Abstract
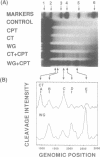
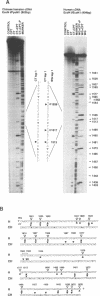
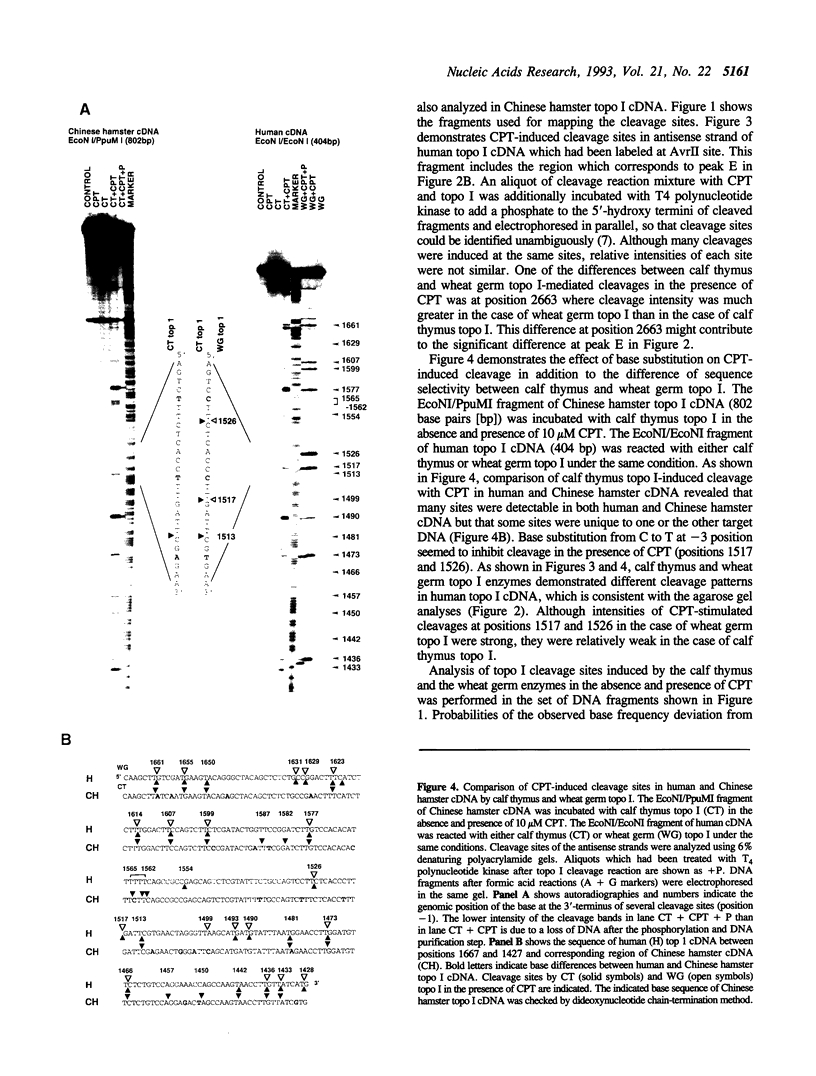
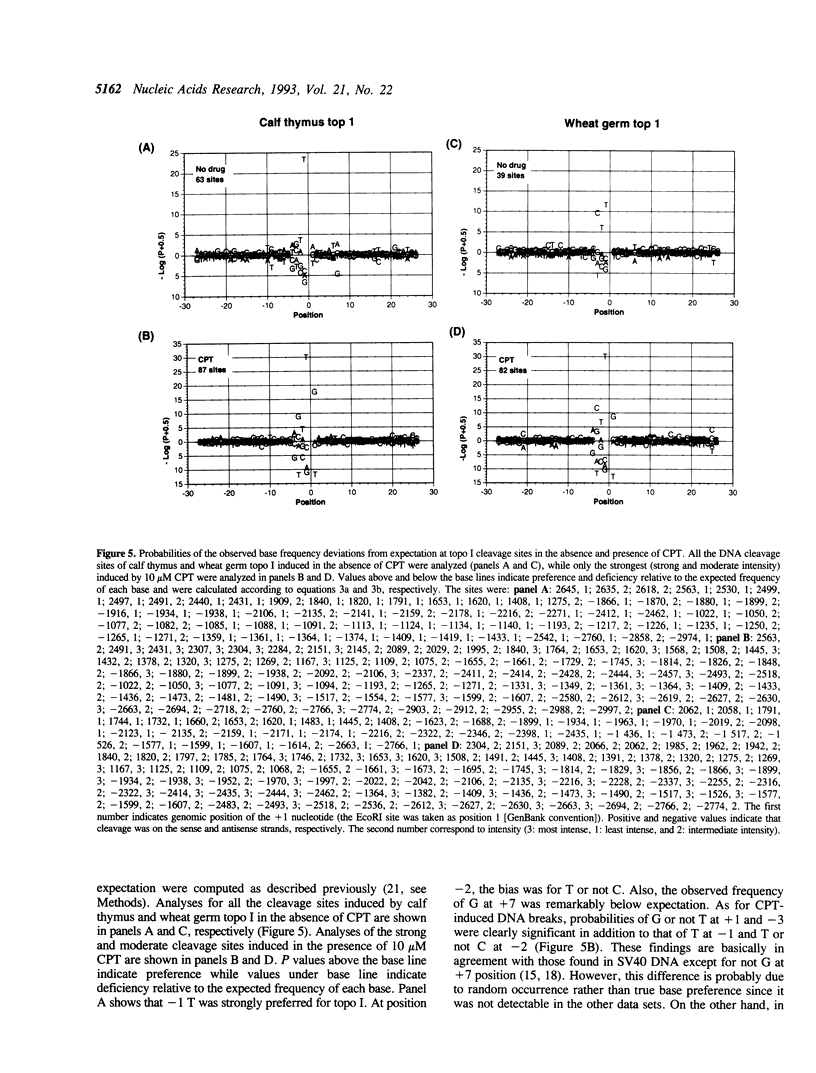
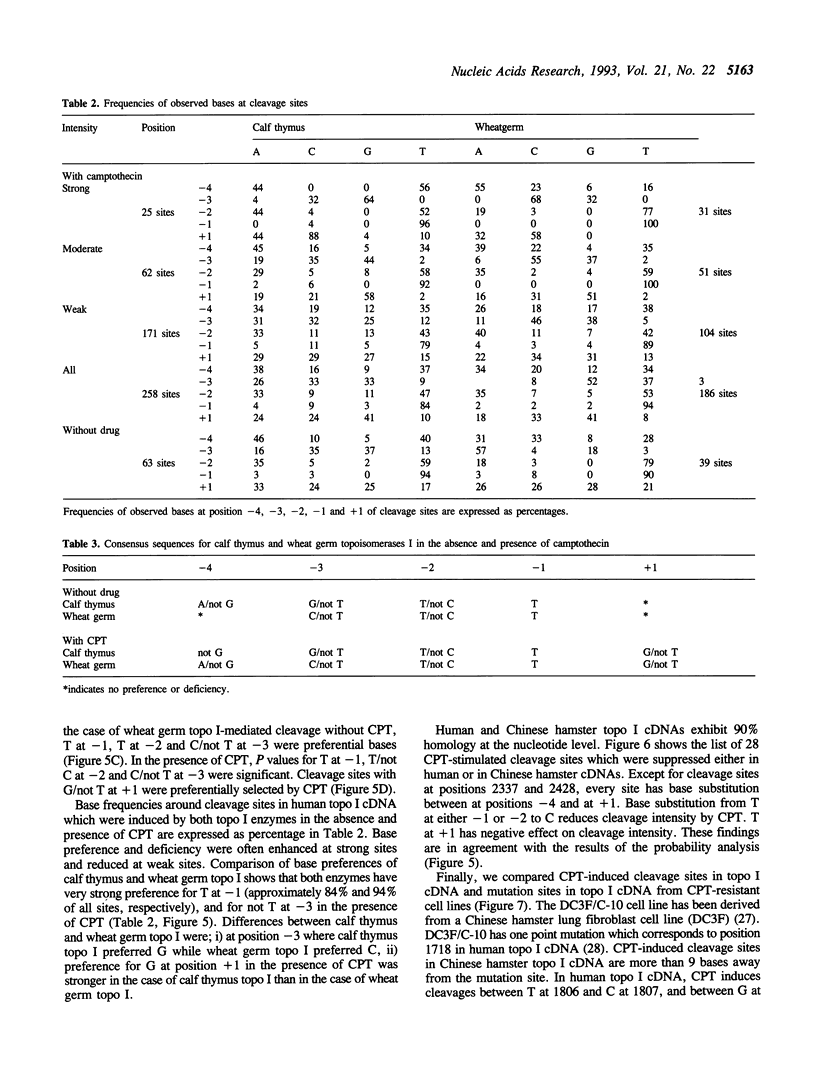
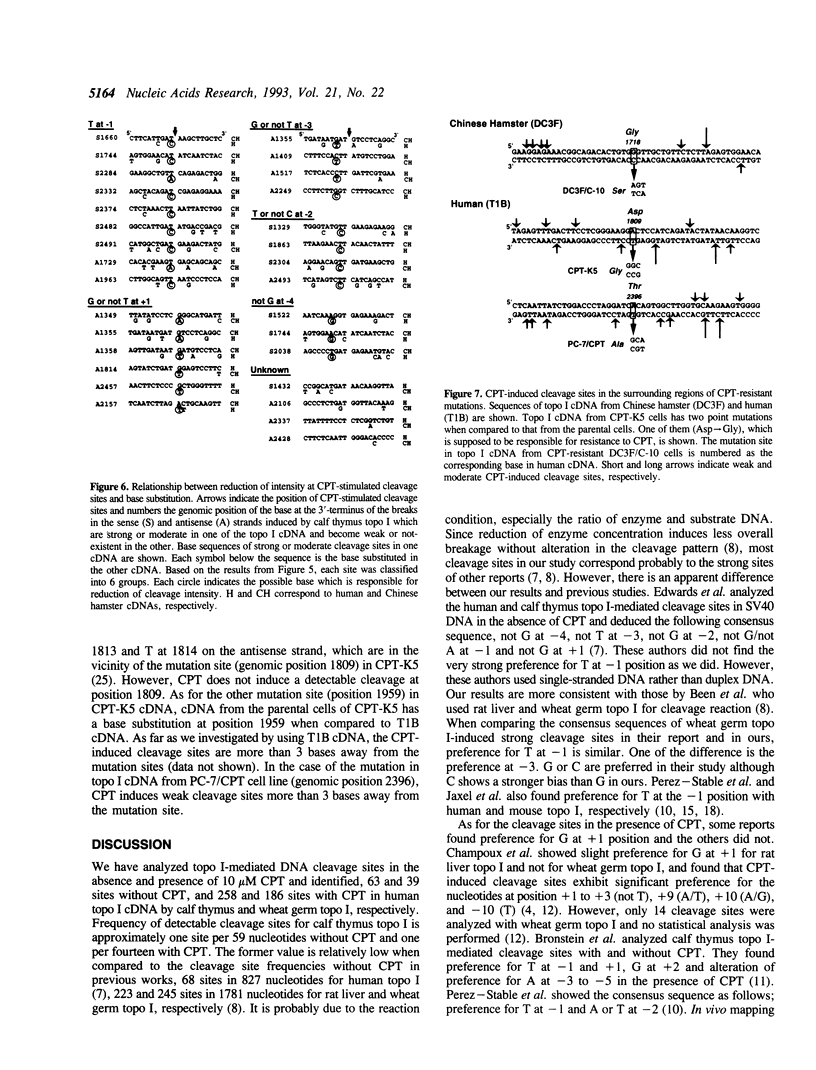
In this study, we further examined the sequence selectivity of camptothecin in mammalian topoisomerase I cDNA from human and Chinese hamster. In the absence of camptothecin, almost all the bases at the 3'-terminus of cleavage sites are T for calf thymus and wheat germ topoisomerase I. In addition, wheat germ topoisomerase I exhibits preference for C (or not T) at -3 and for T at -2 position. As for camptothecin-stimulated cleavage with topoisomerase I, G (or not T) at +1 is an additional strong preference. This sequence selectivity of camptothecin is similar to that previously found in SV40 DNA, suggesting that camptothecin preferentially interacts with topoisomerase I-mediated cleavage sites where G is the base at the 5'-terminus. These results support the stacking model of camptothecin (Jaxel et al. (1991) J. Biol. Chem. 266, 20418-20423). Comparison of calf thymus and wheat germ topoisomerase I-mediated cleavage sites in the presence of camptothecin shows that many major cleavage sites are similar. However, the relative intensities are often different. One of the differences was attributable to a bias at position -3 where calf thymus topoisomerase I prefers G and wheat germ topoisomerase I prefers C. This difference may explain the unique patterns of cleavage sites induced by the two enzymes. Sequencing analysis of camptothecin-stimulated cleavage sites in the surrounding regions of point mutations in topoisomerase I cDNA, which were found in camptothecin-resistant cell lines, reveals no direct relationship between DNA cleavage sites in vitro and mutation sites.