rows) in transverse plane from rostral eye fields to caudal developing pituitary at E11.5 and E13.5. Open bars above indicate the developing SCN within the rostrocaudal axis. Note that, although expression of Six3 and Six6 is consistent along the ventral surface from rostral eye fields to the early pituitary, Lhx1 and Dlx2 mark discrete zones within this region at both E11.5 and E13.5. Please note in D that E11.5 depicts same transverse images as shown in A–C, and E13.5 shows the same Six6 and Fzd5 images shown in A–C now within the rostrocaudal context of the developing brain. Scale bar, 500 μm.
Relationship of early SCN to developmental transcription factors
Identification of the suprachiasmatic neuroepithelium suggested that the SCN was derived from a highly discrete region of the developing neural plate. Our initial reanalysis of brain TF expression identified 58 TFs with discrete expression in the area of the E13.5 anterior hypothalamus (supplemental Table 2, available at www.jneurosci.org as supplemental material). We confirmed the embryonic hypothalamic expression of at least 66% of these 58 TFs using data from Genepaint and Eurexpress expression databases (Visel et al., 2004; Diez-Roux et al., 2011). These TFs included early forebrain genes Six6 and Dlx2. In addition, reduction in the amount of Rx (Rax) present in early neuroepithelium has been shown to disrupt SCN location and cell number by an unknown mechanism (Silver, 1977; Tucker et al., 2001). To clarify the origins of the SCN within the developing telencephalon/diencephalon, we compared the expression of Six3 and Lhx1 with that of early forebrain genes Six6, Dlx2, and Rx.
At E10.5, Rx expression overlapped that of Six3 in the early ventral floor plate (Fig. 6B) from the early eye fields through the early developing pituitary (data not shown), consistent with a previous analysis (Bailey et al., 2004). At E11.5, a rostrocaudal gap was evident in Rx expression. Rx persisted within the eye itself and in ventral floor plate just rostral to the developing pituitary (data not shown). Six6 expression was first seen at E11.5 in a ventrally restricted but overlapping pattern with Six3 within the entire rostrocaudal region examined, including within this Rx-negative gap (Fig. 6B,D). Dlx2 expression was first seen in a small band of ventral cells within the Six3/Six6-expressing and Rx-negative domain (Fig. 6B,D). Lhx1 was expressed in a subset of the Dlx2-expressing domain (Fig. 6D).
By E12.5 and E13.5 as Six3/Six6 expression continued, the Dlx2-expressing region expanded rostrocaudally (Fig. 6D) and dorsally both beneath as well as along the lateral aspects of the third ventricle stratifying the region surrounding the base of the early third ventricle into three subregions (Fig. 6B). The ventralmost portions of Dlx2 expression overlapped with Lhx1 expression. The mid region retained only Six3/Six6/Dlx2, and the most dorsal aspects lacked Dlx2 (Fig. 6B). The overlap between Dlx2 and Lhx1 in the most ventral regions suggested that this combination marked newly postmitotic neurons of the SCN, consistent with findings in embryonic rat. The presence of an Lhx1-negative, Dlx2-positive region suggested that Dlx2 might be expressed during the transition period of cells during the last stages of mitosis to early stages of differentiation similar to its expression within the ventral thalamus (Andrews et al., 2003).
The absence of a single TF specific to the SCN suggested that SCN development might also rely on extrinsic morphogens and other signaling peptides to provide key regionalization signals (Burns et al., 2008; Liu et al., 2010). The expression of Six3, Six6, and Dlx2 in the presumptive suprachiasmatic neuroepithelium can be used to identify the immature SCN within the complex early signaling environment of the developing telencephalon/diencephalon. We found that, at E10.5, the Six3/Rx region also expressed Frizzled5 (Fzd5), a Wnt receptor, in a region surrounding the base of the third ventricle (Fig. 6C). This broad dorsal to ventral Six3/Rx/Fzd5 region was further demarcated by the expression of SHH. Six3/Rx/Fzd5 regions lateral to the ventricle expressed SHH, although it remained absent from the most ventral medial region at the base of the third ventricle (Fig. 6C). Unlike Rx, Fzd5 expression continued to overlap with Six3/Six6 at E11.5, further defining the SCN proliferative zone in both the dorsoventral (Fig. 6C) and rostrocaudal (Fig. 6D) dimensions. Fzd5 expression at E12.5 and E13.5 was still present beneath the third ventricle but was restricted to the dorsal region overlapping with the Six3/Six6 region only (Fig. 6C,D). Fzd5 expression did not overlap with the region containing postmitotic Lhx1 cells and Dlx2, which suggested the expression of Fzd5 likely marked regions of active proliferation. We found that, at E12.5, the most ventral Dlx2- and Lhx1-expressing regions overlapped with TuJ1, a marker of postmitotic neurons, whereas the more dorsal Dlx2-negative region expressed Ki67, a marker of proliferating cells (data not shown). Throughout this period of neurogenesis, the early SCN was completely devoid of bone morphogenetic protein 7 and lacked the common SHH receptor Patched1 (data not shown).
