Abstract
Disruption of autophagy—a key homeostatic process in which cytosolic components are degraded and recycled through lysosomes—can cause neurodegeneration in tissue culture and in vivo. Upregulation of this pathway may be neuroprotective, and much effort is being invested in developing drugs that cross the blood brain barrier and increase neuronal autophagy. One well-recognized way of inducing autophagy is by food restriction, which upregulates autophagy in many organs including the liver; but current dogma holds that the brain escapes this effect, perhaps because it is a metabolically privileged site. Here, we have re-evaluated this tenet using a novel approach that allows us to detect, enumerate and characterize autophagosomes in vivo. We first validate the approach by showing that it allows the identification and characterization of autophagosomes in the livers of food-restricted mice. We use the method to identify constitutive autophagosomes in cortical neurons and Purkinje cells, and we show that short-term fasting leads to a dramatic upregulation in neuronal autophagy. The increased neuronal autophagy is revealed by changes in autophagosome abundance and characteristics, and by diminished neuronal mTOR activity in vivo, demonstrated by a reduction in levels of phosphorylated S6 ribosomal protein in Purkinje cells. The increased abundance of autophagosomes in Purkinje cells was confirmed using transmission electron microscopy. Our data lead us to speculate that sporadic fasting might represent a simple, safe and inexpensive means to promote this potentially therapeutic neuronal response.
Key words: autophagy, fasting, Purkinje cells, cortical neurons, confocal microscopy, electron microscopy, neuroprotection, starvation, CNS, cortex
Introduction
Autophagy is a key homeostatic mechanism whose physiological importance is reflected by its preservation throughout the eukaryotic phylogenetic tree, from yeast to mammals. In recent years, autophagy has been recognized as a crucial defense mechanism against malignancy, infection and neurodegenerative diseases.1–5 Abrogation of autophagy in neurons can lead to neurodegenerative disease1,2,4 and autophagy may play a role in the development of Alzheimer disease; indeed, enhancement of autophagy has been proposed as one possible treatment for this coming plague.6–8 Consequently, substantial research effort is being expended on attempts to develop drugs that can upregulate neuronal autophagy in the intact CNS. Herein we provide evidence to support the notion that the desired physiological goal may be attained by a simple, safe and inexpensive alternative approach: short-term food restriction.
Studies in yeast, and in tissue culture, have contributed significantly to our understanding of the molecular events that are involved in autophagy and its regulation. However, it has been difficult to observe the process in intact organs and tissues, and most especially in the brain.9 This is not due to the lack of in vivo mammalian models. For example, to facilitate the identification and analysis of autophagy in vivo, a transgenic mouse has been generated10,11 that encodes a fusion between GFP and microtubule-associated protein 1 light chain 3 (LC3). The lipidated form of LC3 specifically associates with autophagosomes12 and this protein is, currently, the most widely used marker of autophagy in mammalian cells. Others have used these GFP-LC3 mice to determine the effects of food restriction in vivo. It is well known that food restriction induces autophagy in many organs and tissues,9,10 but autophagy has not previously been detected in the brain even after 48 hours of food restriction; because of this, it has been proposed that the brain is metabolically privileged13,14 and that a more severe insult—e.g., a direct injury mediated by local ischemia or trauma—may be required to trigger an autophagic response within this organ.15,16 Herein, we combine two wellcharacterized techniques to investigate the effect of a brief period of food restriction on neuronal autophagy. We show that this approach identifies autophagosomes at a resolution that allows them to be not only enumerated, but also characterized in some detail. In addition, we have used a standard approach—transmission electron microscopy (TEM)—to confirm the increased abundance of autophagosomes in neurons of food-restricted mice. Our data demonstrate that, contrary to current dogma, food restriction causes a rapid and profound upregulation of autophagy in the brain.
Results
Six- to seven-week old male GFP-LC3 mice were food-restricted for 24 or 48 hours, a process known to induce fatty change and autophagy in the liver.9,10 Water was available ad libitum. These mice, and non-food-restricted control mice, were euthanized (with PBS perfusion) and vibratome sections of liver and brain were prepared and stained as described in Experimental Procedures. Confocal images were captured and analyzed.
Confocal microscopy of vibratome sections identifies autophagosomes in the livers of food-restricted mice.