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andoh T., Ishii K., Suzuki Y., Ikegami Y., Kusunoki Y., Takemoto Y., Okada K. Characterization of a mammalian mutant with a camptothecin-resistant DNA topoisomerase I. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5565–5569. doi: 10.1073/pnas.84.16.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer L. C., Allen J. W., Harrington-Brock K., Campbell J. A., DeMarini D. M., Doerr C. L., Howard D. R., Kligerman A. D., Moore M. M. Genotoxicity of inhibitors of DNA topoisomerases I (camptothecin) and II (m-AMSA) in vivo and in vitro. Mutagenesis. 1990 Nov;5(6):541–547. doi: 10.1093/mutage/5.6.541. [DOI] [PubMed] [Google Scholar]
- Been M. D., Burgess R. R., Champoux J. J. Nucleotide sequence preference at rat liver and wheat germ type 1 DNA topoisomerase breakage sites in duplex SV40 DNA. Nucleic Acids Res. 1984 Apr 11;12(7):3097–3114. doi: 10.1093/nar/12.7.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronshtein I. B., Gromova I. I., Bukhman V. L., Kafiani K. A. Vliianie kamptotetsina na DNK-relaksiruiushchuiu i DNK-rasshchepliaiushchuiu aktivnosti topoizomerazy I iz timusa telenka. Mol Biol (Mosk) 1989 Mar-Apr;23(2):491–501. [PubMed] [Google Scholar]
- Bronstein I. B., Gromova I. I., Razin S. V. Specific cleavage of chicken alpha A-globin and human c-Ha-ras genes by two molecular forms of calf thymus topoisomerase I. Mol Cell Biochem. 1991 Mar 13;101(2):115–124. doi: 10.1007/BF00229529. [DOI] [PubMed] [Google Scholar]
- Bullock P., Champoux J. J., Botchan M. Association of crossover points with topoisomerase I cleavage sites: a model for nonhomologous recombination. Science. 1985 Nov 22;230(4728):954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- Busk H., Thomsen B., Bonven B. J., Kjeldsen E., Nielsen O. F., Westergaard O. Preferential relaxation of supercoiled DNA containing a hexadecameric recognition sequence for topoisomerase I. Nature. 1987 Jun 18;327(6123):638–640. doi: 10.1038/327638a0. [DOI] [PubMed] [Google Scholar]