Our data suggest that the SCN is derived from a proliferative region expressing Six3, Six6, Fzd5, and transient Rx. Furthermore, early postmitotic cells of the SCN express Six3, Six6, Lhx1 and transiently express Dlx2. Figure 7 shows a summary of the relative patterns of developmental-gene expression during SCN development.
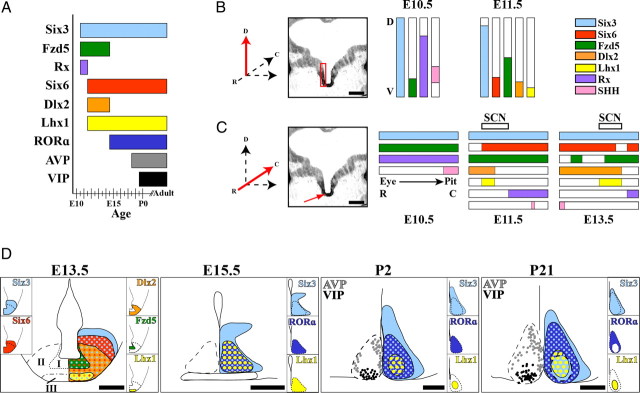
Figure 7.
Summary of SCN development. A, Bar graph indicates onset and duration of transcription factor and peptide mRNA expression from E10.5 to adult in the mouse SCN. B, C, Diagrams summarize dorsoventral and rostrocaudal extent of developmental gene expression within the early telencephalon/diencephalon containing the hypothesized early SCN. Red arrows on orientation axes show directionality of diagram in relation to indicated red regions. B, Summary of dorsoventral (D–V) extent of mRNA expression (colored bars) within the same region of anterior hypothalamus, indicated by red box in image showing Six3 expression, at E10.5 (left) and E11.5 (middle). Scale bar, 250 μm. Colors correspond to genes in legend (right). Note the absence of SHH but presence of Fzd5 within the ventral most domains, i.e., putative early SCN. C, Summary of rostrocaudal (R–C) extent of mRNA expression within the midline (red arrow) of the ventral developing telencephalon/diencephalon at E10.5 (left), E11.5 (middle), and E13.5 (right). Scale bar, 250 μm. The most rostral point is marked by the retina (Eye) and caudal by the developing pituitary (Pit). Colors correspond to genes in legend in B. Open top bars at E11.5 and E13.5 indicate rostrocaudal extent of putative SCN progenitor domain within larger developing brain. D, Diagrams of SCN transcription factor localization indicate expression during neurogenesis (E13.5), within the prenatal postmitotic (E15.5), early postnatal (P2), and functionally mature SCN (P21). Dotted lines on left indicate the margins of the SCN. Insets indicate expression pattern of individual transcription factors. Colored dots indicate regions of coexpression. Note hypothesized zones of developing SCN at E13.5: I, premitotic/proliferative; II, late-mitotic/early-postmitotic; III, postmitotic. Also note the changes in localization of both Lhx1 and RORα over time. Scale bars, 250 μm.
Six3 is required for SCN formation
In the present study, we found that Six3 was continually expressed in the SCN and its progenitors from before neurogenesis to adult when expression was diurnal. This suggested that Six3 has an important role in the SCN and its development. Therefore, we tested whether Six3 is necessary for the formation and maturation of the SCN using a Nestin-cre transgenic to eliminate Six3 (Six3flox/flox) (Liu et al., 2006) specifically in progenitors within the neural plate before specification.
We analyzed the anatomy and expression of Six3 mRNA in Six3flox/flox/Nestin-cre+ mutants (n = 5), Six3flox/flox/Nestin-cre− (n = 3), and Six3flox/+/Nestin-cre+ (n = 2) littermate controls at E18.5–E19.5. We found the Nestin-cre transgenic exhibited a range of Six3 loss in the developing mouse brain, from complete loss to no loss (Fig. 8), likely as a result of variation in the onset and duration of cre-recombinase expression. Six3flox/flox/Nestin-cre+ mice with complete Six3 deletion (n = 4 of 5) showed holoprosencephaly and a noticeable enlargement and/or merger of the lateral and third ventricles not seen in littermate controls (data not shown). Furthermore, Six3 ablations lead to loss of the pituitary (Fig. 8B). In contrast, retina and optic nerves remained intact (data not shown).
Figure 8.
