Fungi are detectable in normal volunteers by molecular means, and therefore the lungs are not sterile from a fungal perspective. There is a highly variable range of fungal load in patients with allergic, chronic and invasive aspergillosis, which may have clinical consequences. Resistance to azoles can be documented directly in clinical samples.
Abstract
Background. Oral triazole therapy is well established for the treatment of invasive (IPA), allergic (ABPA), and chronic pulmonary (CPA) aspergillosis, and is often long-term. Triazole resistance rates are rising internationally. Microbiological diagnosis of aspergillosis is limited by poor culture yield, leading to uncertainty about the frequency of triazole resistance.
Methods. Using an ultrasensitive real-time polymerase chain reaction (PCR) assay for Aspergillus spp., we assessed respiratory fungal load in bronchoalveolar lavage (BAL) and sputum specimens. In a subset of PCR-positive, culture negative samples, we further amplified the CYP51A gene to detect key single-nucleotide polymorphisms (SNPs) associated with triazole resistance.
Results. Aspergillus DNA was detected in BAL from normal volunteers (4/11, 36.4%) and patients with culture or microscopy confirmed IPA (21/22, 95%). Aspergillus DNA was detected in sputum in 15 of 19 (78.9%) and 30 of 42 (71.4%) patients with ABPA and CPA, compared with 0% and 16.7% by culture, respectively. In culture-negative, PCR-positive samples, we detected triazole-resistance mutations (L98H with tandem repeat [TR] and M220) within the drug target CYP51A in 55.1% of samples. Six of 8 (75%) of those with ABPA and 12 of 24 (50%) with CPA had resistance markers present, some without prior triazole treatment, and in most despite adequate plasma drug concentrations around the time of sampling.
Conclusions. The very low organism burdens of fungi causing infection have previously prevented direct culture and detection of antifungal resistance in clinical samples. These findings have major implications for the sustainability of triazoles for human antifungal therapy.
Aspergillus spp. cause diseases ranging from invasive pulmonary aspergillosis (IPA) in immunocompromised patients to chronic pulmonary aspergillosis (CPA) and fungal allergic diseases, including allergic bronchopulmonary aspergillosis (ABPA) and increased severity of asthma (severe asthma with fungal sensitization [SAFS]) [1, 2]. Millions of individuals worldwide are affected or at risk; recent estimates indicate approximately 3 million patients with CPA, 3 million with ABPA and over 10 million with SAFS [3]. Exposure to hundreds of Aspergillus fumigatus conidia is a universal, daily occurrence. Conidia shift from being anergic to the human immune system [4] to producing the largest number of documented allergens of any other living organism on germination [5].
Antifungal therapy with triazoles is recommended for patients with ABPA, CPA, and IPA [6]. There are three licensed triazole compounds highly active against Aspergillus spp.—itraconazole, voriconazole, and posaconazole [7]. However, triazole resistance has emerged as an important factor limiting successful clinical outcome. Itraconazole resistance in A fumigatus was detected in isolates from California in the late 1980s. In the US recently, Martinez et al. [8] found A fumigatus itraconazole minimum inhibitory concentration (MIC) ≥8 mg/L in 13 of 25 (52%) in 2002–2009, compared with 13 of 126 (10%) in 1987–2001; and in Detroit, 18 of 37 (49%) A fumigatus isolates had elevated MICs to the triazoles in 2009, compared with 11 of 45 (24%) in 2003 [9]. Increasing resistance rates have been found since 2004 in the Netherlands and the UK [10–12], with 20% of patients in Manchester in 2009 having triazole-resistant isolates [12]. Extensive use of azoles in agriculture is the putative culprit [13], with the emergence of resistance during treatment documented [11]. Current methodology for resistance detection requires a positive culture but the high frequency of negative cultures greatly limits our ability to detect it.
Oral triazole therapy is given for years to patients with ABPA, SAFS, and CPA, usually safely and effectively [14–17]. Development of resistance results in loss of control of the disease [11, 18]. In these patients, cultures are often negative, and more sensitive means of establishing the reason for loss of disease control are required. For similar reasons, selecting the correct initial therapy as fast as possible is important for good outcomes in IPA [11, 19] with mortality rates ranging from approximately 40–90% with treatment [20, 21].
Mutations in the CYP51A gene, encoding the azole target protein lanosterol 14α-demethylase, are responsible for most instances of resistance [10, 11, 22]. Mutations may result in structural alterations to the enzyme [22]. The most important CYP51A gene resistance mutations are at codons 54, 220, and 98, although we and others have reported several other mutations [10–12, 23, 24]. A strong bias toward key mutations conferring azole resistance enables direct molecular detection without first culturing A fumigatus.