- Champoux J. J., Aronoff R. The effects of camptothecin on the reaction and the specificity of the wheat germ type I topoisomerase. J Biol Chem. 1989 Jan 15;264(2):1010–1015. [PubMed] [Google Scholar]
- D'Arpa P., Machlin P. S., Ratrie H., 3rd, Rothfield N. F., Cleveland D. W., Earnshaw W. C. cDNA cloning of human DNA topoisomerase I: catalytic activity of a 67.7-kDa carboxyl-terminal fragment. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2543–2547. doi: 10.1073/pnas.85.8.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrassi F., De Salvia R., Tanzarella C., Palitti F. Induction of chromosomal aberrations and SCE by camptothecin, an inhibitor of mammalian topoisomerase I. Mutat Res. 1989 Mar;211(1):125–130. doi: 10.1016/0027-5107(89)90112-7. [DOI] [PubMed] [Google Scholar]
- Edwards K. A., Halligan B. D., Davis J. L., Nivera N. L., Liu L. F. Recognition sites of eukaryotic DNA topoisomerase I: DNA nucleotide sequencing analysis of topo I cleavage sites on SV40 DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2565–2576. doi: 10.1093/nar/10.8.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromova I. I., Buchman V. L., Abagyan R. A., Ulyanov A. V., Bronstein I. B. Sequence dependent modulating effect of camptothecin on the DNA-cleaving activity of the calf thymus type I topoisomerase. Nucleic Acids Res. 1990 Feb 11;18(3):637–645. doi: 10.1093/nar/18.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. H., Austin M. J., Pommier Y., Povirk L. F. Small deletion and insertion mutations induced by the topoisomerase II inhibitor teniposide in CHO cells and comparison with sites of drug-stimulated DNA cleavage in vitro. J Mol Biol. 1993 Jan 5;229(1):52–66. doi: 10.1006/jmbi.1993.1007. [DOI] [PubMed] [Google Scholar]