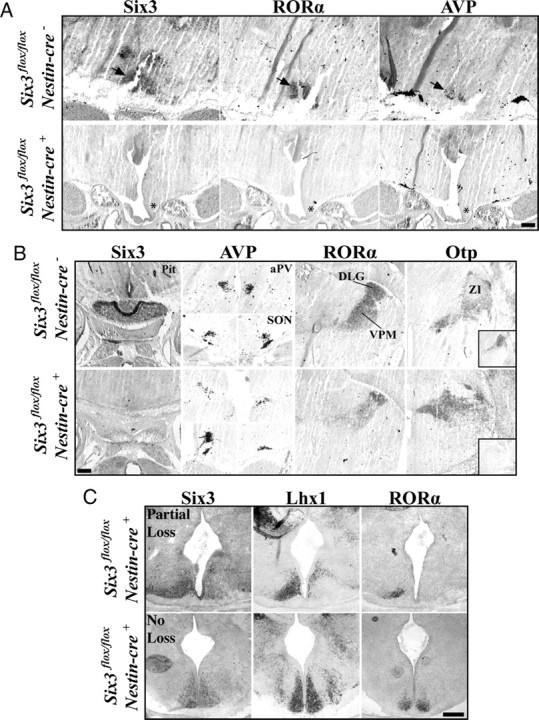
Six3 is necessary for SCN formation. Expression of Six3, RORα, AVP, and Otp in Six3flox/flox/Nestin-cre+ compared with Six3flox/flox/Nestin-cre− littermate controls. A, Expression of Six3, RORα, and AVP in SCN (arrows in top) are completely eliminated in conditional Six3 knock-outs, shown at E18.5 (asterisks). B, Marker analysis of the pituitary, hypothalamic (aPV and SON), and thalamic (dLG, VPM, and ZI) nuclei show that complete disruption of Six3 leads to loss of the pituitary, whereas nuclei surrounding the SCN remain intact in Six3flox/flox/Nestin-cre+ experimental animals. Bottom right inset in Otp column represents adjacent section for Six3 expression. C, Variability in the targeted elimination of floxed Six3 in E15.5 mice. Expression of Six3, Lhx1, and RORα in Six3flox/flox/Nestin-cre+ mice that showed either partial loss (top) or no loss (bottom) of Six3. Top, Unilateral Six3 deletions lead to unilateral loss of RORα and Lhx1. Bottom, Mice that retain Six3 expression still express both Lhx1 and RORα. Scale bars, 250 μm.
Six3flox/flox/Nestin-cre+ E18.5 and E19.5 animals with complete bilateral loss of Six3 expression showed no expression of RORα or AVP in the ventral anterior hypothalamus (Fig. 8A, asterisks). Importantly, loss of Six3 did not lead to the displacement of the SCN because no ectopic nuclei were present along the ventral surface either rostral or caudal to the optic chiasm, as assessed through a DAPI nuclear stain (data not shown). Loss of RORα and AVP was SCN specific because the aPV and SON of Six3flox/flox/Nestin-cre+ mice retained AVP expression and the dorsal lateral geniculate (dLG) and ventroposterior medial (VPM) nuclei in both E18.5 and E19.5 mutants continued to express RORα (Fig. 8B). In addition, in all mutants tested, the zona incerta (ZI), which normally expresses Six3, was unaffected (Fig. 8B).
To assess whether absence of SCN at E18.5 and E19.5 was attributable to dissolution of the nucleus after cells became postmitotic or the complete disruption of neurogenesis, we analyzed Six3flox/flox/Nestin-cre+ mice at E15.5 (n = 5) for early postmitotic SCN markers Lhx1 and RORα as well as for dense cell clusters. We found that loss of Six3 leads to absence of both early postmitotic genes, consistent with a disruption in specification. E15.5 Six3flox/flox/Nestin-cre+ mice displaying partial Six3 deletion showed a loss of RORα and Lhx1 only in regions lacking Six3 (Fig. 8C). Six3flox/flox/Nestin-cre+ mice that retained expression of Six3 showed prominent expression of both RORα and Lhx1 (Fig. 8C). Together, these results show the necessity of a transcription factor for proper SCN formation and development.
Discussion
Recently, several groups have directly analyzed the expression of large numbers of genes (>1000) within the developing and/or mature hypothalamus by in situ hybridization (Gray et al., 2004; Lein et al., 2007; Shimogori et al., 2010), looking for patterns of localization capable of defining small populations of neurons that may correspond to unique functional classes. All of these analyses have shown broad genetic distinctions between hypothalamic nuclei. Other studies have also provided detailed analyses of the roles of several developmental genes in hypothalamic and pituitary ontogeny (Sládek et al., 2004; Davis et al., 2010). However, large anatomical screens, because of their breadth, often lack the spatial detail necessary for a more thorough analysis of specific brain regions. Here we provide a detailed analysis of developmental gene expression in the SCN, describe a specific gene cascade from which SCN neurons are derived, and identify a highly conserved transcription factor necessary for SCN formation.
Altman and Bayer (1986) hypothesized that the early suprachiasmatic neuroepithelium formed from a region just caudal to the optic recess. Our data narrow this description showing that the SCN is derived from a distinct region of neuroepithelium expressing a combination of the developmental genes Six3, Six6, Fzd5, and transient Rx, allowing us to pinpoint the suprachiasmatic neuroepithelium within the broader developing telencephalon/diencephalon (for summary, refer to Fig. 7A–C).
ZRDCT anopthalmic mice have decreased expression of Rx protein, and 30% of mutant mice show defects in SCN anatomy and function (Silver, 1977; Tucker et al., 2001). The defects vary widely with respect to disruptions in location and/or cell number, but dense clusters of cells are always seen, indicating formation of nuclei. These data along with our observation of transient Rx before SCN neurogenesis (Figs. 6D, 7A,B) suggests that Rx is not necessary for specification of SCN neurons but that loss may disrupt early progenitor domains (Liu et al., 2010).