We first validated the method by analyzing the liver, an organ in which autophagy is known to be induced by food restriction. Representative confocal images of liver are shown in Figure 1A. As expected, essentially no GFP signal was present in the liver of a C57BL/6 mouse. In normal-fed GFP-LC3 mice, GFP signal was detectable in hepatocytes, and was distributed throughout the cytosol. Following 24 hours of food restriction, individual autophagosomes could be detected as discrete regions of high fluorescence, and autophagosomes were even more abundant after 48 hours. The high resolution of the approach allowed quantitative analyses of individual autophagosomes. We were able to measure not only the number of vesicles per cell, but also their area, perimeter, feret (the longest diameter within an enclosed irregular shape), and circularity (Fig. 1B). The first response to food restriction, observed at 24 hours, was a significant increase in number. Autophagosome characteristics also changed, with increases in perimeter and in feret, a decreased circularity and a slight increase in area. All of these changes were magnified after 48 hours of food restriction. Importantly, the physical characteristics of these autophagosomes are similar to those previously reported using conventional electron microscopy methods, which have established the average diameters of autophagosomes as 0.5–1.5 µm; this validates the use of our new approach for quantitative analyses. These findings can be visualized at high resolution in three dimensions in Supplementary videos 1 through 4. The approach next was applied to two regions of the brain in normalfed and food-restricted GFP-LC3 mice.
Figure 1.
Identification and quantitation of autophagosomes in the liver of food-restricted mice. GFP-LC3 transgenic mice were food-restricted (food-rest) to activate autophagy, and 24 or 48 hours later, the liver was harvested and the induction of autophagy was analyzed in vibratome-cut sections by confocal microscopy. (A) representative flattened images of GFP-LC3 signal in hepatocytes are shown; sections were stained with phalloidin and Hoechst 33342 to label F-actin and nuclei, respectively. A merged fluorescent image for each mouse is shown in the right-hand column; GFP-LC3 (green), F-actin (red), nuclei (blue). Two sets of control mice were included: wild-type C57BL/6 mice were used to determine the background level of green fluorescence, which was very low (one advantage of vibratome sections); and normal-fed GFP-LC3 mice provided a baseline for autophagic activity in the liver. (B) Quantitative analysis of autophagosomes in hepatocytes, including the number of autophagosomes/cell, as well as their area, perimeter, feret and circularity [defined as 4π(area)/(perimeter)2]. Data are shown as the average + se of 80–134 cells from 3 or 4 mice per group; ap < 0.001, bp < 0.01, cp < 0.05.
Autophagosomes are detectable in cortical neurons of normal-fed mice, and their abundance and characteristics are markedly altered by short-term food restriction.
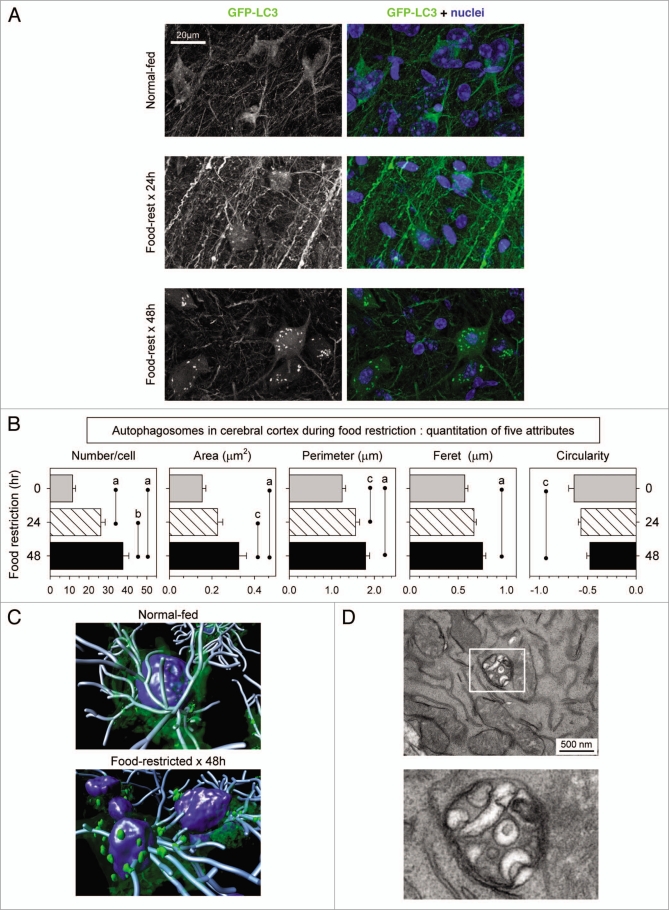