- Hsiang Y. H., Hertzberg R., Hecht S., Liu L. F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985 Nov 25;260(27):14873–14878. [PubMed] [Google Scholar]
- Jaxel C., Capranico G., Kerrigan D., Kohn K. W., Pommier Y. Effect of local DNA sequence on topoisomerase I cleavage in the presence or absence of camptothecin. J Biol Chem. 1991 Oct 25;266(30):20418–20423. [PubMed] [Google Scholar]
- Jaxel C., Kohn K. W., Pommier Y. Topoisomerase I interaction with SV40 DNA in the presence and absence of camptothecin. Nucleic Acids Res. 1988 Dec 9;16(23):11157–11170. doi: 10.1093/nar/16.23.11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzawa F., Sugimoto Y., Minato K., Kasahara K., Bungo M., Nakagawa K., Fujiwara Y., Liu L. F., Saijo N. Establishment of a camptothecin analogue (CPT-11)-resistant cell line of human non-small cell lung cancer: characterization and mechanism of resistance. Cancer Res. 1990 Sep 15;50(18):5919–5924. [PubMed] [Google Scholar]
- Kjeldsen E., Mollerup S., Thomsen B., Bonven B. J., Bolund L., Westergaard O. Sequence-dependent effect of camptothecin on human topoisomerase I DNA cleavage. J Mol Biol. 1988 Jul 20;202(2):333–342. doi: 10.1016/0022-2836(88)90462-7. [DOI] [PubMed] [Google Scholar]
- Kubota N., Kanzawa F., Nishio K., Takeda Y., Ohmori T., Fujiwara Y., Terashima Y., Saijo N. Detection of topoisomerase I gene point mutation in CPT-11 resistant lung cancer cell line. Biochem Biophys Res Commun. 1992 Oct 30;188(2):571–577. doi: 10.1016/0006-291x(92)91094-7. [DOI] [PubMed] [Google Scholar]
- Masurekar M., Kreuzer K. N., Ripley L. S. The specificity of topoisomerase-mediated DNA cleavage defines acridine-induced frameshift specificity within a hotspot in bacteriophage T4. Genetics. 1991 Mar;127(3):453–462. doi: 10.1093/genetics/127.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Stable C., Shen C. C., Shen C. K. Enrichment and depletion of Hela topoisomerase I recognition sites among specific types of DNA elements. Nucleic Acids Res. 1988 Aug 25;16(16):7975–7993. doi: 10.1093/nar/16.16.