Altman and Bayer further hypothesized that the suprachiasmatic neuroepithelium was divided into two regions, dorsal and ventral, from which the corresponding dorsomedial/shell and ventrolateral/core were derived. Our data are not consistent with the presence of two genetically distinct regions contained within the suprachiasmatic neuroepithelium. We suggest that the proposed regions outlined by Altman and Bayer are a consequence of temporal proliferation or differential response to non-cell-autonomous cues, such as morphogens or other early-signaling gradients, rather than distinct genetic differences in progenitor type.
We demonstrated that the onset of Dlx2 and Lhx1 expression at E11.5 is coincident with the documented onset of SCN neurogenesis (Kabrita and Davis, 2008). This suggests that Dlx2 and Lhx1 mark neurons as they exit the proliferative zone that will form the SCN. At E12.5/E13.5, the region that gives rise to the SCN contains both proliferating and postmitotic neurons. We demonstrate three zones of overlapping gene expression in the SCN at this age and propose that these distinct TF-expressing zones represent phases of SCN neuronal development: (I) dorsal Six3/Six6/Fzd5: premitotic/proliferative, (II) mid Six3/Six6/Dlx2: late mitotic/early postmitotic, and (III) ventral Six3/Six6/Dlx2/Lhx1: postmitotic (for summary, refer to Fig. 7D). Our proposed late-mitotic/early-postmitotic Six3/Six6/Dlx2 zone is similar to the region of migratory “late-forming neurons” outlined by Altman and Bayer (1986). In addition, our data support the notion of a ventrodorsal gradient in SCN neurogenesis (Kabrita and Davis, 2008) because the expression of Lhx1, a postmitotic marker, was first seen at the most ventral aspects of the early SCN and progressively expanded dorsally as neurogenesis proceeded.
We found that TF expression within the postmitotic SCN was not static but rather showed specific temporal and spatial changes during both prenatal development and postnatal maturation. This was indicated by dynamic expression of Lhx1 prenatally and RORα postnatally, in which expression of both genes spanned the entire SCN at early stages but became differentially regionalized through development. We propose that changes in localization of gene expression may indicate underlying structural or functional maturation of the SCN. This is supported by the central regionalization of Lhx1 at approximately E17.5, coincident with the perinatal onset of peptide expression. Furthermore, dorsomedial/shell localization of RORα, evident by P15, is concurrent with the proposed termination of retinal fibers into the SCN (Takatsuji et al., 1995) and onset of photoperiodic entrainment of clock genes (Kováciková et al., 2005). It would be reasonable to presume that neurons or areas within the SCN may undergo additional specification or specialization as external innervation into the nucleus, such as postnatal retinal innervation (McNeill et al., 2011) and synaptogenesis progress. The identification of TFs with discrete localization within the SCN may reflect additional subdivisions and/or subsets of neurons with distinct functions.
Regional specificity of gene expression, similar to RORα, has also been reported for Period1 and Period2, with expression mainly limited to the dorsomedial/shell during subjective day in rat and hamster (Hamada et al., 2001). Interestingly, some evidence suggests functional division of the SCN into regions of endogenous rhythmicity (dorsomedial/shell) or induced rhythms (ventrolateral/core) (Sumova et al., 1998; Guido et al., 1999; Hamada et al., 2001; Yan and Okamura, 2002; Hamada et al., 2004; Ramanathan et al., 2006). Given the role of RORα in stability aspects of the clock through regulation of Bmal1, it may be advantageous for the clock to lack stabilizing elements in an inducible region. Furthermore, the pan-SCN expression of Six3 in phase with Per1 may suggest a role for Six3 in maintenance of clock gene expression or underlying circadian rhythmicity.
RORα is one member of related orphan nuclear receptors that serve not only in the differentiation and development of multiple tissue types, including the cerebellum and thalamus (Becker-André et al., 1993; Carlberg et al., 1994; Hamilton et al., 1996; Matysiak-Scholze and Nehls, 1997; Nakagawa et al., 1997, 1998; Dussault et al., 1998; Vogel et al., 2000; Sato et al., 2004), but also in circadian function (Schaeren-Wiemers et al., 1997; Sumi et al., 2002). We found that loss of RORα had no effect on either the presence or location of AVP and VIP neurons, indicating that it was not necessary for peptide expression or development of peptidergic segregation.
The role of Six3 in eye development is highly conserved across vertebrates and in Drosophila (Simeone et al., 1994; Seo et al., 1999). Six3 has been proposed to play a role in proliferation of retinal precursors (Del Bene et al., 2004), lens formation (Liu et al., 2006), neuroretina specification (Liu et al., 2010), regulation of rhodopsin expression (Manavathi et al., 2007), and early anterior patterning (Kobayashi et al., 2002; Lagutin et al., 2003; Gestri et al., 2005; Lavado et al., 2008). Six3 has been shown widely expressed throughout the anterior brain at early stages, narrowing expression over time and remaining transient for many neuronal populations. We found that Six3 was expressed within the suprachiasmatic neuroepithelium before the onset of neurogenesis into adulthood, suggesting an important role for Six3 in the SCN. Moreover, expression of Six3 in adult mice displayed diurnal variation. Although our data cannot discriminate between whether this variation is a response to rhythmic photic input or reveals an underlying circadian rhythm in Six3 expression, they suggest a link between Six3 and day/night changes.