Figure 2, and Supplementary videos 5 and 6, show analyses of the cerebral cortex of normal-fed and food-restricted GFP-LC3 mice. Figure 2A shows representative flattened images; (B) presents quantitative evaluations of autophagosomes in neuronal cell bodies using the five criteria described above; and (C) shows an isosurface rendering of the nuclei, autophagosomes and dendritic processes of several cortical neurons. Autophagosomes were readily detectable in cortical neurons of normal-fed mice (Fig. 2A, top row). To our knowledge, this is the first in vivo demonstration of autophagosomes in the cortical neurons of normal-fed mammals. These vesicles were few in number and small in size (Fig. 2B). Food restriction caused a marked increase in both the number and the size of neuronal autophagosomes in the cell body of cortical neurons; these changes were present after 24 hours of food restriction (Fig. 2A, middle row), and were even more dramatic at 48 hours (Fig. 2A and bottom row). As was observed in the liver (Fig. 1), when compared to autophagosomes in normal-fed mice, those present in the cortical neurons of mice that had been food-restricted for 48 hours showed reduced circularity, with concomitantly increased feret and perimeter (Fig. 2B). Changes in GFP-LC3 signal also were observed in the neuronal cell processes (Fig. 2A, middle row); compared to normal-fed mice, the signal was increased by >3-fold in neurites at 24 hours (p = 0.004, data not shown), but diminished to resting levels after 48 hours. Others have proposed that neuronal autophagosomes are present in neurites and that they undergo retrograde translocation to the cell body, where they fuse with neuronal lysosomes that reside in a juxtanuclear location.17,18 We speculate that food restriction enhances this retrograde traffic, and that the appearance after 24 hours represents an in vivo snapshot of this event; and that it may be completed by 48 hours. To confirm that the cortical neurons of food-restricted mice did, indeed, contain autophagosomes, we evaluated the cells by TEM. A representative image is shown in Figure 2D. These data show clearly that, contrary to current dogma, food restriction increases autophagosome formation and/or accumulation within the brain; and, specifically, within cortical neurons. We considered it important to determine if these changes were limited to cortical neurons or, conversely, were more widespread within the brain. To this end, we evaluated the effects of food restriction on Purkinje cells of the cerebellum.
Figure 2.
Identification and quantitation of autophagosomes in the cerebral cortex of food-restricted mice. GFP-LC3 transgenic mice were food-restricted for 24 or 48 hours, and then the brain was isolated and sagittal vibratome-cut sections were analyzed by confocal microscopy. (A) Representative flattened images of GFP-LC3 signal in cortical neurons are shown. A merged fluorescent image for each mouse is shown in the right-hand column; GFP-LC3 (green), nuclei (blue). (B) Quantitative analysis of autophagosomes in cortical neurons. Data are shown as the average + SE of 50 cells from 2 or 3 mice per group; ap < 0.001, bp < 0.01, cp < 0.05. A 3D rendering of these neurons in normal-fed and food-restricted mice is shown in (C). The isosurface tool was used to demarcate the nuclei (blue) and autophagosomes (green), and neurites were traced with filament tracker, using IMARIS software (Bitplane, Inc.). (D) A TEM image from a section of cortical neuron from a 48-hour food-restricted mouse is shown; the boxed area is enlarged to better demonstrate that the enclosed structure is double-membraned.
Purkinje cells in the cerebella of food-restricted mice contain greater numbers of autophagosomes and show reduced mTOR function.