METHODS
Processing of Normal Volunteer BAL Samples
All volunteers gave written informed consent and the study was approved by the local Ethical Review Committee. Each volunteer underwent a standard bronchoscopy and bronchoalveolar lavage (BAL). Up to 25 mL BAL fluid was centrifuged and the pellets subjected to DNA extraction using the MycXtra fungal DNA extraction kit (Myconostica Ltd). Up to 5 mL was centrifuged for culture and from the resuspended pellet was streaked on two Sabouraud plates and incubated at 30°C and 37°C for 7 days, according to the UK national methodology [25].
Processing of Invasive Aspergillosis BAL Samples
All BAL specimens had been collected from at-risk and infected patients as part of a standard diagnostic workup in Innsbruck over 3 years. All samples were processed prospectively in the same way when received in the clinical laboratory, and excess samples stored for retrospective PCR analysis. The criteria for the diagnosis of IPA are consistent with modified European Organization for Research and Treatment of Cancer/Mycoses Study Group (EORTC/MSG) criteria [26]. Fungal culture and microscopy of 1–5 mL BAL fluid (volume dependent) followed centrifugation (15,000g, 5 min) and resuspension in 0.5 mL. One drop of resuspended sample was inspected under fluorescent microscopy with Calcofluor. The remainder was plated on 3 fungal media (Sabouraud with chloramphenicol, brain heart infusion, and malt extract) and cultured at 30°C and 37°C. Positive cultures were identified by conventional means. Bacterial culture and gram stain were done with 100–150 μL of BAL fluid. Residual sample was stored at –20°C or (usually) –80°C prior to DNA extraction with the MycXtra fungal DNA extraction kit and real-time PCR. All clinical data were anonymized.
Processing of Sputum Samples
Sputum samples were collected from CPA and ABPA patients in Manchester. Patients gave written informed consent. The diagnosis of CPA was based on antibody and radiology data [16], ABPA on clinical and serological data [27, 28], and SAFS as described previously [15]. Samples were split for fungal culture, microscopy, and DNA extraction. Sputum was digested with Sputasol (ratio 1:1), vortexed, and 10 μL-streaked on two Sabouraud plates [25], which were incubated at 30°C and 37°C for 7 days. DNA extraction was performed from 0.5–3 mL of sample immediately after the samples were received, following the MycXtra kit instructions. DNA was eluted in 40 μL of buffer S5 and 10 μL was used for quantitative PCR (qPCR).
Aspergillus PCR Assay
We used the commercially available real-time PCR diagnostic assay MycAssay Aspergillus (Myconostica) for the detection of Aspergillus spp. At least 15 different Aspergillus spp. are detected with the assay, including the 5 most frequent pathogenic species, and Penicillium spp. It uses molecular beacons and targets an area of the 18S ribosomal RNA (rRNA) genome [29]. An internal control sequence of plant origin was included to detect any PCR inhibitors in the sample. On the Cepheid SmartCycler, the assay limit of blank is a cycle threshold (Ct) of 38 cycles with a target sensitivity of 50 18S copies, approximately 1 genome, given a range of 18S copy number from 37 to 90 copies per A fumigatus genome [30]. The assay is CE-marked with a nonquantitative endpoint but we chose to use it (off-label) in a quantitative manner to assess fungal load.
Direct Detection of Key Azole Resistance Mutations
As the quantity of Aspergillus DNA was modest in most samples (and absent in the IPA samples), a nested PCR approach was used to obtain maximum sensitivity. We partially amplified the CYP51A gene in two ∼900 bp fragments. Fragment 1 (876 bp) covered the promoter tandem repeat region to codon 98. The second amplicon (748 bp) covered codons 54 to 266. The amplified products were evaluated in a real-time assay with allele-specific molecular beacon directed at key single-nucleotide polymorphisms (SNPs) linked with azole resistance (G54, L98 + promoter tandem repeat [TR], G138, and M220). All results were confirmed by DNA sequencing. Patients’ notes were reviewed for their antifungal treatment. Resistance data were not used for clinical decision making.
RESULTS
Extraction of Aspergillus DNA from Respiratory Samples
Extraction of sufficient fungal DNA for molecular detection is the most challenging technical aspect of PCR for fungi. The combination of very few fungal cells in a clinical sample and a sturdy cell wall requiring fracture for DNA release is problematic. We utilized an optimized bead-beating approach to break open cells, preceded by a digestion step. Overall, 10% efficiency from unswollen conidia was demonstrated (Supplementary Figure S1).
Detection of Aspergillus DNA in Volunteers with PCR
To better understand Aspergillus burdens in the lungs of healthy individuals, we tested BAL from 11 normal adults who underwent bronchoscopy. Of these, 4 culture-negative samples (36.4%) had detectable signals in the PCR assay (Table 1). No signal was detected in 7 samples (63.6%), of which one grew Penicillium spp. (3 morphologies) and 1 Paecilomyces spp. The positive Ct values ranged from 36.2 to 34.3 (Figure 1), consistent with Aspergillus spp. being present in normal lungs.
Table 1.