We demonstrated that Six3 is crucial to the development of the SCN. Using a Nestin-cre transgene to limit loss of Six3 to neural progenitors, we tested the role of Six3 in SCN specification. We showed that the loss of Six3 during development leads to elimination of RORα and AVP expression as well as the absence of dense cell clustering perinatally, consistent with the elimination of the SCN. In contrast, extra-SCN structures, including hypothalamic SON and aPV, as well as thalamic VPM and Six3-expressing ZI, were still present. This suggests that Six3 may not be acting solely in global early patterning but may have an additional role in specification of the SCN. We propose that the absence of gene expression is a consequence of the failure to form the SCN initially and not a role for Six3 in the maintenance of SCN cell survival, because SCN-specific Lhx1 expression and cell density were also absent at E15.5. These data suggest a necessary role for Six3 in SCN development and that Lhx1 and RORα represent lineage-specific SCN genes.
Overall, these data demonstrate that TF expression can provide insight into both the specification and development of the SCN. We find that TF expression can describe the early suprachiasmatic neuroepithelium and demonstrate that the SCN undergoes maturation both prenatally and postnatally. Maturation is evident by the changes in localization of gene expression within the SCN over time. It is currently unclear whether the specific loss of gene expression in distinct areas of the SCN is necessary for proper circadian function. However, we find that disruption of at least one TF, Six3, can have large-scale effects on SCN development. Profiling of genes with SCN-specific expression for spatial and temporal onset allows researchers, for the first time, to begin to understand the roles of these genes in the anatomical and functional development of this complex nucleus.
Footnotes
This work was supported by National Institute of General Medical Sciences Grant GM T32 GM009151 (C.V.) and National Heart, Lung, and Blood Institute Grant R01 HL089742 (C.V., L.A.H., P.A.G.). We thank George McMurray and Guang Y. Ling for expert technical assistance. We thank the following for advice, assistance, discussion, and biological resources that made this work possible: Guillermo Oliver (St. Jude Children's Research Hospital, Memphis, TN) for Six3flox mice, Monica Vetter (University of Utah, Salt Lake City, UT) for providing Fzd5 probe, Kristen Kroll (Washington University School of Medicine, St. Louis, MO) for providing TuJ1 and Ki67 antibodies, and Erik Herzog, Matt Thimgan, and Alexis Webb for their expertise and helpful discussions.
References
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the diencephalon in the rat. I. Autoradiographic study of the time of origin and settling patterns of neurons of the hypothalamus. J Comp Neurol. 1978;182:945–971. doi: 10.1002/cne.901820511. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. The development of the rat hypothalamus. Adv Anat Embryol Cell Biol. 1986;100:1–178. [PubMed] [Google Scholar]
- Andrews GL, Yun K, Rubenstein JL, Mastick GS. Dlx transcription factors regulate differentiation of dopaminergic neurons of the ventral thalamus. Mol Cell Neurosci. 2003;23:107–120. doi: 10.1016/s1044-7431(03)00016-2. [DOI] [PubMed] [Google Scholar]
- Antle MC, LeSauter J, Silver R. Neurogenesis and ontogeny of specific cell phenotypes within the hamster suprachiasmatic nucleus. Brain Res Dev Brain Res. 2005;157:8–18. doi: 10.1016/j.devbrainres.2005.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TJ, El-Hodiri H, Zhang L, Shah R, Mathers PH, Jamrich M. Regulation of vertebrate eye development by Rx genes. Int J Dev Biol. 2004;48:761–770. doi: 10.1387/ijdb.041878tb. [DOI] [PubMed] [Google Scholar]
- Ban Y, Shigeyoshi Y, Okamura H. Development of vasoactive intestinal peptide mRNA rhythm in the rat suprachiasmatic nucleus. J Neurosci. 1997;17:3920–3931. doi: 10.1523/JNEUROSCI.17-10-03920.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker-André M, André E, DeLamarter JF. Identification of nuclear receptor mRNAs by RT-PCR amplification of conserved zinc-finger motif sequences. Biochem Biophys Res Commun. 1993;194:1371–1379. doi: 10.1006/bbrc.1993.1976. [DOI] [PubMed] [Google Scholar]
- Bendová Z, Sumová A, Illnerová H. Development of circadian rhythmicity and photoperiodic response in subdivisions of the rat suprachiasmatic nucleus. Brain Res Dev Brain Res. 2004;148:105–112. doi: 10.1016/j.devbrainres.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Burns CJ, Zhang J, Brown EC, Van Bibber AM, Van Es J, Clevers H, Ishikawa TO, Taketo MM, Vetter ML, Fuhrmann S. Investigation of Frizzled-5 during embryonic neural development in mouse. Dev Dyn. 2008;237:1614–1626. doi: 10.1002/dvdy.21565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlberg C, Hooft van Huijsduijnen R, Staple JK, DeLamarter JF, Becker-André M. RZRs, a new family of retinoid-related orphan receptors that function as both monomers and homodimers. Mol Endocrinol. 1994;8:757–770. doi: 10.1210/mend.8.6.7935491. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Davis FC, Boada R, LeDeaux J. Neurogenesis of the hamster suprachiasmatic nucleus. Brain Res. 1990;519:192–199. doi: 10.1016/0006-8993(90)90077-o. [DOI] [PubMed] [Google Scholar]
