Abstract
A critical issue during the 2009 influenza A (H1N1) pandemic was determining the appropriate duration of time individuals with influenza-like illness (ILI) should remain isolated to reduce onward transmission while limiting societal disruption. Ideally this is based on knowledge of the relative infectiousness of ill individuals at each point during the course of the infection. Data on 261 clinically apparent pH1N1 infector-infectee pairs in households, from 7 epidemiological studies conducted in the United States early in 2009, were analyzed to estimate the distribution of times from symptom onset in an infector to symptom onset in the household contacts they infect (mean, 2.9 days, not correcting for tertiary transmission). Only 5% of transmission events were estimated to take place >3 days after the onset of clinical symptoms among those ill with pH1N1 virus. These results will inform future recommendations on duration of isolation of individuals with ILI.
Between 15 April and 17 April 2009, the Centers for Disease Control and Prevention (CDC) identified a novel influenza A virus (now known as 2009 pandemic influenza A [H1N1], or pH1N1) detected from 2 children with febrile respiratory illness in southern California [1]. The virus spread rapidly, and by 5 May, individuals with laboratory-confirmed pH1N1 infection had been reported from 41 US states and 21 countries worldwide [2, 3]. Determining the appropriate duration of isolation for clinically affected individuals to control the outbreak was a key policy challenge that required a balance between disease prevention and societal disruption. On 28 April 2009, the CDC issued an interim recommendation that people with influenza-like illness (ILI; defined to be fever or feverishness with either cough or sore throat) should be strongly encouraged to self-isolate in their home for 7 days after the onset of illness or ≥24 hours after symptoms had resolved, whichever was longer, on the basis of historical data regarding viral shedding duration that indicate immune-naive people are likely to shed longer than those with pre-existing immunity to seasonal influenza strains [4]. Over the course of the pandemic, as new information was gathered, particularly about severity of illness, the guidelines changed. Currently, the CDC recommends that people with ILI remain at home until ≥24 hours after they are free of fever (temperature, 100° F [37.8°C]), or signs of a fever, without the use of fever-reducing medications [5, 6].
Determining the appropriate period of isolation is, ideally, based on an understanding of the relative infectiousness of clinically affected individuals at each point during the course of the infection, as measured by the temporal distribution of secondary infections, relative to the onset of symptoms in the index case. Because the transmissibility characteristics of pH1N1 were unknown, the initial recommendations issued by CDC during the 2009 pandemic were based on studies of seasonal influenza infections in otherwise healthy adults that showed that viral shedding, through nasal and oropharyngeal secretions, peaks during the first 24 to 72 hours of symptomatic illness and generally lasts up to 5 days [7–9]. Immunologically naive persons, children, and immunocompromised patients have been found to shed virus for substantially longer [10–11]. Recently published studies are yielding valuable shedding data on the pH1N1 virus [12–14].
The duration and quantitative level of viral shedding are only 2 of the many factors that are expected to influence the likelihood of transmission. Other factors that may be associated with transmission include the presence of symptoms such as sneezing and coughing that disperse virus, the behavioral characteristics of ill persons (eg, the types and duration of interactions between people and whether someone stays in bed or at least away from work [15–17]), and demographic factors (eg, age and the number of individuals in a household [18–20]). It is precisely because of this complex interplay between viral shedding, symptoms, and behavior that epidemiological studies of detailed outbreak data are needed to characterize how transmission risk varies as a function of time since infection or the onset of symptoms. This study presents a summary analysis of household data collated by the CDC in 2009, focusing on estimating the temporal distribution of secondary infections within households. The serial interval of an infectious disease is defined as the duration of time between the onset of symptoms of an index individual and the onset of symptoms of an infected contact [21–22]. Previous studies have estimated the mean or median serial interval for pH1N1 to be 2.5–2.8 days [20, 23, 24, 25], comparable with most past estimates of seasonal influenza.
In this study, we combine data from 7 studies of ILI and acute respiratory illness (ARI) in households that were conducted during community outbreaks of pH1N1 in the United States early in the 2009 pandemic to estimate the serial interval distribution and thereby, through the use of the incubation period distribution, to quantify the distribution over time of secondary infections within households, relative to the onset of symptoms in the index case. The incubation period (the time from infection to the onset of symptoms) is required because the serial interval is from dates of symptom onset in the index case until symptom onset in the infected contact, whereas the temporal distribution of secondary infections (not onsets) is quantified relative to the onset of symptoms in the index case. These results will inform future recommendations on duration of isolation of individuals with ILI.