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y., Capranico G., Orr A., Kohn K. W. Distribution of topoisomerase II cleavage sites in simian virus 40 DNA and the effects of drugs. J Mol Biol. 1991 Dec 20;222(4):909–924. doi: 10.1016/0022-2836(91)90585-t. [DOI] [PubMed] [Google Scholar]
- Pommier Y., Capranico G., Orr A., Kohn K. W. Local base sequence preferences for DNA cleavage by mammalian topoisomerase II in the presence of amsacrine or teniposide. Nucleic Acids Res. 1991 Nov 11;19(21):5973–5980. doi: 10.1093/nar/19.21.5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter S. E., Champoux J. J. Mapping in vivo topoisomerase I sites on simian virus 40 DNA: asymmetric distribution of sites on replicating molecules. Mol Cell Biol. 1989 Feb;9(2):541–550. doi: 10.1128/mcb.9.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter S. E., Champoux J. J. The basis for camptothecin enhancement of DNA breakage by eukaryotic topoisomerase I. Nucleic Acids Res. 1989 Nov 11;17(21):8521–8532. doi: 10.1093/nar/17.21.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A., Kapitza M. Histone H1 inhibits eukaryotic DNA topoisomerase I. FEBS Lett. 1991 Dec 2;294(1-2):125–128. doi: 10.1016/0014-5793(91)81357-e. [DOI] [PubMed] [Google Scholar]
- Ripley L. S., Dubins J. S., deBoer J. G., DeMarini D. M., Bogerd A. M., Kreuzer K. N. Hotspot sites for acridine-induced frameshift mutations in bacteriophage T4 correspond to sites of action of the T4 type II topoisomerase. J Mol Biol. 1988 Apr 20;200(4):665–680. doi: 10.1016/0022-2836(88)90479-2. [DOI] [PubMed] [Google Scholar]
- Shuman S. DNA strand transfer reactions catalyzed by vaccinia topoisomerase I. J Biol Chem. 1992 Apr 25;267(12):8620–8627. [PubMed] [Google Scholar]
- Shuman S., Prescott J. Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J Biol Chem. 1990 Oct 15;265(29):17826–17836. [PubMed] [Google Scholar]