- Davis SW, Castinetti F, Carvalho LR, Ellsworth BS, Potok MA, Lyons RH, Brinkmeier ML, Raetzman LT, Carninci P, Mortensen AH, Hayashizaki Y, Arnhold IJ, Mendonça BB, Brue T, Camper SA. Molecular mechanisms of pituitary organogenesis: in search of novel regulatory genes. Mol Cell Endocrinol. 2010;323:4–19. doi: 10.1016/j.mce.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bene F, Tessmar-Raible K, Wittbrodt J. Direct interaction of geminin and Six3 in eye development. Nature. 2004;427:745–749. doi: 10.1038/nature02292. [DOI] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, Lin-Marq N, Koch M, Bilio M, Cantiello I, Verde R, De Masi C, Bianchi SA, Cicchini J, Perroud E, Mehmeti S, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussault I, Fawcett D, Matthyssen A, Bader JA, Giguère V. Orphan nuclear receptor ROR alpha-deficient mice display the cerebellar defects of staggerer. Mech Dev. 1998;70:147–153. doi: 10.1016/s0925-4773(97)00187-1. [DOI] [PubMed] [Google Scholar]
- Gestri G, Carl M, Appolloni I, Wilson SW, Barsacchi G, Andreazzoli M. Six3 functions in anterior neural plate specification by promoting cell proliferation and inhibiting Bmp4 expression. Development. 2005;132:2401–2413. doi: 10.1242/dev.01814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gray PA. Transcription factors and the genetic organization of brain stem respiratory neurons. J Appl Physiol. 2008;104:1513–1521. doi: 10.1152/japplphysiol.01383.2007. [DOI] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk DI, Tsung EF, Cai Z, Alberta JA, Cheng LP, Liu Y, Stenman JM, Valerius MT, Billings N, Kim HA, Greenberg ME, McMahon AP, Rowitch DH, Stiles CD, Ma Q. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Guido ME, de Guido LB, Goguen D, Robertson HA, Rusak B. Daily rhythm of spontaneous immediate-early gene expression in the rat suprachiasmatic nucleus. J Biol Rhythms. 1999;14:275–280. doi: 10.1177/074873099129000687. [DOI] [PubMed] [Google Scholar]
- Hamada T, LeSauter J, Venuti JM, Silver R. Expression of Period genes: rhythmic and nonrhythmic compartments of the suprachiasmatic nucleus pacemaker. J Neurosci. 2001;21:7742–7750. doi: 10.1523/JNEUROSCI.21-19-07742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, Antle MC, Silver R. Temporal and spatial expression patterns of canonical clock genes and clock-controlled genes in the suprachiasmatic nucleus. Eur J Neurosci. 2004;19:1741–1748. doi: 10.1111/j.1460-9568.2004.03275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton BA, Frankel WN, Kerrebrock AW, Hawkins TL, FitzHugh W, Kusumi K, Russell LB, Mueller KL, van Berkel V, Birren BW, Kruglyak L, Lander ES. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. 1996;379:736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Schwartz WJ. A neural clockwork for encoding circadian time. J Appl Physiol. 2002;92:401–408. doi: 10.1152/japplphysiol.00836.2001. [DOI] [PubMed] [Google Scholar]
- Isobe Y, Nakajima K, Nishino H. Arg-vasopressin content in the suprachiasmatic nucleus of rat pups: circadian rhythm and its development. Brain Res Dev Brain Res. 1995;85:58–63. doi: 10.1016/0165-3806(94)00192-3. [DOI] [PubMed] [Google Scholar]
- Jean D, Bernier G, Gruss P. Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/hypothalamic axis and the ventral optic stalk. Mech Dev. 1999;84:31–40. doi: 10.1016/s0925-4773(99)00068-4. [DOI] [PubMed] [Google Scholar]
- Kabrita CS, Davis FC. Development of the mouse suprachiasmatic nucleus: determination of time of cell origin and spatial arrangements within the nucleus. Brain Res. 2008;1195:20–27. doi: 10.1016/j.brainres.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatsoreos IN, Yan L, LeSauter J, Silver R. Phenotype matters: identification of light-responsive cells in the mouse suprachiasmatic nucleus. J Neurosci. 2004;24:68–75. doi: 10.1523/JNEUROSCI.1666-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi D, Kobayashi M, Matsumoto K, Ogura T, Nakafuku M, Shimamura K. Early subdivisions in the neural plate define distinct competence for inductive signals. Development. 2002;129:83–93. doi: 10.1242/dev.129.1.83. [DOI] [PubMed] [Google Scholar]