Aspergillus Culture, qPCR, and A fumigatus Resistance Mutation Detection in 4 Study Populations
| Laboratory result | ABPA | CPA | IPA | Normals |
| Culture positive for Aspergillus spp. | 0/19 | 7/42 (16.7%) | 20/22 (90.9%) | 0/11 |
| Culture positive for A fumigatus | 0/19 | 7/42 (16.7%) | 10/22 (45.5%) | 0/11 |
| qPCR positive for Aspergillus spp | 15/19 (78.9%) | 30/42 (71.4%) | 21/22 (95.5%) | 4/11 (36.4%) |
| A. fumigatus CYP51A mutation detected directly from qPCR-positive sample | 6/8 (75%) | 12/24 (50%) | NTa | NTa |
NOTE. qPCR indicates quantitative polymerase chain reaction; ABPA, allergic bronchopulmonary aspergillosis; CPA, chronic pulmonary aspergillosis; IPA, invasive pulmonary aspergillosis.
NT indicates not tested (insufficient sample remaining).
Figure 1.
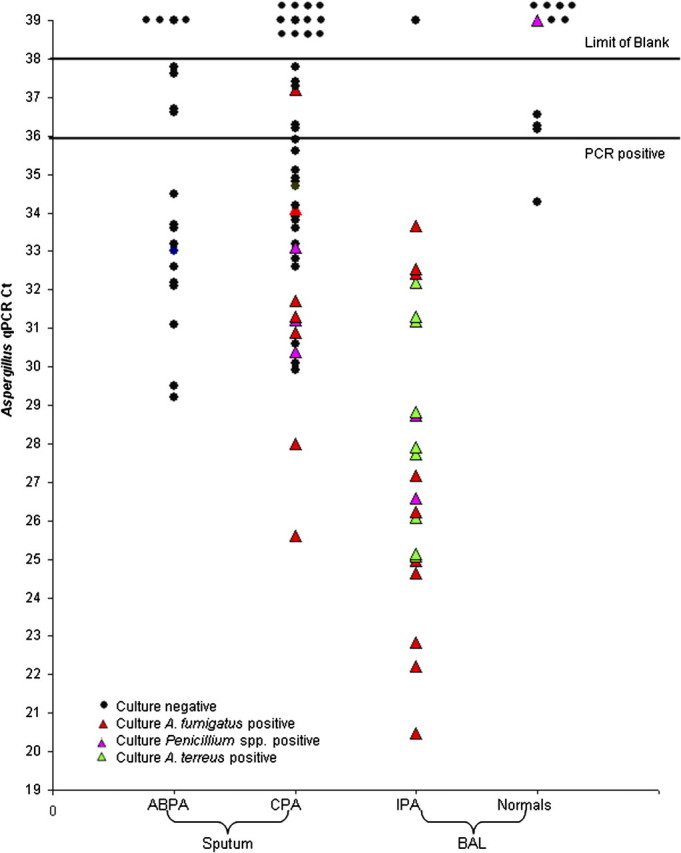
Aspergillus load measured by quantitative polymerase chain reaction (qPCR) in respiratory samples from 3 patient groups and 1 volunteer group. Spontaneously produced sputum in clinic from patients with allergic bronchopulmonary aspergillosis (ABPA) (including one patient with severe asthma with fungal sensitization [SAFS]) and chronic pulmonary aspergillosis (CPA) were split for culture and DNA extraction before qPCR. Ct, cycle threshold.
PCR in Invasive Pulmonary Aspergillosis
We analyzed 22 samples from patients with IPA with mycological confirmation. Of the 22 samples, 20 (90.9%) had hyphae consistent with Aspergillus spp. visible on microscopy. All 22 (100%) were culture-positive for a filamentous fungus, 10 for A fumigatus, 9 for A terreus, and 2 for Penicillium spp., and 1 grew A niger, Rhizopus oryzae, and Lichtheimia corymbifera (PCR-negative). Five of the patients had proven and 17 probable IPA in the context of typical immunocompromising conditions, including organ transplant (n = 10) and acute leukemia. Using the normal volunteer data as negative controls and a Ct cut-off of 36, the sensitivity was 94%, specificity 91%, positive predictive value 97%, and negative predictive value 83%. Seventeen patients (77.3%) had received some antifungal prophylaxis or therapy. Aspergillus DNA was detected by PCR in 21 (95.5%) samples (Table 1) with Ct values ranging from 20.5 to 33.7 (Figure 1). Both samples that grew Penicillium were PCR-positive. Furthermore, in these 22 samples, the signal strength was generally much stronger than that in the normal volunteers, indicative of a greater load of Aspergillus in IPA than in normal people.
