Abstract
The clinical potential of siRNAs for silencing genes critical to disease progression is clear, but a fail-proof method for delivering siRNAs to the cytoplasm of diseased tissues or cells has yet to be identified. A variety of delivery approaches have been explored to directly or indirectly couple siRNAs to delivery vehicles. This review explores the use of synthetic single-stranded DNA and RNA aptamers as a means to deliver siRNAs, shRNAs and antisense oligonucleotides for therapeutic intervention. Topics covered include: the advantages and challenges of using aptamers as delivery tools; current aptamer-mediated siRNA delivery platforms for the treatment of cancer and HIV; and emerging methodologies for the identification of aptamers capable of internalizing into target cell types.
With our ever-growing understanding of the intricate molecular mechanisms of diseases, we are becoming increasingly aware of the advantages of personalized medicine: determining the molecular signature of each patient’s specific disease and tailoring treatments to maximize efficacy and avoid off-target effects [1]. In a recent article, Hamburg et al. outline the steps that are being taken by the US FDA and NIH to expedite translating basic science research to patient treatment [2].
To truly and effectively implement the concept of personalized medicine, we must first understand the molecular fingerprint of each patient’s disease. This understanding is likely to enable the development of tailored therapies with increased efficacy and safety profiles. Use of siRNAs to selectively turn off disease-driving genes is one promising technology being applied to personalized medicine. However, efficient delivery to the cytoplasm of target cells represents the primary challenge in implementing siRNA technology as a bona fide therapeutic. This review article focuses on an emerging platform technology to deliver siRNAs and other RNA-based therapeutics, including synthetic single-stranded nucleic acid ligands such as aptamers (Figure 1).
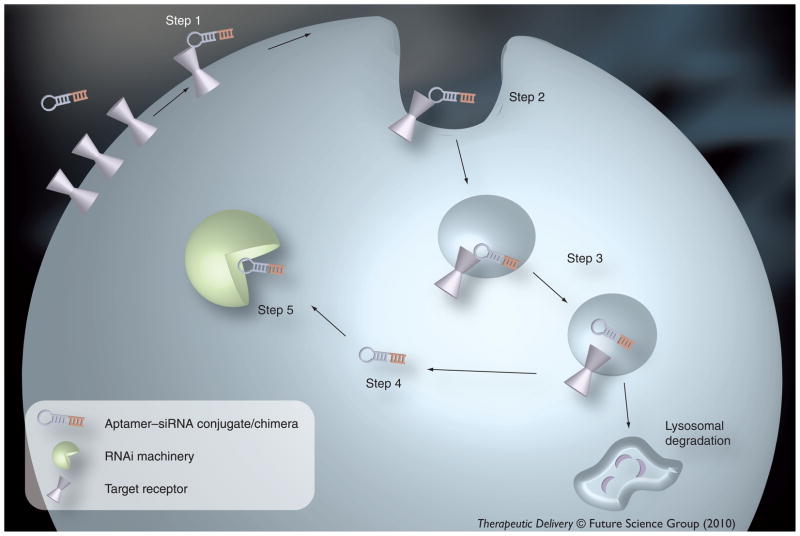
Figure 1. Aptamer-mediated siRNA internalization.
Upon binding of the aptamer portion of the conjugate to the target T cell-surface receptor (Step 1), the complex is internalized (Step 2), likely through clathrin-mediated endocytosis. The complex is presumably shuttled through the early, sorting and late endosome (Step 3). To be loaded into the RNAi machinery, the aptamer–siRNA conjugate must escape the endosome (Step 4). The aptamer–siRNA conjugate is recognized by the RNAi machinery (Step 5) and mediates silencing of the target mRNA.
Types of RNA-based therapeutics
RNA has long been evaluated for its clinical potential. Several different types of RNA molecules have been extensively researched for this purpose, including siRNAs, short hairpin (sh)RNAs, miRNAs, antisense oligonucleotides (AS OGNs) and ribozymes. siRNAs, shRNAs and miRNAs accomplish gene-specific silencing through the RNAi pathway [3]. AS OGNs (both DNA- and RNA-based oligos) are also used to inhibit gene expression. In the case of RNA oligos, their mode of action includes inhibition of translation, regulation of alternative splicing decisions, or transcriptional gene silencing (TGS) [4]. Ribozymes are catalytic RNAs that possess the ability to cleave phosphodiester bonds, such as those found in target mRNA sequences, and can thus modulate the function of a given RNA substrate [5].
Recently, substantial effort and resources have been placed on developing effective siRNA-based drugs with an emphasis on devising ways to better shield siRNAs from serum nucleases, retard their renal clearance, and efficiently target the siRNAs to the desired tissues and cell types.