- Kováciková Z, Sládek M, Laurinová K, Bendová Z, Illnerová H, Sumová A. Ontogenesis of photoperiodic entrainment of the molecular core clockwork in the rat suprachiasmatic nucleus. Brain Res. 2005;1064:83–89. doi: 10.1016/j.brainres.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Kováciková Z, Sládek M, Bendová Z, Illnerová H, Sumová A. Expression of clock and clock-driven genes in the rat suprachiasmatic nucleus during late fetal and early postnatal development. J Biol Rhythms. 2006;21:140–148. doi: 10.1177/0748730405285876. [DOI] [PubMed] [Google Scholar]
- Lagutin OV, Zhu CC, Kobayashi D, Topczewski J, Shimamura K, Puelles L, Russell HR, McKinnon PJ, Solnica-Krezel L, Oliver G. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003;17:368–379. doi: 10.1101/gad.1059403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavado A, Lagutin OV, Oliver G. Six3 inactivation causes progressive caudalization and aberrant patterning of the mammalian diencephalon. Development. 2008;135:441–450. doi: 10.1242/dev.010082. [DOI] [PubMed] [Google Scholar]
- Leak RK, Moore RY. Topographic organization of suprachiasmatic nucleus projection neurons. J Comp Neurol. 2001;433:312–334. doi: 10.1002/cne.1142. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Liu W, Lagutin OV, Mende M, Streit A, Oliver G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006;25:5383–5395. doi: 10.1038/sj.emboj.7601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Lagutin O, Swindell E, Jamrich M, Oliver G. Neuroretina specification in mouse embryos requires Six3-mediated suppression of Wnt8b in the anterior neural plate. J Clin Invest. 2010;120:3568–3577. doi: 10.1172/JCI43219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavathi B, Peng S, Rayala SK, Talukder AH, Wang MH, Wang RA, Balasenthil S, Agarwal N, Frishman LJ, Kumar R. Repression of Six3 by a corepressor regulates rhodopsin expression. Proc Natl Acad Sci U S A. 2007;104:13128–13133. doi: 10.1073/pnas.0705878104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matysiak-Scholze U, Nehls M. The structural integrity of ROR alpha isoforms is mutated in staggerer mice: cerebellar coexpression of ROR alpha1 and ROR alpha4. Genomics. 1997;43:78–84. doi: 10.1006/geno.1997.4757. [DOI] [PubMed] [Google Scholar]
- McNeill DS, Sheely CJ, Ecker JL, Badea TC, Morhardt D, Guido W, Hattar S. Development of melanopsin-based irradiance detecting circuitry. Neural Dev. 2011;6:8. doi: 10.1186/1749-8104-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moga MM, Moore RY. Organization of neural inputs to the suprachiasmatic nucleus in the rat. J Comp Neurol. 1997;389:508–534. doi: 10.1002/(sici)1096-9861(19971222)389:3<508::aid-cne11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Watanabe M, Inoue Y. Prominent expression of nuclear hormone receptor ROR alpha in Purkinje cells from early development. Neurosci Res. 1997;28:177–184. doi: 10.1016/s0168-0102(97)00042-4. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Watanabe M, Isobe T, Kondo H, Inoue Y. Cytological compartmentalization in the staggerer cerebellum, as revealed by calbindin immunohistochemistry for Purkinje cells. J Comp Neurol. 1998;395:112–120. doi: 10.1002/(sici)1096-9861(19980525)395:1<112::aid-cne8>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, O'Leary DD. Dynamic patterned expression of orphan nuclear receptor genes RORalpha and RORbeta in developing mouse forebrain. Dev Neurosci. 2003;25:234–244. doi: 10.1159/000072271. [DOI] [PubMed] [Google Scholar]
- Oishi K, Fukui H, Ishida N. Rhythmic expression of BMAL1 mRNA is altered in Clock mutant mice: differential regulation in the suprachiasmatic nucleus and peripheral tissues. Biochem Biophys Res Commun. 2000;268:164–171. doi: 10.1006/bbrc.1999.2054. [DOI] [PubMed] [Google Scholar]
- Okamura H, Fukui K, Koyama E, Tsutou HL, Tsutou T, Terubayashi H, Fujisawa H, Ibata Y. Time of vasopressin neuron origin in the mouse hypothalamus: examination by combined technique of immunocytochemistry and [3H]thymidine autoradiography. Brain Res. 1983;285:223–226. doi: 10.1016/0165-3806(83)90055-x. [DOI] [PubMed] [Google Scholar]
- Oliver G, Mailhos A, Wehr R, Copeland NG, Jenkins NA, Gruss P. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- Ramanathan C, Nunez AA, Martinez GS, Schwartz MD, Smale L. Temporal and spatial distribution of immunoreactive PER1 and PER2 proteins in the suprachiasmatic nucleus and peri-suprachiasmatic region of the diurnal grass rat (Arvicanthis niloticus) Brain Res. 2006;1073–1074:348–358. doi: 10.1016/j.brainres.2005.11.082. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Schwartz WJ. The suprachiasmatic nuclei of the fetal rat: characterization of a functional circadian clock using 14C-labeled deoxyglucose. J Neurosci. 1984;4:1677–1682. doi: 10.1523/JNEUROSCI.04-07-01677.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivkees SA, Weaver DR, Reppert SM. Circadian and developmental regulation of Oct-2 gene expression in the suprachiasmatic nuclei. Brain Res. 1992;598:332–336. doi: 10.1016/0006-8993(92)90203-l. [DOI] [PubMed] [Google Scholar]