METHODS
Study Designs – Case Finding and Data Collection
The data analyzed are clinically apparent serial intervals identified in 7 studies with independently collected data on household transmission. The sample size given below for each study is the number of clinically apparent serial intervals recorded, excluding any >10 days. (Data from studies >10 days in length become less comparable because of variable durations of follow-up. This threshold was judged sufficiently long on the basis of the results subsequently obtained.) The observations in which the contact experienced the onset of symptoms on the same day as the onset of symptoms in the index individual (sometimes termed co-primary infections) were also excluded.
An ARI was defined by the presence of ≥2 of the following symptoms: fever or feverishness, cough, sore throat, and runny nose. There was no systematic laboratory confirmation of either ARI or ILI among ill contacts. Given that infection with the pH1N1 virus often produced mild illness, the primary analysis was of ARI, with supplementary analyses performed for the data with the stricter ILI definition. (Note that on the basis of our definitions, ILI is a subset of ARI.)
Study 1: Household Transmission Study Based on Case Report Forms Submitted to CDC (ILI, n = 66; non-ILI ARI, n = 18).
Early in the epidemic (29 April to 25 May 2009), US state health departments were asked to report all laboratory-confirmed cases of pH1N1 influenza to the CDC. Persons with illness that met the definition of a confirmed or probable (as acute febrile respiratory illness in a person who tested positive for influenza A virus but negative for human H1 and H3 serotypes, as assessed with the use of reverse-transcriptase polymerase chain reaction) case were interviewed on the basis of the standardized case-report form that included requests for information on the (index) individual's symptoms and date of the onset of symptoms. Information on symptoms and date of symptom onset (that occurred within 7 days before or after the onset of symptoms in the index individual) was also requested for household members, defined to include any person who had stayed overnight in the house ≥1 night within 7 days before or after the date of symptom onset in the index individual. A detailed analysis of these data has been published previously [20].
Study 2: New York City Study of Households of School Students, Faculty, and Staff with Confirmed pH1N1 or ILI (ILI, n = 73).
Household contacts of index cases were surveyed to characterize transmission of pH1N1 within households and to identify potential risk factors for transmission [26]. Index cases were school students, faculty, and staff who had laboratory-confirmed pH1N1 infection with onset between 22 April and 30 April or who had an ILI with onset during the peak of the school outbreak, 22 April to 25 April. Letters were sent from the school inviting all households of index cases to complete an online survey about illness and risk factors. The letter requested that a single individual complete the survey on behalf of all household members. Because of poor response to the invitation letters, nonresponding households were called by study staff and invited to complete the survey by phone.
Study 3: Southeastern Pennsylvania Study of Households of School Students with ILI (ILI, n = 37).
An initial telephone survey was conducted during 16 May to 21 May among parents or guardians of school students by means of a structured questionnaire. A second, more in-depth structured questionnaire was administered by telephone from 26 May to 2 June to parents and guardians of school students. Respondents gave information about household composition, demographics, and the presence and onset date of ILI among any household member(s) since 1 May. Data from the 2 surveys were merged for this analysis. Onset dates and clinical information from the first survey were used for persons who participated only in this survey and for those whose ILI onsets were on or before the first survey was conducted. Onset dates and clinical information from the second survey were used for individuals who participated only in the second survey as well as those who became ill after the first survey.
Study 4: San Antonio, Texas, Household Study (Laboratory-Confirmed, n = 17; ILI, n = 10; non-ILI ARI, n = 8).
Individuals with laboratory-confirmed pH1N1 infection were identified through the review of local laboratory records from 10 April to 8 May 2009, and testing of local high school students reported as absent between 9 April and 28 April and who reported an ARI [27]. Laboratory-confirmed individuals and all members of their household were interviewed about the occurrence of illness.
Study 5: Imperial County, California, Household Study (ILI, n = 30) and Study 6: San Diego, California, Household Study (ILI, n = 9; non-ILI ARI, n = 5).
Individuals with laboratory-confirmed pH1N1 infection were contacted by phone by staff of the Public Health Department of Imperial County or San Diego or by the CDC and asked about date of onset and symptoms. In addition, confirmed cases were asked whether household contacts became ill, their dates of illness onset, and their age and symptoms.
Study 7: Houston, Texas, Study of Households of School Students, Faculty, and Staff with Laboratory- Confirmed pH1N1 Infection or ILI (ILI, n = 2; non-ILI ARI, n = 2).
All students with laboratory-confirmed pH1N1 infection were investigated, and the parents or guardians of these students were contacted. An online questionnaire was developed and included questions about dates and symptoms of illness that occurred since 12 April 2009 in school students, staff, and their household contacts. The questionnaire was available online in May 2009, and responses were stored on secure web servers.
All field studies were part of the emergency public health practice response to the pandemic. They were reviewed by a human subjects coordinator at CDC and deemed not to be research, in accordance with the federal human subjects protection regulations at 45 Code of Federal Regulations 46.101c and 46.102d and CDC's Guidelines for Defining Public Health Research and Public Health Non-Research.