Analyses of the cerebellum (Fig. 3A and Suppl. videos 7 and 8) identified small autophagosomes in the Purkinje cells of normal-fed mice, perhaps indicative of the basal level of autophagy that is thought to occur in this cell type.1,2,19 Furthermore, we observed marked alterations in the accumulation of GFP-LC3 signal in Purkinje cells in response to food restriction; there was a substantial increase in signal in the cell bodies of Purkinje neurons at both 24 and 48 hours. Some of the increased GFP-LC3 signal in Purkinje cells was more diffuse than had been observed in cortical neurons, forming a reticular perinuclear pattern that occupied a substantial proportion of the cell body. These findings are consistent with fusion of autophagosomes with lysosomes. The perinuclear distribution of autophagosomes induced by food restriction differs markedly from the changes that have been reported in degenerating Purkinje neurons of lurcher mice.19 In addition to this perinuclear pattern, discrete green puncta could be identified within the cell somata (Fig. 3A and Suppl. video 8). Next, we evaluated the in vivo activity of the protein mTOR (mammalian target of rapamycin), which plays a central role in the regulation of autophagy. mTOR exerts an inhibitory effect on autophagy, and mTOR activity is inversely correlated with autophagy. mTOR is highly active in cells in a nutrient-rich environment (in which autophagy is at a basal level), but nutrient deprivation decreases mTOR activity, leading to increased autophagy. Therefore, we investigated the possibility that food restriction might reduce mTOR activity in Purkinje cells in vivo. The measurement of mTOR activity in living cells is indirect, relying on the evaluation of downstream targets. mTOR activates the p70 S6 kinase which, in turn, phosphorylates its ribosomal substrate, S6RP; consequently, the abundance of phospho-S6RP in a living cell is directly related to mTOR activity, and is inversely related to the level of cellular autophagy. Figure 3B compares the cerebellum of a normal-fed mouse to that of a 48-hour food-restricted animal. The increase in GFP-LC3 signal following food restriction is again evident (left column) and there is a dramatic reduction in the level of phospho-S6RP in the Purkinje cell bodies of food-restricted mice (middle column). The merged image (right column) highlights the starvation-induced increase in autophagy that occurs in these neurons.
Figure 3.
Identification of autophagosomes and reduction of mTOR activity in the cerebellum of food-restricted mice. (A) Low magnification flattened images of GFP-LC3 fluorescence were re-assembled to generate a complete sagittal cross-section of cerebellum from normal and food-restricted mice (left column). Higher magnification images of the white boxed regions show GFP-LC3 fluorescence in purkinje cells along the boundary between the molecular and granular layers (second column). Additionally magnified representative images of the (boxed) Purkinje cell bodies reveal finer detail and localization of the GFP-LC3 signal (far right column). A merged fluorescent image for each mouse is shown in the third column; GFP-LC3 (green), nuclei (blue), pan-neuronal marker (orange), GFAP/astrocyte marker (red). (B) Cerebellar sections were stained with an antibody specific for phospho-S6RP, a protein whose abundance varies directly with mTOR activity, and inversely with autophagy (see text).
The increase in number and in size of autophagosomes in cerebellar Purkinje cells of food-restricted mice are confirmed by TEM.
Finally, we used TEM to assess starvation-induced changes in autophagosomes in cerebellar Purkinje cells. In normal-fed mice, the autophagosomes were small (∼0.2 µm diameter, Fig. 4A left part), perhaps explaining the somewhat reticular appearance that was observed by confocal microscopy (Fig. 3). In contrast, the double-membraned structures in 48 hour foodrestricted mice were larger (∼0.5 µm in diameter; Fig. 4A middle & right parts); this size is similar to that of the green puncta that are visible in Figure 3 and in Supplementary video 8. Thus, food restriction appears to change the size of autophagosomes in Purkinje cells; does it also alter their abundance? TEM sections of the somata of Purkinje cells were examined and autophagosomes were enumerated. A representative section (Fig. 4B) shows the soma of a single Purkinje neuron from a mouse that had been food-restricted for 48 hours. Four autophagosomes (all ∼0.5 µm in diameter) are visible in this section; they are enclosed in colored boxes, and higher-magnification views are provided; white arrows indicate the autophagosomes. For both normal-fed and food-restricted mice, ∼20 different Purkinje cells were examined in this way, and autophagosomes were enumerated. As shown in Figure 4B (graph at lower right), there was a 3- to 4-fold increase in the number of autophagosomes in Purkinje cells from food-restricted animals (p < 0.0002). This increase in abundance is similar in extent to that observed in cortical neurons using our novel approach of vibratome sectioning and confocal microscopy (Fig. 2B).
Figure 4.
TEM identification and quantitation of autophagosomes in Purkinje cells of normal-fed and food-restricted mice. (A) Images from Purkinje cells of normal-fed (left) or 48 hour food-restricted (middle & right) mice are shown. (B) A single Purkinje cell from a 48 hour food-restricted mouse is shown. Autophagosomes are enclosed in colored boxes, and higher-magnification images of each are provided, with autophagosomes indicated by white arrows. For both normal-fed and food-restricted mice, autophagosomes were enumerated in TEM sections of ∼20 different Purkinje cells; sections from food-restricted mice showed a ∼4-fold increase in autophagosome number (graph, bottom right; p < 0.0002 by Student's t test).