- Shuman S. Recombination mediated by vaccinia virus DNA topoisomerase I in Escherichia coli is sequence specific. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10104–10108. doi: 10.1073/pnas.88.22.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup J. Q., Christiansen K., Gromova I. I., Andersen A. H., Westergaard O. New technique for uncoupling the cleavage and religation reactions of eukaryotic topoisomerase I. The mode of action of camptothecin at a specific recognition site. J Mol Biol. 1991 Dec 5;222(3):669–678. doi: 10.1016/0022-2836(91)90503-x. [DOI] [PubMed] [Google Scholar]
- Tamura H., Kohchi C., Yamada R., Ikeda T., Koiwai O., Patterson E., Keene J. D., Okada K., Kjeldsen E., Nishikawa K. Molecular cloning of a cDNA of a camptothecin-resistant human DNA topoisomerase I and identification of mutation sites. Nucleic Acids Res. 1991 Jan 11;19(1):69–75. doi: 10.1093/nar/19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa A., Pommier Y. Topoisomerase I alteration in a camptothecin-resistant cell line derived from Chinese hamster DC3F cells in culture. Cancer Res. 1992 Apr 1;52(7):1848–1854. [PubMed] [Google Scholar]
- Tsui S., Anderson M. E., Tegtmeyer P. Topoisomerase I sites cluster asymmetrically at the ends of the simian virus 40 core origin of replication. J Virol. 1989 Dec;63(12):5175–5183. doi: 10.1128/jvi.63.12.5175-5183.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Recent studies of DNA topoisomerases. Biochim Biophys Acta. 1987 Jun 6;909(1):1–9. doi: 10.1016/0167-4781(87)90040-6. [DOI] [PubMed] [Google Scholar]
- Yamashita Y., Kawada S., Fujii N., Nakano H. Induction of mammalian DNA topoisomerase I and II mediated DNA cleavage by saintopin, a new antitumor agent from fungus. Biochemistry. 1991 Jun 18;30(24):5838–5845. doi: 10.1021/bi00238a005. [DOI] [PubMed] [Google Scholar]