PCR in Chronic and Allergic Aspergillosis
In spontaneously produced sputum from patients with ABPA, SAFS, and CPA, we detected Aspergillus DNA much more frequently than cultures were positive. In the ABPA patients, all cultures were negative despite strongly positive immunoglobin E (IgE) serology for A fumigatus. Aspergillus DNA was detectable by PCR in 15 of 19 (78.9%) ABPA patients (Table 1), 11 of these samples having strong PCR signals (Figure 1). Among the 42 patients with CPA, all of whom had detectable Aspergillus IgG antibodies and grossly abnormal chest radiographs, 7 (16.7%) had a positive culture for A fumigatus and 30 (71.4%) had Aspergillus DNA detectable by PCR (Table 1). In patients with CPA, stronger PCR signals were generally seen in those with positive cultures.
Direct Detection of Azole Resistance
We selected DNA from the first 25 sputum samples obtained from ABPA and CPA patients that were PCR-positive, culture-negative, as well as 4 culture-positive, PCR-positive samples patients with CPA (Supplementary Table S1). No G54 or M138 mutations were found. Four samples had M220 mutations: 2 were M220K and 2 M220R cyp51A substitutions on sequencing. Twenty-seven of 29 (93.1%) had an L98H mutation, and 16 (55.2%) also had an upstream 34 bp TR, the combination conferring itraconazole and voriconazole resistance [10]. The TR was found without the L98H mutation in 2 samples. Two samples had an M220R mutation with both the TR and L98H mutation. Of the 4 culture-positive samples, 2 yielded susceptible isolates, and in one of these clinical samples, no resistance mutations were detected in the other TR + L98H was detected by molecular screening. One patient’s isolate was azole-resistant (itraconazole MIC >8 mg/L, voriconazole MIC 2.0 mg/L, and posaconazole 0.25 mg/L), but no resistance mutation was found in the corresponding clinical sample from which this isolate was cultured or on sequencing the CYP51A gene from the isolate. The fourth patient isolate was multi-azole–resistant and an M220K mutation was found directly in the clinical sample and confirmed by CYP51A sequencing of the isolate. Overall, 16 of 29 (55.1%) samples had evidence of azole resistance and 2 of 4 (50%) isolates.
Prior and Concurrent Azole Therapy
Of the 28 patients sampled (one CPA was sampled both prior to starting therapy and while on posaconazole), resistance markers were found in 6 of 8 (75%) with ABPA or SAFS, and 10 of 20 (50%) with CPA. The time of sampling and azole exposure could be relevant to finding resistance markers; in Table 2 and Supplementary Table S1, these relationships are shown. Low plasma concentrations were seen in 2 of 7 (29%), 0 of 10, and 2 of 5 (40%) patients taking itraconazole, posaconazole, and voriconazole, respectively, but many had received prior therapy. Overall numbers are too small to draw many conclusions other than prior azole exposure and current therapy at the time of sampling does not reliably predict azole resistance marker detection.
Table 2.
Interrelationship Between Azole Therapy, Sampling Time, and Frequency of Azole Resistance Marker Detected
| Azole treatment experience | Number of patients with azole resistance marker/total tested (%) |
||||
| Sample collected on azole therapy |
Totals | ||||
| Itra | Vori | Posa | None | ||
| Azole naive | – | – | – | 2/3 (67) | 2/3 (67) |
| Itra only | 2/5 (40)a | – | – | 2/2 (100)a | 4/7 (57) |
| Posa only | – | – | 1/4 (25)a | 0/1 (0) | 1/4 (25) |
| Itra + vori | 1/1 (100) | 2/4 (50) | - | 2/2 (100) | 5/7 (71) |
| Itra + Posa | – | – | 1/2 (50) | – | 1/3 (33) |
| Itra + vori + posa | – | – | 3/5 (60)a | – | 3/5 (40) |
| Totals | 3/6 (50) | 2/4 (50) | 5/11 (45) | 6/8 (75) | 16/29 (55) |
NOTE. Itra indicates itraconazole; vori, voriconazole; posa, posaconazole.
M220 mutation (n = 4).
Resistance and Therapeutic Outcome
Two of three patients who had never received azoles had L98H and TR detected; both were culture-negative. One was treated with itraconazole and died of progressive CPA within 3 months. The other was treated with posaconazole and remained stable over the following 12 months. Three of the 4 patients with the M220 marker detected failed therapy immediately (itraconazole 1, posaconazole 3), and the fourth failed therapy 12 months later. Of the 14 patients with L98H and TR markers only, 3 were unevaluable, 6 had failed itraconazole or voriconazole treatment, and 5 had stabilized or improved on posaconazole (n = 3), itraconazole, or voriconazole. Rescue therapy for those with pan-azole–resistant infections includes thrice weekly liposomal amphotericin B or six times weekly micafungin or caspofungin, through a Port-A-Cath.
DISCUSSION
Aspergillus spp. can be detected in sputum samples and lung tissue in ‘uninfected’ patients by optimized culture in 38–42% of people sampled [31, 32]. Our data showing that 36.3% of normal healthy volunteers have detectable Aspergillus (or Penicillium) spp. DNA in BAL samples clearly indicates that the lungs are not typically sterile from a fungal perspective, in contrast to bacteria. This might be expected from daily inhalation of spores and hyphal fragments [33] from the environment.