Calculation of Serial Interval
The clinically apparent serial intervals were calculated from the date of onset in the (confirmed) index case to the time of symptom onset in the ill household member (secondary case). The key difference among the studies was the definition of secondary cases. This was addressed with parallel analyses using ARI and ILI case definitions.
Statistical Analysis
We observed clinically apparent serial intervals---that is, the time lags from onset in the first case of the household to onsets in subsequent cases, from data on clinically apparent infector-infectee pairs. However, we cannot know which are, in fact, true serial intervals (in which the index individual was the source of the infection in the contact) and which are false serial intervals. False serial intervals arise when the contact was either a tertiary case (ie, infected by a household member who was not the index case) or a community case (ie, infected by an unidentified source outside the household). Both types of false serial intervals will on average have longer duration than a true serial interval. The tertiary cases arise from secondary cases and thus, on average, experience clinical onset later than the secondary cases. The community cases will appear to have, on average, longer apparent serial intervals if the risk from sources outside the household was constant or increasing for the period under study. It is highly unlikely that the risk from communities decreased during the study periods, because the studies were performed early in the pH1N1 pandemic. These longer-than-average false longer intervals will thus cause the mean of the clinically apparent serial interval distribution to overestimate the mean of the true serial interval distribution. In this study, we do not attempt to model the possibility of tertiary transmission within households, because only aggregate data on observed serial intervals were available from some studies (rather than data on every contact in every household regardless of illness status), but our estimates are corrected for the occurrence of community cases, assuming a constant risk of transmission from sources outside the household.
We modeled the distribution of serial intervals by fitting, via maximum likelihood, mixtures of parametric distributions (γ, lognormal, and Weibull, representing true serial intervals) and uniform distributions (representing a constant risk of infections from sources outside the household) to the observed clinically apparent serial intervals. We used likelihood ratio tests to determine whether the datasets from the different studies were well fitted by the same distribution. We let ni be the number of ill contacts with clinical onset i days after onset in the index individual where ni is observed from day 1 to Dmax. Let p(i) be the probability of contacts experiencing the onset of clinical symptoms on day i conditional on i being between 1 and Dmax inclusive, based on the assumed parametric form. Thus, the log likelihood of the observed data is given by:
Maximum likelihood estimates were obtained by maximizing the log likelihood, individually for each data set and simultaneously for multiple data sets (which might have different Dmax values). This approach explicitly excludes observations in which the contact experienced the onset of symptoms on the same day as the onset of symptoms in the index individual.
The fitting of continuous distributions, such as the γ, lognormal, and Weibull distributions, requires the conversion of a continuous probability density function to a discrete distribution, because we have data on the dates, but not times, of the onset of clinical symptoms in index cases and their household contracts. We let F(i) be the cumulative distribution function for the assumed parametric distribution. We define pS(i) to equal [FS(i + .5) – FS(i – .5) ]/[FS(Dmax + .5) – FS(.5) ], such that the sum of pS(i) from i = 1 to Dmax equals 1. The means and standard deviations of the fitted distribution reported here were calculated on the basis of this discretized distribution (ie, the fitted mean was obtained calculating the sum of i × p(i) from i = 1 to Dmax). The goodness-of-fit of the parametric mixture distribution was assessed through the comparison of its maximized likelihood value to that obtained from the nonparametric fit to the data in which a separate parameter was fitted for each day (from 1 to Dmax).
In the primary analysis, we assumed that Dmax was equal to 7 days, thus excluding all data for which there were >7 days between the onset of clinical symptoms in the index individual and the onset of clinical symptoms in the contact. To examine the sensitivity of our results to our assumptions, we performed our analyses with Dmax assumed to equal 10 days instead of 7 days, excluding study 1 from this analysis because, by design, it collected data only on illness in contacts ≤7 days after the onset of the index individual.
Confidence intervals (CIs) were obtained by bootstrapping the observed serial interval data, assuming all data points were independent and arose from a multinomial distribution. The bootstrap samples were obtained through repeated sampling, with replacement, from the observed serial interval distributions. The model fitting procedure was then performed independently for each of these bootstrap samples.
Characterization of the Temporal Distribution of Secondary Infections Within Households
Consider an index case with symptom onset on day 0 and one of his or her secondary cases with symptom onset on day i (i > 0). Here, we estimated the probability pT(t) that transmission between the 2 individuals took place on day t, conditional on transmission having taken place. This relied on the fact that the distribution of the serial interval pS(.) is the convolution of the distribution of transmission events pT(.) and distribution of the incubation period pI(.)---that is, the time interval between infection and symptom onset:
In other words, the distribution of the serial interval is obtained by summing over all values of pT(t) and pI(i-t) where t ≤ i.