Discussion
Sporadic short-term fasting, driven by religious and spiritual beliefs, is common to many cultures and has been practiced for millennia, but scientific analyses of the consequences of caloric restriction are more recent. Published studies indicate that the brain is spared many of the effects of short-term food restriction, perhaps because it is a metabolically privileged site that, relative to other organs, is protected from the acute effects of nutrient deprivation, including autophagy.17 We show here that this is not the case: short-term food restriction induces a dramatic upregulation of autophagy in cortical and Purkinje neurons. To our knowledge, this is the first demonstration that food restriction leads to in vivo neuronal autophagy. Our data extend recent reports in tissue culture systems,18,20 and have implications for individuals who, by choice or necessity, have limited food intake. Our findings also may have therapeutic implications, as outlined below. Autophagy is sometimes referred to as cellular “cleansing”, and our observations provide an attractive neuronal parallel to the organismal benefits that, historically, are perceived to derive from fasting.
In this study we describe a novel technique to identify and characterize autophagosomes in the tissues of GFP-LC3 mice, using vibratome sections and confocal microscopy. The changes that we report using this method are confirmed using the standard technique of TEM; this both validates the new approach, and confirms the conclusions that we have drawn therefrom. Herein, we make at least three novel observations. First, we identify and localize autophagosomes in cortical and Purkinje neurons of normal-fed mammals. Second, we show that, in these CNS neurons, short-term food restriction leads to a marked increase in the number of autophagosomes, together with changes in their physical characteristics. In principle, the observed changes could result either from increased production of autophagosomes, or from reduced fusion of autophagosomes with lysosomes. We believe that the former explanation is the more likely, because the activity of mTOR, a key protein that restrains autophagy, is reduced in the Purkinje cells of food-restricted mice (Fig. 3B). Thus, our data favor the interpretation that the increased abundance of autophagosomes results from upregulation of the autophagy pathway in neurons, rather than from the blockade of autophagosome maturation, although some contribution from the latter cannot be excluded.
Third, we demonstrate that the changes that are induced by food restriction differ somewhat depending on the neuronal cell type. This is consistent with the recent proposal that autophagy pathways may differ among cell types, as a result of evolutionary adaptation to the specific physiological needs of the cell.14 Published studies in which autophagy was inhibited have shown that the outcome varies with cell or tissue type. For example, interruption of neuronal autophagy can lead to degenerative disease,1,2 but an equivalent block in pancreatic acinar cells causes no overt dysfunction.21 However, until now it has been thought that increased autophagy would result in phenotypically-similar changes in closely-related cell types. Here we show that this appears not to be the case in vivo. Following food restriction, we observed both similarities and differences in the size and distribution of autophagosomes in cortical neurons and Purkinje cells. The fact that the observed changes in autophagy differed between these two neuronal cell types suggests that a more extensive analysis is warranted, to determine the extent to which short-term food restriction induces autophagy—and, possibly, other changes—in the mammalian CNS.
Our observation that a brief period of food restriction can induce widespread upregulation of autophagy in CNS neurons may have clinical relevance. As noted above, disruption of autophagy can cause neurodegenerative disease, and the converse also may hold true: upregulation of autophagy may have a neuroprotective effect. For example, in vitro models have shown that starvation in neuronal cell lines can remove toxic molecules and damaged mitochondria from neurons.22–24 Other tissue culture studies, of mutant huntingtin and α-synuclein proteins (which are associated with Huntington disease and familial Parkinson disease respectively), have identified autophagy substrates that can be removed by drug-induced enhancement of autophagy. Most importantly, some neuroprotective effects of drug-enhanced autophagy also have been observed in vivo, in a D. melanogaster model of Huntington disease.25 Finally, it has been suggested that intermittent fasting might improve neuronal function by means that are entirely independent of caloric intake, and may instead reflect an intrinsic neuronal response that is triggered by fasting;26,27 we speculate that the reported improvement of neuronal function may be related to the upregulation of autophagy that we show here. The above findings have encouraged the development of drugs that might enhance neuronal autophagy, thereby protecting against disease. Such drugs must: (i) be able to cross the intact blood-brain barrier; (ii) upregulate neuronal autophagy; and (iii) be harmless to the recipient. Food restriction is a simple, reliable, inexpensive and harmless alternative to drug ingestion and, therefore, we propose that short-term food restriction may represent an attractive alternative to the prophylaxis and treatment of diseases in which candidate drugs are currently being sought. However, caution is counseled, because studies in rat brain have suggested that chronic starvation might inhibit autophagy,28 an outcome that could damage, rather than protect, neurons.28,29
Materials and Methods
Mice.