Culture-positive rates in confirmed IPA are typically ∼30% or less [34–36]. Few culturable conidia or hyphae are present in a typical airway sample from those with disease. In one small study of culture-positive cases, less than 20 colony-forming units (CFUs) were cultured from 75% of patients, most with invasive aspergillosis [37]. In 3 series of CPA patients, cultures were positive in 10 of 18 (56%), 15 of 24 (65%), and 34 of 42 (81%) [16, 17, 38]. In ABPA, rates of culture positivity were 58% if 3 specimens were examined [39] and 60% in another study [40]. In our recent study of SAFS (same laboratory methodology), only 2 of 58 (3.5%) grew Aspergillus spp., despite frequent requests [15]. In a recently published study using induced sputum and direct plating onto potato dextrose agar, 7% of normal volunteers and 31–63% of asthmatics grew A fumigatus [41]. Clearly, studies of optimal culture methods and sampling are needed, including the impact of antifungal therapy on yield.
Some laboratories have employed PCR-based detection of Aspergillus to improve sensitivity with varying degrees of success [42]. Variability among labs has prompted calls for standardized and validated PCR tests for Aspergillus spp. As untreated IPA is 100% fatal without complete resolution of immunosuppression (and usually rapidly so) [20, 21], a key target for a molecular diagnostic assay is the early, sensitive diagnosis of IPA. Almost all prior molecular detection studies have focused on BAL or blood from immunocompromised patients [42, 43] and very few on spontaneously produced sputum. In addition, quantification of the fungal load in the airways with qPCR offers much greater precision and dynamic range, facilitating greatly improved understanding of fungal infectious and allergic syndromes, including the time course of infection.
The rapid identification of azole resistance in culture-negative samples provides compelling evidence that PCR offers a more sensitive methodology than culture for the detection of Aspergillus spp. Given that azoles are critical components of antifungal therapy, the emergence of azole resistance will clearly impact clinical outcomes. The high prevalence of resistance markers detected among patients with chronic Aspergillus infections in the absence of confirmed cultures has important implications for clinical care. If patients harbor resistant strains, as suggested by this molecular marker data, they would be expected to respond poorly to therapy or would show diminished response if a mixed (susceptible and resistant) population was present. We found response rates to antifungal therapy of <50% in ABPA and CPA. Thus, the molecular resistance data may help explain a long-standing clinical conundrum of modest response rates to azole therapy, despite apparently adequate therapy and favorable immune status. Alterative treatments should be considered when resistance markers are identified. The loss of disease control with the emergence of resistance is most consistent with antifungal activity being mediated directly and not by immunological or steroid-boosting effects [15, 44].
A key consideration is whether the molecular approach to finding or excluding resistance demonstrated here is more comprehensive than culture, or too sensitive. In conventional microbiology, only a single colony is typically selected for susceptibility testing, whereas sampling all the Aspergillus DNA in a clinical specimen allows detection of resistance genotypes present in only a subpopulation of infecting strains. We suspect this is the reason why we have found both a higher frequency of resistance overall and a much higher proportion of L98H mutations than we find in isolates that have grown in culture [11, 12]. Reduced culturability on agar (but not necessarily virulence) of certain resistant strains is also a possible explanation of this disparity. The unexpected finding of multiple mutations in the same sample (but not necessarily the same strain) (M220K and L98H) is novel. Likewise, the dissociation of the TR and L98H has not been described to date. Prospective studies with greater statistical power and careful patient monitoring are required.
Enabling direct detection of resistance using rapid molecular methods greatly facilitates optimal therapy for individual patients. Not only will ineffective therapy be avoided, but alternative strategies utilizing combination therapy become directly testable, with microbiological endpoints, instead of relying on imperfect clinical and surrogate biomarker endpoints, as is currently the case. However, translation of the finding of a specific cyp51A mutation into a treatment decision requires additional work for all second-generation triazoles, as our understanding of cross-resistance is currently limited. Furthermore, not all resistance is mediated by cyp51A mutations [9], so while detection of a key SNP is helpful in detecting resistance (and is more sensitive than culture), a negative screen does not rule out resistance, as demonstrated in one patient in this series. Likewise, new mutations conferring resistance are likely to continue to arise, and might be missed without a sequence-based approach.