We assumed the incubation period distribution to be a discretized γ distribution Γ (mean, 2 days; variance, .25 days) truncated after day 6, which gave a discretized distribution with mean of 1.5 days and variance of .3 days. The mean and variance were chosen to be similar to incubation period data from past seasonal outbreaks [19, 28–29] as well as data from pH1N1 influenza [24, 25]. Estimation of the pT(.) distribution was performed via least-squares fitting.
pT(.) is the distribution of transmission events, relative to the onset of clinical symptoms on the index case, that yields estimates of the proportion of total within-household transmission that occurs more than a certain time following the onset of clinical symptoms. This interpretation of the deconvolution requires the assumption that the impact of tertiary transmission (that is from the secondarily infected contacts within the household, rather than just from the index individual) is negligible.
RESULTS
Combining data from the 7 studies, there were data on 277 clinically apparent ARI serial intervals, of which 261 were ≤7 days in length. There were 244 clinically apparent ILI serial intervals, of which 230 were ≤7 days in length. Figures 1A, 1B, and 1C show the frequency distributions of the clinically apparent serial intervals observed in the 7 studies.
Figure 1.
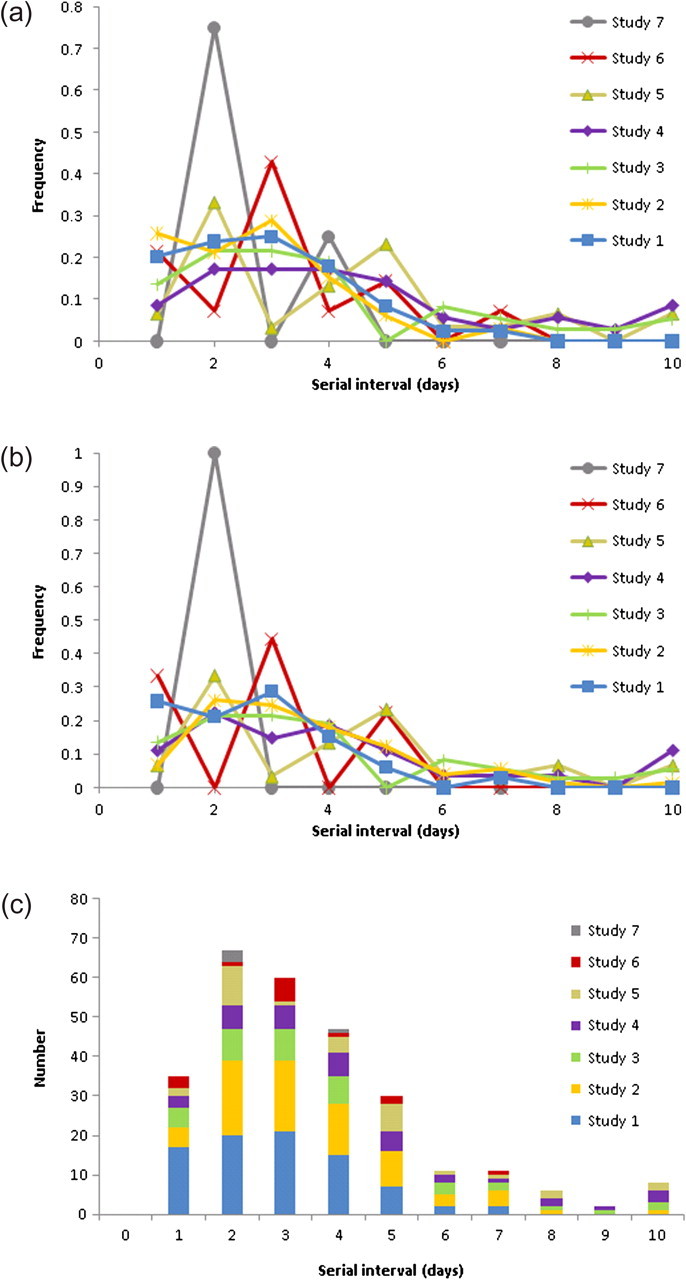
The distribution of the clinically apparent serial interval by study based on acute respiratory illness (ARI) (a) and on influenza-like illness (ILI) (b), and the combined distribution of the ARI clinically apparent serial interval based on all studies (c).
Primary Analysis: ARI Assuming Dmax = 7 Days
Likelihood-based goodness-of-fit tests indicated that all 3 parametric forms fitted the observed data well, compared to a single nonparametric model fitted to all the data sets simultaneously (P > .3). Assuming the Weibull parametric form (which produced a greater maximized likelihood value than did the γ or lognormal distributions), the likelihood ratio tests indicated no significant pairwise difference between the studies. The proportion of transmission that arose from sources outside the household was estimated to be 23% (95% CI, 0–38%; P = .11). The maximum likelihood estimate of the distribution of true serial intervals (conditional on them being ≥1 day), simultaneously fitted to the 7 ARI datasets, is presented in Figure 2A. The mean serial interval was 2.9 days (95% CI, 2.6–3.3 days).