GFP-LC3 mice contain a transgene encoding an eGFP-rat LC3 fusion protein.10 GFP-LC3 transgenic mice, back-crossed to the C57BL/6 background, were obtained from Riken BioResource Center, and maintained as homozygous transgenic (Tg/Tg). Homozygous transgenic mice were bred with wild-type C57BL/6 mice to generate heterozygous transgenic (Tg/+) mice. Pathogen-free C57BL/6J mice were purchased from the Scripps Research Institute (TSRI) breeding facility or from Jackson Laboratories. Mice were housed in a temperature-controlled environment with 12-hr light/dark cycles where they received food and water. For studies on the effects of food restriction, 6–7 week old male C57BL/6J and GFP-LC3 (Tg/+) mice were deprived of food for 24 or 48 hr. These mice had free access to drinking water. All experimental procedures with mice were conducted in accordance with the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All animal protocols were approved by TSRI Animal Care and Use Committee.
Perfusion, fixation and sectioning of tissues.
Mice were perfused with Dulbecco's PBS (DPBS, Invitrogen, 14040-141), and tissues were harvested and fixed using buffered zinc formalin (Z-FIX; Annatech Ltd., 171) at room temperature (RT) overnight. The inclusion of zinc is important, because it limits the extensive cross-linking that would occur with formalin alone, thereby preserving protein antigenicity and enhancing reagent penetration into the tissue section; but the residual cross-linking is sufficient to prevent the leaching of small molecules from cells.30 After overnight fixation, the fixative was replaced with PBS containing 0.1% (w/v) sodium azide on the following day, and the organs were preserved at 4°C. Sixty µm sections of brain were cut with a Leica VT 1000S vibratome.
Immunostaining of liver and brain vibratome sections.
Free-floating vibratome-cut liver sections (60 µm) were washed 3X in PBS (5 min each) and permeabilized in PBS with 0.5% Triton X-100 for 2 hrs at RT. Next, the liver sections were washed 3X in PBS and then incubated with Alexa Fluor 568 phalloidin (1:40; Invitrogen, A12380) in PBS with 1% normal goat serum (NGS; Invitrogen, PCN5000) and 0.3% Triton X-100 at 4°C overnight to label F-actin. After incubation, the sections were washed 5X in PBS and then counterstained with Hoechst 33342 (1:1,000; Invitrogen, H3570) for 1 hr at RT to label nuclei, and mounted with Vectashield (Vector labs, H1000) for confocal microscopy. Sagittal free-floating brain sections were washed 3X in PBS, permeabilized (similarly to the liver), and then blocked in PBS with 5% NGS and 0.3% Triton X-100 for 2 hrs at RT. Sections were washed 3X in PBS with 0.3% Triton, and incubated for ∼42–44 hrs at 4°C with the following primary antibodies diluted in 1% NGS and 0.3% Triton: mouse Milli-Mark Pan Neuronal Marker (1:50, Millipore, MAB2300); rabbit antibody against phospho- S6 ribosomal protein (phospho-S6RP; 1:100, Cell Signaling Technology, 2211); and rabbit anti-glial fibrillary acidic protein (GFAP; 1:500, DAKO, Z0334). The sections were then washed 3X in PBS with 0.3% Triton, and incubated for 2 hrs at RT with the following secondary antibodies (1:250, all from Invitrogen): goat anti-mouse IgG rhodamine red (R6393), donkey anti-rabbit IgG AlexaFluor 647 (A31573), and/or goat anti-rabbit IgG rhodamine red (R6394) diluted in 1% NGS and 0.3% Triton. After incubation, the sections were washed 5X in PBS. All brain sections were counterstained with Hoechst 33342 (1:1,000) for 1 hr at RT and mounted with Vectashield for confocal microscopy. As much as possible, all sections were protected from light throughout these procedures.
Confocal laser scanning microscopy analysis: high resolution imaging of autophagy in tissues.