Our remarkably high rate of resistance (55.1%) needs confirmation from other groups. Triazole resistance rates in A fumigatus cultures of this magnitude have been seen in 2 US institutions [8, 9], and lower (but highly significant) rates in Europe [12, 13]. The environmental presence of azole-resistant A fumigatus strains has been documented in Europe. Resistant strains have been found in the environment in numerous countries, including Denmark and Belgium; 10 of 570 (1.8%) A fumigatus cells found in the air in Belgium were resistant to itraconazole [45]. Thus, the human lung may be a filter of Aspergilli in the air, and what is found in samples reflects recent exposure, especially in patients who cannot clear this fungus from their respiratory tract. Clearly, rapidly expanding antifungal therapy worldwide will favor persistence of resistant subpopulations.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://www.oxfordjournals.org/our_journals/cid/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Acknowledgments
We are indebted to Chris Harris for sourcing patient clinic notes, and to Sarah Follett, Adrian Moody, and Gillian Morgan at Myconostica for their guidance and support.
Financial support.This work was supported by grants from Myconostica and the National Institute Health Research Translational Research Facility in Respiratory Medicine, and National Institutes of Health grant AI066561 to D.S.P. Saudi Arabia’s Ministry of Health funds A.B. The Chronic Granulomatous Disorder Research Trust partially funds M.K. The work of the Mycology Reference Centre Manchester has been underwritten by the Fungal Research Trust since 1991.
Potential conflicts of interest.D.W.D. holds founder shares in F2G Ltd and Myconostica Ltd, both University of Manchester spin-out companies, and has received grant support from F2G as well as the Fungal Research Trust, the Wellcome Trust, the Moulton Trust, The Medical Research Council, The Chronic Granulomatous Disease Research Trust, the National Institute of Allergy and Infectious Diseases, National Institute of Health Research and the European Union, AstraZeneca, and Basilea. He continues to act as an advisor/consultant to F2G and Myconostica as well as other companies over the last 5 years, including Basilea, Vicuron (now Pfizer), Pfizer, Schering Plough, Nektar, Daiichi, Astellas, Gilead, and York Pharma. He has been paid for talks on behalf of Schering, Astellas, Merck, Dainippon, and Pfizer. C.L.-F. acts as consultant to Pfizer, Astellas, and Schering Plough, and was paid for talks on behalf of Pfizer, Astellas, Schering Plough, Merck, and Gilead. C.B.M. holds a grant from Pfizer and is a shareholder in Myconostica. She has been paid for talks on behalf of Pfizer. P.B. .holds grants from the EU, Fungal Research Trust, AstraZeneca, and Alergenitica, and is a shareholder in Myconostica. D.S.P. receives support from the US National Institute of Allergy and Infectious Diseases; he has received past support from Pfizer and Merck and participates in expert panels for these companies, and is a shareholder in Myconostica. All other authors report no potential conflilcts.
References
- 1.Hope WW, Walsh TJ, Denning DW. The invasive and saprophytic syndromes due to Aspergillus spp. Med Mycol. 2005;43(Suppl 1):S207–38. doi: 10.1080/13693780400025179. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal R, Aggarwal AN, Gupta D, Jindal SK. Aspergillus hypersensitivity and allergic bronchopulmonary aspergillosis in patients with bronchial asthma: systematic review and meta-analysis. Int J Tuberc Lung Dis. 2009;13:936–44. [PubMed] [Google Scholar]
- 3.Denning D, Pleuvry A, Cole D. Global burden of allergic bronchopulmonary aspergillosis (ABPA) complicating asthma. Thorax 2010; 65(Suppl 4):A155–A155. [Google Scholar]
- 4.Aimanianda V, Bayry J, Bozza S, et al. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature. 2009;460:1117–21. doi: 10.1038/nature08264. [DOI] [PubMed] [Google Scholar]
- 5.Fedorova ND, Khaldi N, Joardar VS, et al. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 2008;4:e1000046. doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh TJ, Anaissie EJ, Denning DW, et al. Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America (IDSA) Clin Infect Dis. 2008;46:327–60. doi: 10.1086/525258. [DOI] [PubMed] [Google Scholar]
- 7.Denning DW, Hope WW. Therapy for fungal diseases: opportunities and priorities. Trends Microbiol. 2010;18:195–204. doi: 10.1016/j.tim.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Martinez M, Cloud G, Chen V, Stevens DA. 4th Advances Against Aspergillosis. Rome: Itraconazole and amphotericin B resistance 1987–2009 in clinical Aspergillus fumigatus in northern California [Abstract 30] February 4–6th 2010. [Google Scholar]