Figure 2.
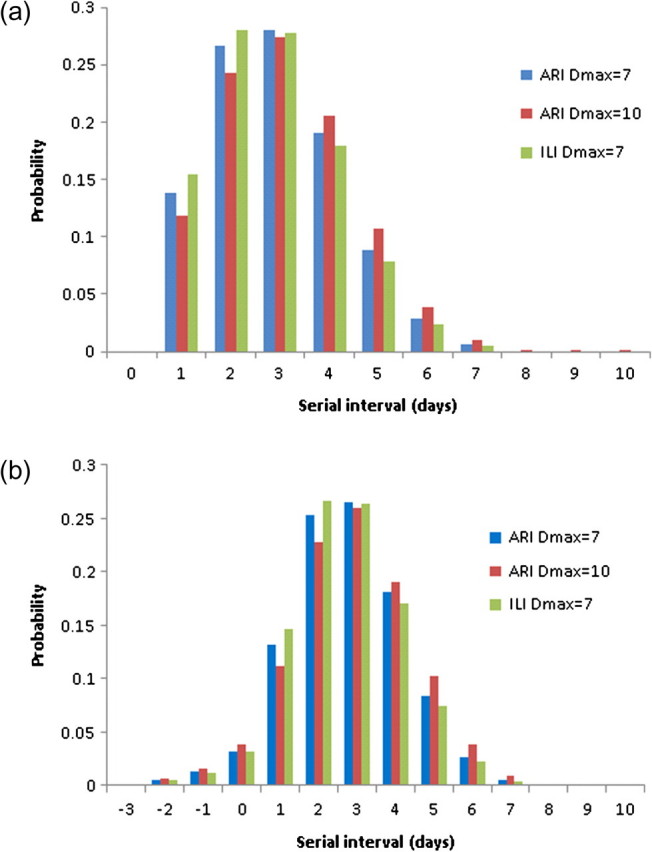
The maximum likelihood Weibull distributions for the true serial interval conditional on it being between 1 and Dmax inclusive (a) and the corresponding unconditional distributions (b): for the primary analysis (based on acute respiratory illness [ARI] assuming Dmax = 7 days) and accompanying analyses (based on ARI assuming Dmax = 10 days and based on influenza-like illness [ILI] assuming Dmax = 7 days).
Temporal Distribution of Secondary Infections within Households
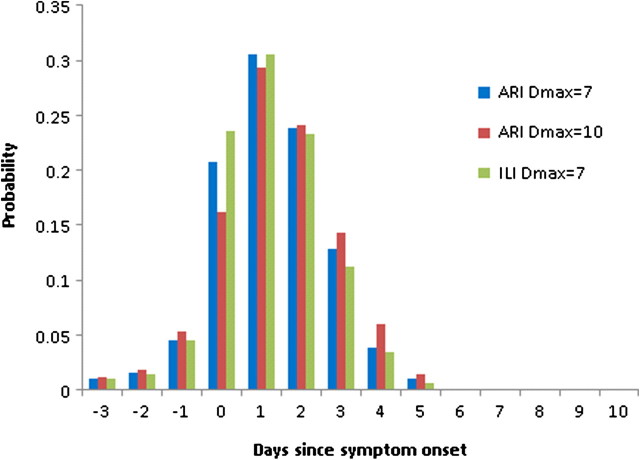
When deconvoluted using a discretized incubation period distribution, the resulting distribution of transmission events by day, relative to symptom onset in the index case, demonstrated that only 18% of within-household transmission happens >2 days after the onset of clinical symptoms and only 5% happens >3 days after onset (Figure 3). Finally, the unconditional distribution of the true serial intervals (ie, the distribution allowing for secondary cases that onset on the same day as the index case or even possibly before the index case) was reconstructed from convolution of the distribution of transmission events and the assumed incubation period (Figure 2B). The mean of the unconditional serial interval distribution was 2.8 days.
Figure 3.
The estimated distribution of transmission events, relative to the onset of clinical symptoms on the index case obtained from the deconvolution of the maximum likelihood Weibull serial interval distribution: for the primary analysis (based on acute respiratory illness [ARI] assuming Dmax = 7 days), and accompanying analyses (based on ARI assuming Dmax = 10 days and based on influenza-like illness [ILI] assuming Dmax = 7 days).