Confocal images were captured using a Zeiss LSM 710 Laser Confocal Scanning Microscope running Zen 2009 Zeiss software suite. Representative regions within each vibratome section of brain were scanned as 8-bit optical sections (either 512 × 512 or 1,024 × 1,024 image sizes) and reconstructed for analysis. Exposure and image acquisition settings were identical for all sections. The observed green (GFP-LC3) signals did not result from autofluorescence because: (i) narrow band filters were used and the signals were observed only in the green channel and (ii) the signals were absent from non-transgenic mice (See Fig. 1).
Image processing, 3D reconstruction and quantitative analysis of GFP-LC3 vesicles.
All serial optical image sections (obtained at 0.3–0.5 µm interval step slices) were imported and spatially re-assembled using Imaris (BitPlane Inc.) or Zen 2009 (Carl Zeiss Inc.) software to generate a 3-dimensional representation of the tissue and rotational movies. All images were then maximum projected for 2D analysis in Image Pro Plus (Media Cybernetics Inc.) or Image J (NIH, http://rsbweb.nih.gov/ij/) software for quantitative image analysis. For all quantitation, serial confocal stacks were first maximum projected/flattened into one 8-bit 2D image per scanned region, and then further processed in Image Pro-Plus/3DS and Image J. Processing consisted of automatically outlining each cell with a tracing tool and then defining the background level of GFP-LC3 fluorescent signal using the threshold tool. Once traced, the fluorescent areas within each cell were individually outlined by the software and used to calculate 2D parameters including: the number of vesicles, area (assuming a uniform height), perimeter, feret (the longest diameter within an irregular shape), and circularity (a deviation from roundness of a sphere). To ensure valid comparisons of signal intensities among various images, all images and all colors were processed using identical software settings.
Transmission electron microscopy.
Mice were perfused with 0.9% saline followed by 4% paraformaldehyde, 1.5% glutaraldehyde in 0.1 M cacodylate buffer with 1 mM CaCl2. The brain was removed and immersed in the above fixative on ice for 6 hrs, and then was transferred to 2.5% glutaraldehyde in 0.1 M cacodylate buffer + 1 mM CaCl2 for overnight fixation. After a buffer wash, the tissue was further fixed in 1% OsO4 with 1.5% potassium ferricyanide in 0.1 M Na cacodylate and again washed in cacodylate buffer, dehydrated in graded ethanols and transitioned in propylene oxide. Tissue sections were embedded in Embed 812/Araldite (Electron Microscopy Sciences). Thick sections (1–2 µm) were cut, mounted on glass slides and stained in toluidine blue for general assessment in the light microscope. Subsequently, 70 nm thin sections were cut, mounted on copper slot grids coated with parlodion and stained with uranyl acetate and lead citrate for examination on a Philips CM100 electron microscope (FEI, Hillsbrough OR) at 80 kV. Images were collected using either an SIA-12C camera (Scientific Instruments and Applications) or a Megaview III CCD camera (Olympus Soft Imaging Solutions).
Statistics.
Statistical significance was determined by: (i) a one-way analysis of variance (ANOVA), and Tukey's post-hoc test was used for multiple comparisons within each data set (GraphPad Prism, GraphPad Software, Inc.) or (ii) Student's t test (Microsoft Excel 2007). In all cases, a p value <0.05 was considered statistically significant.