- 9.Krishnan-Natesan S, Swaminathan S, Cutright J, et al . 50th Interscience Conference of Antimicrobial Agents Chemotherapy. Boston, MA: Antifungal susceptibility pattern of Aspergillus fumigatus isolated from clinical specimens in Detroit Medical Center (DMC): rising frequency of high MIC of azoles (2003–06) [Abstract M-389] American Society of Microbiology. September 12–14th 2010. [Google Scholar]
- 10.Snelders E, van der Lee HA, Kuijpers J, et al. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 2008;5:e219. doi: 10.1371/journal.pmed.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard SJ, Cerar D, Anderson MJ, et al. Frequency and evolution of Azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis. 2009;15:1068–76. doi: 10.3201/eid1507.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bueid A, Howard SJ, Moore CB, et al. Azole antifungal resistance in Aspergillus fumigatus—2008 and 2009. J Antimicrob Chemother. 2010;65:2116–8. doi: 10.1093/jac/dkq279. [DOI] [PubMed] [Google Scholar]
- 13.Verweij PE, Snelders E, Kema GH, Mellado E, Melchers WJ. Azole resistance in Aspergillus fumigatus: a side-effect of environmental fungicide use? Lancet Infect Dis. 2009;9:789–95. doi: 10.1016/S1473-3099(09)70265-8. [DOI] [PubMed] [Google Scholar]
- 14.Stevens DA, Schwartz HJ, Lee JY, et al. A randomized trial of itraconazole in allergic bronchopulmonary aspergillosis. N Engl J Med. 2000;342:756–62. doi: 10.1056/NEJM200003163421102. [DOI] [PubMed] [Google Scholar]
- 15.Denning DW, O'Driscoll BR, Powell G, et al. Randomized controlled trial of oral antifungal treatment for severe asthma with fungal sensitisation (SAFS), the FAST study. Am J Respir Crit Care Med. 2009;179:11–8. doi: 10.1164/rccm.200805-737OC. [DOI] [PubMed] [Google Scholar]
- 16.Denning DW, Riniotis K, Dobrashian R, Sambatakou H. Chronic cavitary and fibrosing pulmonary and pleural aspergillosis: case series, proposed nomenclature and review. Clin Infect Dis. 2003;37(Suppl 3):S265–80. doi: 10.1086/376526. [DOI] [PubMed] [Google Scholar]
- 17.Camuset J, Nunes H, Dombret MC, et al. Treatment of chronic pulmonary aspergillosis by voriconazole in non-immunocompromised patients. Chest. 2007;131:1435–41. doi: 10.1378/chest.06-2441. [DOI] [PubMed] [Google Scholar]
- 18.Howard SJ, Pasqualotto AC, Denning DW. Azole resistance in ABPA and Aspergillus bronchitis. Clin Microbiol Infect. 2010;16:683–8. doi: 10.1111/j.1469-0691.2009.02911.x. [DOI] [PubMed] [Google Scholar]
- 19.van der Linden JW, Jansen RR, Bresters D, et al. Azole-resistant central nervous system aspergillosis. Clin Infect Dis. 2009;48:1111–3. doi: 10.1086/597465. [DOI] [PubMed] [Google Scholar]
- 20.Pagano L, Caira M, Picardi M, et al. Invasive Aspergillosis in patients with acute leukemia: update on morbidity and mortality–SEIFEM-C Report. Clin Infect Dis. 2007;44:1524–5. doi: 10.1086/517849. [DOI] [PubMed] [Google Scholar]
- 21.Bulpa P, Dive A, Sibille Y. Invasive pulmonary aspergillosis in patients with chronic obstructive pulmonary disease. Eur Respir J. 2007;30:782–800. doi: 10.1183/09031936.00062206. [DOI] [PubMed] [Google Scholar]
- 22.Warrilow AG, Melo N, Martel CM, et al. Expression, purification and characterization of Aspergillus fumigatus sterol 14-{alpha} demethylase (CYP51) isoenzymes A and B. Antimicrob Agents Chemother. 2010;54:4920–3. doi: 10.1128/AAC.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balashov SV, Gardiner R, Park S, Perlin DS. Rapid, high-throughput, multiplex, real-time PCR for identification of mutations in the cyp51A gene of Aspergillus fumigatus that confer resistance to itraconazole. J Clin Microbiol. 2005;43:214–22. doi: 10.1128/JCM.43.1.214-222.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howard SJ, Webster I, Moore CB, et al. Multi-azole resistance in Aspergillus fumigatus. Int J Antimicrob Agents. 2006;28:450–3. doi: 10.1016/j.ijantimicag.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Standards Unit, Evaluations and Standards Laboratory, Centre for Infections. Investigation of bronchoalveolar lavage, sputum and associated specimens; BSOP57. www.hpa-standardmethods.org.uk/documents/bsop/pdf/bsop57.pdf. Accessed 4 September 2010. [Google Scholar]