- Romero MT, Silver R. Time Course of peptidergic expression in fetal suprachiasmatic nucleus transplanted into adult hamster. Brain Res Dev Brain Res. 1990;57:1–6. doi: 10.1016/0165-3806(90)90177-z. [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, André E, Kapfhammer JP, Becker-André M. The expression pattern of the orphan nuclear receptor RORbeta in the developing and adult rat nervous system suggests a role in the processing of sensory information and in circadian rhythm. Eur J Neurosci. 1997;9:2687–2701. doi: 10.1111/j.1460-9568.1997.tb01698.x. [DOI] [PubMed] [Google Scholar]
- Seo HC, Curtiss J, Mlodzik M, Fjose A. Six class homeobox genes in drosophila belong to three distinct families and are involved in head development. Mech Dev. 1999;83:127–139. doi: 10.1016/s0925-4773(99)00045-3. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Shibata S, Moore RY. Development of neuronal activity in the rat suprachiasmatic nucleus. Brain Res. 1987;431:311–315. doi: 10.1016/0165-3806(87)90220-3. [DOI] [PubMed] [Google Scholar]
- Shimada M, Nakamura T. Time of neuron origin in mouse hypothalamic nuclei. Exp Neurol. 1973;41:163–173. doi: 10.1016/0014-4886(73)90187-8. [DOI] [PubMed] [Google Scholar]
- Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, Qi L, Qian J, Blackshaw S. A genomic atlas of mouse hypothalamic development. Nat Neurosci. 2010;13:767–775. doi: 10.1038/nn.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J. Abnormal development of the suprachiasmatic nuclei of the hypothalamus in a strain of genetically anophthalmic mice. J Comp Neurol. 1977;176:589–606. doi: 10.1002/cne.901760409. [DOI] [PubMed] [Google Scholar]
- Simeone A, D'Apice MR, Nigro V, Casanova J, Graziani F, Acampora D, Avantaggiato V. Orthopedia, a novel homeobox-containing gene expressed in the developing CNS of both mouse and Drosophila. Neuron. 1994;13:83–101. doi: 10.1016/0896-6273(94)90461-8. [DOI] [PubMed] [Google Scholar]
- Sládek M, Sumová A, Kováciková Z, Bendová Z, Laurinová K, Illnerová H. Insight into molecular core clock mechanism of embryonic and early postnatal rat suprachiasmatic nucleus. Proc Natl Acad Sci U S A. 2004;101:6231–6236. doi: 10.1073/pnas.0401149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi Y, Yagita K, Yamaguchi S, Ishida Y, Kuroda Y, Okamura H. Rhythmic expression of ROR beta mRNA in the mice suprachiasmatic nucleus. Neurosci Lett. 2002;320:13–16. doi: 10.1016/s0304-3940(02)00011-3. [DOI] [PubMed] [Google Scholar]
- Sumová A, Trávnícková Z, Mikkelsen JD, Illnerová H. Spontaneous rhythm in c-Fos immunoreactivity in the dorsomedial part of the rat suprachiasmatic nucleus. Brain Res. 1998;801:254–258. doi: 10.1016/s0006-8993(98)00619-2. [DOI] [PubMed] [Google Scholar]
- Sumová A, Bendová Z, Sládek M, El-Hennamy R, Laurinová K, Jindráková Z, Illnerová H. Setting the biological time in central and peripheral clocks during ontogenesis. FEBS Lett. 2006;580:2836–2842. doi: 10.1016/j.febslet.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Sunkin SM. Towards the integration of spatially and temporally resolved murine gene expression databases. Trends Genet. 2006;22:211–217. doi: 10.1016/j.tig.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Takatsuji K, Senba E, Mantyh PW, Tohyama M. A relationship between substance P receptor and retinal fibers in the rat suprachiasmatic nucleus. Brain Res. 1995;698:53–61. doi: 10.1016/0006-8993(95)00802-w. [DOI] [PubMed] [Google Scholar]
- Tucker P, Laemle L, Munson A, Kanekar S, Oliver ER, Brown N, Schlecht H, Vetter M, Glaser T. The eyeless mouse mutation (ey1) removes an alternative start codon from the Rx/rax homeobox gene. Genesis. 2001;31:43–53. doi: 10.1002/gene.10003. [DOI] [PubMed] [Google Scholar]
- Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MW, Sinclair M, Qiu D, Fan H. Purkinje cell fate in staggerer mutants: agenesis versus cell death. J Neurobiol. 2000;42:323–337. doi: 10.1002/(sici)1097-4695(20000215)42:3<323::aid-neu4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Wang W, Lufkin T. The murine Otp homeobox gene plays an essential role in the specification of neuronal cell lineages in the developing hypothalamus. Dev Biol. 2000;227:432–449. doi: 10.1006/dbio.2000.9902. [DOI] [PubMed] [Google Scholar]
- Webb AB, Angelo N, Huettner JE, Herzog ED. Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc Natl Acad Sci U S A. 2009;106:16493–16498. doi: 10.1073/pnas.0902768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Okamura H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur J Neurosci. 2002;15:1153–1162. doi: 10.1046/j.1460-9568.2002.01955.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Fuhrmann S, Vetter ML. A nonautonomous role for retinal frizzled-5 in regulating hyaloid vitreous vasculature development. Invest Ophthalmol Vis Sci. 2008;49:5561–5567. doi: 10.1167/iovs.08-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]