Secondary Analysis: ARI Assuming Dmax = 10 Days
The likelihood-based goodness-of-fit tests indicated that only the Weibull distribution fitted the observed data well, compared to a single nonparametric model fitted to all the data sets simultaneously (P = .23). Assuming the Weibull parametric form, the likelihood ratio tests indicated no significant pairwise difference between the studies. The proportion of transmission that arose from sources outside the household was estimated to be 28% (95% CI, 13%– 40%; P < .001). The maximum likelihood estimate of the distribution of true serial intervals (conditional on them being ≥1 day), simultaneously fitted to the 7 ARI datasets, is presented in Figure 2A. The mean serial interval was 3.1 days (95% CI, 2.8–3.6 days). The deconvoluted distribution of transmission events (Figure 3) and the unconditional distribution of true serial intervals (Figure 2B) were very similar to that obtained assuming Dmax was equal to 7 days.
Secondary Analysis: ILI Assuming Dmax = 7 Days
Likelihood-based goodness-of-fit tests indicated that all 3 parametric forms fitted the observed data well, compared to a single nonparametric model fitted to all the datasets simultaneously (P > .2). The proportion of transmission that arose from sources outside the household was estimated to be 25% (95% CI, 0–39%; P = .09). The maximum likelihood estimate of the distribution of true serial intervals (conditional on them being ≥1 day), simultaneously fitted to the 7 ARI datasets, is presented in Figure 2A. The mean serial interval was 2.8 days (95% CI, 2.5–3.1 days). The deconvoluted distribution of transmission events (Figure 3) and the unconditional distribution of true serial intervals (Figure 2B) were very similar to that obtained on the basis of the analysis of ARI data.
DISCUSSION
We describe the combined analysis of 7 household infection studies of pH1N1 influenza conducted in early 2009 to determine the serial interval and the temporal distribution of secondary infections within households. The mean serial interval was 2.9 days. Only 18% of transmission events were estimated to take place >2 days after the onset of clinical symptoms among those ill with pH1N1 virus, and only 5% >3 days after onset. To our knowledge, this is the largest analysis of its type to date and the first to estimate the temporal distribution of secondary infections within households.
The gold standard data for the estimation of the serial interval distribution of a disease are linked pairs of dates of the onset of clinical symptoms in infectors and infectees. Although it cannot typically be proven that within-household pairs are in fact infector-infectee (particularly within communities affected by ongoing disease transmission), rigorous analysis of clinically apparent serial intervals–in which unrealistically long serial intervals are excluded and external introductions of infection are allowed for–was implemented to infer the underlying distribution of true serial intervals. Our approach may lead to slight overestimation of the true serial interval because it did not correct for tertiary cases. However, such bias would be expected to be relatively small, given that studies of pH1N1 infections have shown low secondary attack rates in households, implying relatively few tertiary infections [20]. An indication of the level of bias can be derived by comparing the estimates obtained from study 1 in the current analyses with those derived from an analysis that corrected for tertiary transmission [20]. In the latter analysis, the mean serial interval was estimated to be 2.6 days, whereas our methods, applied only to study 1, yielded an estimate of 2.8 days. We therefore conclude that our estimates of a mean clinically apparent serial interval of 2.9 days is consistent with estimates of 2.5–2.7 days obtained from analyses that corrected for tertiary transmission [20, 23, 24, 25].
From the point of view of providing information relevant to the recommended duration of case isolation, our estimates are conservative, in that not correcting for tertiary transmission means we slightly overestimate the proportion of within-household transmission occurring after a particular number of days after symptom onset. However, depending on the occupation of an infected individual, the risks of onward transmission associated with return to work after a particular number of days after symptom onset may be much greater than those implied by the household data---for example, in the case of an individual caring for immunocompromised patients.
The distributions of within-household transmissions (following the onset of clinical symptoms), implied by these estimated serial interval distributions, consistently indicate that only approximately 15% to 20% of within-household transmission happens >2 days after symptoms appear. This supports the view that requiring isolation of those with ILI for a full week after the onset of clinical symptoms may not be necessary to reduce substantially the risks of transmission from clinically affected individuals to communities and workplaces. However, reducing transmission substantially at a population level will not be, of course, the only consideration, because there will be a desire to reduce the risk of transmission from individuals working in high-risk settings to virtually zero. Furthermore, if a future influenza pandemic were associated with increased severity, the balance between preventing onward transmission and the need to limit societal disruption might be such that isolation guidelines would be more restrictive. Interpretation of the results should be cautious because of the absence of information on the periods of isolation undertaken by the index individuals under study; it is possible that within-household transmission was somewhat reduced following the return of the index individual to his or her usual activities (eg, work, school, and shopping).