- 26.De Pauw B, Walsh TJ, Donnelly JP, et al. Defining invasive fungal diseases for clinical research: revised definitions of the EORTC/MSG Consensus Group. Clin Infect Dis. 2008;46:181–21. doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ricketti AJ, Greenberger PA, Mintzer RA, Patterson R. Allergic bronchopulmonary aspergillosis. Arch Intern Med. 1983;143:1553–7. [PubMed] [Google Scholar]
- 28.Patterson R, Greenberger PA, Harris KE. Allergic bronchopulmonary aspergillosis. Chest. 2000;118:7–8. doi: 10.1378/chest.118.1.7. [DOI] [PubMed] [Google Scholar]
- 29.Khot PD, Ko DL, Hackman RC, Fredricks DN. Development and optimization of quantitative PCR for the diagnosis of invasive aspergillosis with bronchoalveolar lavage fluid. BMC Infect Dis. 2008;8:73. doi: 10.1186/1471-2334-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herrera ML, Vallor AC, Gelfond JA, Patterson TF, Wickes BL. Strain-dependent variation in 18S ribosomal DNA copy numbers in Aspergillus fumigatus. J Clin Microbiol. 2009;47:1325–32. doi: 10.1128/JCM.02073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullins J, Seaton A. Fungal spores in lung and sputum. Clin Allergy. 1978;8:525–33. doi: 10.1111/j.1365-2222.1978.tb01506.x. [DOI] [PubMed] [Google Scholar]
- 32.Lass-Flörl C, Salzer GM, Schmid T, Rabl W, Ulmer H, Dierichi MP. Pulmonary Aspergillus colonization in humans and its impact on management of critically ill patients. Br J Haematol. 1999;104:745–7. doi: 10.1046/j.1365-2141.1999.01260.x. [DOI] [PubMed] [Google Scholar]
- 33.Green BJ, Tovey ER, Beezhold DH, et al. Surveillance of fungal allergic sensitization using the fluorescent halogen immunoassay. J Mycol Med. 2009;19:253–61. doi: 10.1016/j.mycmed.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarrand JJ, Lichterfeld M, Warraich I, et al. Diagnosis of invasive septate mold infections. A correlation of microbiological culture and histologic or cytologic examination. Am J Clin Pathol. 2003;119:854–8. doi: 10.1309/EXBV-YAUP-ENBM-285Y. [DOI] [PubMed] [Google Scholar]
- 35.Hope WW, Walsh TJ, Denning DW. Laboratory diagnosis of invasive aspergillosis. Lancet Infect Dis. 2005;9:609–22. doi: 10.1016/S1473-3099(05)70238-3. [DOI] [PubMed] [Google Scholar]
- 36.Meersseman W, Lagrou K, Maertens J, et al. Galactomannan in bronchoalveolar lavage fluid: a tool for diagnosing aspergillosis in intensive care unit patients. Am J Respir Crit Care Med. 2008;177:27–34. doi: 10.1164/rccm.200704-606OC. [DOI] [PubMed] [Google Scholar]
- 37.Greub G, Bille J. Aspergillus species isolated from clinical specimens: suggested clinical and microbiological criteria to determine significance. Clin Microbiol Infect. 1998;4:710–6. doi: 10.1111/j.1469-0691.1998.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 38.Nam HS, Jeon K, Um SW, et al. Clinical characteristics and treatment outcomes of chronic necrotizing pulmonary aspergillosis: a review of 43 cases. Int J Infect Dis. 2010;14:e479–82. doi: 10.1016/j.ijid.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 39.McCarthy DS, Pepys J. Allergic bronchopulmonary aspergillosis. Clinical immunology. 2. Skin, nasal and bronchial tests. Clin Allergy. 1971;1:415–32. doi: 10.1111/j.1365-2222.1971.tb00793.x. [DOI] [PubMed] [Google Scholar]
- 40.Chakrabarti A, Sethi S, Raman DS, Behera D. Eight-year study of allergic broncho-pulmonary aspergillosis in an Indian teaching hospital. Mycoses. 2002;45:295–9. doi: 10.1046/j.1439-0507.2002.00738.x. [DOI] [PubMed] [Google Scholar]
- 41.Fairs A, Agbetile J, Hargadon B, et al. IgE Sensitisation to Aspergillus fumigatus is associated with reduced lung function in asthma. Am J Respir Crit Care Med. 2010;182:1362–8. doi: 10.1164/rccm.201001-0087OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuon FF. A systematic literature review on the diagnosis of invasive aspergillosis using polymerase chain reaction (PCR) from bronchoalveolar lavage clinical samples. Rev Iberoam Micol. 2007;24:89–94. [PubMed] [Google Scholar]
- 43.Mengoli C, Cruciani M, Barnes RA, Loeffler J, Donnelly JP. Use of PCR for diagnosis of invasive aspergillosis: systematic review and meta-analysis. Lancet Infect Dis. 2009;9:89–96. doi: 10.1016/S1473-3099(09)70019-2. [DOI] [PubMed] [Google Scholar]
- 44.Pasqualotto AC, Powell G, Niven R, Denning DW. The effects of antifungal therapy on severe asthma with fungal sensitisation and allergic bronchopulmonary aspergillosis. Respirology. 2009;14:1121–7. doi: 10.1111/j.1440-1843.2009.01640.x. [DOI] [PubMed] [Google Scholar]
- 45.Vanhee LM, Perman D, Nelis HJ, Coenye T. Rapid quantification of itraconazole-resistant Aspergillus fumigatus in air. J Microbiol Methods. 2010;81:197–9. doi: 10.1016/j.mimet.2010.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