Acknowledgements
We are grateful to Annette Lord for excellent secretarial support, and to Janet Weber and Hanna Lewicki for technical assistance and advice. This work was supported by NIH R01 awards AI-027028, AI-042314 and AI-077607 (J.L.W.); and by NIH T32 award NS41219 (C.C.K.). This is manuscript number 19985 from the Scripps Research Institute.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/12376
Supplementary Material
References
- 1.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 2.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alirezaei M, Kiosses WB, Flynn CT, Brady NR, Fox HS. Disruption of neuronal autophagy by infected microglia results in neurodegeneration. PLoS ONE. 2008;3:2906. doi: 10.1371/journal.pone.0002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orvedahl A, Levine B. Eating the enemy within: autophagy in infectious diseases. Cell Death Differ. 2009;16:57–69. doi: 10.1038/cdd.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizushima N. Aβ generation in autophagic vacuoles. J Cell Biol. 2005;171:15–17. doi: 10.1083/jcb.200508097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nixon RA. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci. 2007;120:4081–4091. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- 8.Yu WH, et al. Macroautophagy—a novel β-amyloid peptide-generating pathway activated in Alzheimer's disease. J Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinet W, De Meyer GR, Andries L, Herman AG, Kockx MM. In situ detection of starvation-induced autophagy. J Histochem Cytochem. 2006;54:85–96. doi: 10.1369/jhc.5A6743.2005. [DOI] [PubMed] [Google Scholar]
- 10.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N, Kuma A. Autophagosomes in GFP-LC3 Transgenic Mice. Methods Mol Biol. 2008;445:119–124. doi: 10.1007/978-1-59745-157-4_7. [DOI] [PubMed] [Google Scholar]
- 12.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boland B, Nixon RA. Neuronal macroautophagy: from development to degeneration. Mol Aspects Med. 2006;27:503–519. doi: 10.1016/j.mam.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Yue Z, Wang QJ, Komatsu M. Neuronal autophagy: going the distance to the axon. Autophagy. 2008;4:94–96. doi: 10.4161/auto.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koike M, et al. Inhibition of autophagy prevents hippocampal pyramidal neuron death after hypoxic-ischemic injury. Am J Pathol. 2008;172:454–469. doi: 10.2353/ajpath.2008.070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai Y, et al. Autophagy is increased after traumatic brain injury in mice and is partially inhibited by the antioxidant γ-glutamylcysteinyl ethyl ester. J Cereb Blood Flow Metab. 2008;28:540–550. doi: 10.1038/sj.jcbfm.9600551. [DOI] [PubMed] [Google Scholar]
- 17.Komatsu M, et al. Constitutive autophagy: vital role in clearance of unfavorable proteins in neurons. Cell Death Differ. 2007;14:887–894. doi: 10.1038/sj.cdd.4402120. [DOI] [PubMed] [Google Scholar]
- 18.Du L, et al. Starving neurons show sex difference in autophagy. J Biol Chem. 2009;284:2383–2396. doi: 10.1074/jbc.M804396200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang QJ, et al. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci. 2006;26:8057–8068. doi: 10.1523/JNEUROSCI.2261-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young JE, Martinez RA, La Spada AR. Nutrient deprivation induces neuronal autophagy and implicates reduced insulin signaling in neuroprotective autophagy activation. J Biol Chem. 2009;284:2363–2373. doi: 10.1074/jbc.M806088200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashimoto D, et al. Involvement of autophagy in trypsinogen activation within the pancreatic acinar cells. J Cell Biol. 2008;181:1065–1072. doi: 10.1083/jcb.200712156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaeger PA, Wyss-Coray T. All-you-can-eat: autophagy in neurodegeneration and neuroprotection. Mol Neurodegener. 2009;4:16. doi: 10.1186/1750-1326-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marazziti D, et al. Induction of macroautophagy by overexpression of the Parkinson's disease-associated GPR37 receptor. FASEB J. 2009;23:1978–1987. doi: 10.1096/fj.08-121210. [DOI] [PubMed] [Google Scholar]
- 24.Hung SY, Huang WP, Liou HC, Fu WM. Autophagy protects neuron from Aβ-induced cytotoxicity. Autophagy. 2009;5:502–510. doi: 10.4161/auto.5.4.8096. [DOI] [PubMed] [Google Scholar]
- 25.Sarkar S, et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat Chem Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anson RM, et al. Intermittent fasting dissociates beneficial effects of dietary restriction on glucose metabolism and neuronal resistance to injury from calorie intake. Proc Natl Acad Sci USA. 2003;100:6216–6220. doi: 10.1073/pnas.1035720100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duan W, et al. Dietary restriction normalizes glucose metabolism and BDNF levels, slows disease progression, and increases survival in huntingtin mutant mice. Proc Natl Acad Sci USA. 2003;100:2911–2916. doi: 10.1073/pnas.0536856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanahisa Y, Yamaguchi M. Characterization of calcium accumulation in the brain of rats administered orally calcium: the significance of energy-dependent mechanism. Mol Cell Biochem. 1996;158:1–7. doi: 10.1007/BF00225876. [DOI] [PubMed] [Google Scholar]
- 29.Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- 30.Ott SR. Confocal microscopy in large insect brains: zinc-formaldehyde fixation improves synapsin immunostaining and preservation of morphology in whole-mounts. J Neurosci Methods. 2008;172:220–230. doi: 10.1016/j.jneumeth.2008.04.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.