This study demonstrates the value of using detailed epidemiological data obtained through household studies to determine the serial interval and the temporal distribution of secondary infections within households. By analyzing multiple studies simultaneously, we were able to increase precision and to show the consistency of the observed data. To our knowledge, this is the largest analysis to date of data from linked pairs of dates of the onset of ILI or ARI clinical symptoms in clinically apparent infectors and infectees. Our methods provide the estimated proportion of within-household transmission risk that remains following a particular number of days after symptoms, which, when combined with information on severity of the virus, informs policymakers of the impact of specified periods of isolation. Future methodological research priorities for this type of analysis include developing approximate methods for correcting for tertiary transmission in the absence of household-level data.
The pH1N1 Household Investigations Working Group
The pH1N1 Household Investigations Working Group: Adebowale Awosika-Olumo, Crystal Beasley, Achuyt Bhattarai, Matthew Biggerstaff, Dianna Blau, Karen Chronister, Fatimah Dawood, Ryan P. Fagan, Anne Marie France, Gale Galland, Michele Ginsberg, Matt Gladden, James Hadler, Jenifer Jaeger, Annie Kao, Paula Kriner, Hyewon Lee-Han, Karla Lopez, Michael Lynch, Azarnoush Maroufi, Christine Mattson, Ma'ria E. Moll, Stephen Ostroff, Byron Oujesky, Diane Rexin, Margarita Rios, Carol Roach, Trudi Shim, Kirstin Short, and David Sugerman.
Acknowledgments
We thank Conrad Weiser Area School District and the Houston Independent School District. We also thank the involved state and local health department staff for their assistance in this investigation.
Financial Support. This study was supported by the Medical Research Council (N. M. F., C. A. D., S. C.), the Bill and Melinda Gates Foundation (49276 to N. M. F.), the National Institute of General Medical Sciences (NIGMS) Models of Infectious Disease Agent Study (MIDAS) initiative (1U54GM088491-01 to N. M. F., C. A. D., and S. C.), the European Community's Seventh Framework Programme (201601 to N. M. F.), the Research Councils UK (to S. C.), an appointment to the Applied Epidemiology Fellowship Program administered by the Council of State and Territorial Epidemiologists (to T. L. M.), and the CDC Cooperative Agreement (U60/CCU007277 to T. L. M.). Funders took no part in the design or interpretation of the study. The content of this article is solely the responsibility of the authors and does not necessarily represent the views of the funding sources.
Supplement sponsorship. Published as part of a supplement entitled “The 2009 H1N1 Influenza Pandemic: Field and Epidemiologic Investigations,” sponsored by the Centers for Disease Control and Prevention.
Potential conflicts of interest. S.C. has received consulting fees from Sanofi Pasteur for advising them on modelling the spread of varicella and varicella herpes zoster. All other authors: no conflicts.
References
- 1.Ginsberg M, Hopkins J, Maroufi A, et al. Swine influenza A (H1N1) infection in two children–-Southern California, March–April 2009. MMWR. 2009;58:400–402. [PubMed] [Google Scholar]
- 2.Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team, Dawood F, Jain S, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. New Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. Global alert and response: influenza A(H1N1) - update 16. 2009 http://www.who.int/csr/don/2009_05_05a/en/index.html. Accessed 2 November 2010. [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC) Interim CDC guidance for nonpharmaceutical community mitigation in response to human infections with swine influenza (H1N1) virus. 2009 http://www.thebody.com/content/news/art51596.html. Accessed 2 November 2010. [Google Scholar]
- 5.Centers for Disease Control and Prevention (CDC) CDC recommendations for the amount of time persons with influenza-like illness should be away from others. 2009 http://www.cdc.gov/H1N1flu/guidance/exclusion.htm. Accessed 2 November 2010. [Google Scholar]
- 6.Centers for Disease Control and Prevention (CDC) CDC guidance for state and local public health officials and school administrators for school (K-12) responses to influenza during the 2009-2010 school year. 2009 http://www.cdc.gov/h1n1flu/schools/schoolguidance.htm. Accessed 2 November 2010. [Google Scholar]
- 7.Bell DM. Non-pharmaceutical interventions for pandemic influenza, international measures. Emerg Infect Dis. 2006;12:81–87. doi: 10.3201/eid1201.051370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrat F, Vergu E, Ferguson NM, et al. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol. 2008;167:775–785. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
- 9.Aoki FY, Boivin G. Influenza virus shedding: excretion patterns and effects of antiviral treatment. J Clin Virol. 2009;44:255–261. doi: 10.1016/j.jcv.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Sato M, Hosoya M, Kato K, Suzuki H. Viral shedding in children with influenza virus infections treated with neuraminidase inhibitors. Pediatr Infect Dis J. 2005;24:931–932. doi: 10.1097/01.inf.0000180976.81055.ce. [DOI] [PubMed] [Google Scholar]
- 11.Weinstock DM, Gubareva LV, Zuccotti G. Prolonged shedding of multidrug-resistant influenza A virus in an immunocompromised patient. N Engl J Med. 2003;348:867–868. doi: 10.1056/NEJM200302273480923. [DOI] [PubMed] [Google Scholar]
- 12.De Serres G, Rouleau I, Hamelin ME, Quach C, Skowronski D, Flamand L. Contagious period for pandemic (H1N1) 2009. Emerg Infect Dis. 2010 doi: 10.3201/eid1605.091894. http://www.cdc.gov/EID/content/16/5/783.htm. Accessed 2 November 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling LM, Chow AL, Lye DC, et al. Effects of early oseltamivir therapy on viral shedding in 2009 pandemic influenza A (H1N1) virus infection. Clin Infect Dis. 2010;50:963–969. doi: 10.1086/651083. [DOI] [PubMed] [Google Scholar]
- 14.Witkop CT, Duffy MR, Macias EA, et al. Novel influenza A (H1N1) outbreak at the US Air Force Academy - epidemiology and viral shedding duration. Am J Prev Med. 2010;38:121–126. doi: 10.1016/j.amepre.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 15.flusurvey. Internet-based monitoring system for influenza surveillance – social contact patterns. 2009 http://flusurvey.org.uk. Accessed 2 November 2010. [Google Scholar]
- 16.Kamate S, Agrawal A, Chaudhary H, Singh K, Mishra P, Asawa K. Public knowledge, attitude and behavioural changes in an Indian population during the influenza A (H1N1) outbreak. J Infect Dev Ctries. 2010;4:7–14. doi: 10.3855/jidc.501. [DOI] [PubMed] [Google Scholar]
- 17.Bish A, Michie S. Demographic and attitudinal determinants of protective behaviours during a pandemic: a review. Br J Health Psychol. 2010;15(Pt 4):797–824. doi: 10.1348/135910710X485826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cauchemez S, Carrat F, Viboud C, Valleron AJ, Boelle PY. A Bayesian MCMC approach to study transmission of influenza: application to household longitudinal data. Stat Med. 2004;23:3469–3487. doi: 10.1002/sim.1912. [DOI] [PubMed] [Google Scholar]
- 19.Ferguson NM, Cummings DA, Cauchemez S, et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005;437:209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 20.Cauchemez S, Donnelly CA, Reed C, et al. Household transmission of 2009 pandemic influenza A (H1N1) virus in the United States. N Engl J Med. 2009;361:2619–2627. doi: 10.1056/NEJMoa0905498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fine PEM. The interval between successive cases of an infectious disease. Am J Epidemiol. 2003;158:1039–1047. doi: 10.1093/aje/kwg251. [DOI] [PubMed] [Google Scholar]
- 22.Cowling BJ, Fang VJ, Riley S, Peiris JSM, Leung GM. Estimation of the serial interval of influenza. Epidemiology. 2009;20:344–347. doi: 10.1097/EDE.0b013e31819d1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White LF, Wallinga J, Finelli L, et al. Estimation of the reproductive number and the serial interval in early phase of the 2009 influenza A/H1N1 pandemic in the USA. Influenza Other Respir Viruses. 2009;3:267–276. doi: 10.1111/j.1750-2659.2009.00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghani AC, Baguelin M, Griffin J, et al. The early transmission dynamics of H1N1pdm influenza in the United Kingdom. PLoS Curr. 2009 doi: 10.1371/currents.RRN1130. 1;RRN1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lessler J, Reich NG, Cummings DAT New York City Department of Health and Mental Hygiene Swine Influenza Investigation Team. Outbreak of 2009 pandemic influenza A (H1N1) at a New York City school. 2009;361:2628–2636. doi: 10.1056/NEJMoa0906089. [DOI] [PubMed] [Google Scholar]
- 26.France AM, Jackson M, Schrag S, et al. Household transmission of 2009 influenza A (H1N1) virus after a school-based outbreak in New York City, April–May 2009. J Infect Dis. 2010;201:984–992. doi: 10.1086/651145. [DOI] [PubMed] [Google Scholar]
- 27.Morgan OW, Parks S, Shim T, et al. Household transmission of pandemic (H1N1) 2009, San Antonio, Texas, USA, April-May, 2009. Emerg Infect Dis. 2010;16:631–637. doi: 10.3201/eid1604.091658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moser MR, Bender TR, Margolis HS, Noble GR, Kendal AP, Ritter DG. An outbreak of influenza aboard a commercial airliner. Am J Epidemiol. 1979;110:1–6. doi: 10.1093/oxfordjournals.aje.a112781. [DOI] [PubMed] [Google Scholar]
- 29.Lessler J, Reich NG, Brookmeyer R, Perl TM, Nelson KE, Cummings DA. Incubation periods of acute respiratory viral infections: a systematic review. Lancet Infect Dis. 2009;9:291–300. doi: 10.1016/S1473-3099(09)70069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]