In its native state, RNA is an inherently unstable molecule, requiring chemical modifications of the phosphate backbone (e.g., boranophosphate, phosphorothioate, phosphoroamidate and methylphosphonate modifications) and/or sugar moieties (e.g., 2′-fluoro, 2′-O-methyl, locked nucleic acids [LNAs]) to prevent degradation of the RNA serum nucleases when used in in vivo applications [6]. However, in the case of siRNAs, chemical modifications have the potential to adversely affect processing by Dicer and/or loading into the RNA-induced silencing complex (RISC), and thus must be tested empirically [6]. Another challenge for RNA-based therapeutics is avoidance of nonspecific innate immune activation through the Toll-like receptors [7], although this effect can also largely be reduced or evaded by chemical modification of the RNA [3,6,8]. Once these initial technical hurdles are overcome, the therapeutic RNAs must be delivered to the desired cell types and, in the case of siRNAs, find their way to the cytoplasm of target cell-surface receptors where the siRNAs can enter into the RNAi pathway to mediate gene-specific silencing. Currently, several approaches have been explored for delivery of siRNA duplexes and other therapeutic RNAs.
Current delivery approaches
Current delivery approaches can be broadly grouped into either noncovalent or covalent assemblies. Examples of noncovalent assemblies include electrostatic interactions between an siRNA and a cationic molecule (i.e., protamine, arginine-rich peptides) [9–11] or encapsulation of an siRNA into a nanoparticle (i.e., a cationic lipid) [12–14]. Targeting moieties (antibodies, peptides or aptamers) are often included to facilitate delivery of the siRNAs to specific cell types [10,15–17]. For covalent assemblies, siRNAs have been directly conjugated to aptamers [18–24], lipids (i.e., cholesterol) [25], vitamins (i.e., α-tocopherol) [26], antibody–protamine fusion proteins [10] and peptides [27,28]. Some of these types of covalent linkages involve a targeting moiety (e.g., aptamers, antibodies, peptides or ligands) that directs the siRNA to a specific cell type, whereas others (lipophilic molecules or cholesterol) nonspecifically facilitate cellular uptake. In order to minimize off-target effects resulting from delivery to nontarget (healthy) tissues, targeted delivery is desired and aptamers are emerging as promising tools in this regard.
Advantages of aptamers as delivery tools
By definition, aptamers are synthetic, single-stranded DNA or RNA oligonucleotides that recognize targets with a similar specificity and affinity as antibody/antigen interactions. However, aptamers are selected through an in vitro process of systematic evolution of ligands by exponential enrichment (SELEX) [29,30]. For therapeutic applications, this process relies on DNA or RNA polymerases that can accept modified nucleotides as substrates (e.g., 2′-fluoro pyrimidines) yielding nuclease-resistant aptamers that are partially or completely substituted [31–36]. In addition, aptamers can be chemically synthesized, thus reducing batch-to-batch variability. The process of chemical synthesis also allows for substituting native nucleotides with a variety of additional modified nucleotides (2′-fluoro/2′-O-methyl ribose purines or pyrimidines and LNA) or terminal modifications (e.g., terminal caps such as inverted nucleotides, high-molecular mass polyethylene glycol [PEG] or cholesterol) to promote nuclease resistance and improve the in vivo properties of aptamers [21,36,37,38]. Chemical modifications introduced during synthesis (post-selection) may or may not disrupt the function of the aptamer. Meticulous scanning of nuclease-resistant modifications introduced at each site is ultimately necessary for the identification of aptamers with improved stability/pharmacokinetics for therapeutic use [35,39,40]. For a more detailed review of aptamer modifications see [41,42].
Importantly, Phase I clinical trials with chemically modified aptamers have so far yielded no evidence of toxicity or activation of an innate immune response, such as interferon-γ activation [43]. One aptamer targeting vascular endothelial growth factor (VEGF) is currently FDA-approved for the treatment of macular degeneration, and several others are in the clinical pipeline. For a list of aptamers that are currently in the clinical pipeline see [43]. In addition, for a comprehensive account of aptamers to targets of therapeutic interest see [41].
As well as activating [31] or inhibiting [43,44] the function of their protein targets, aptamers have recently been used to deliver siRNAs (Figure 1) [16,18–23,45,46], chemotherapeutics/toxins [47–49], enzymes [50] and radioisotopes [51,52] to target cells. Several criteria must be met in order for aptamers to act as proficient delivery tools to transport RNA molecules intracellularly. First, the aptamer must recognize a cell-surface entity (usually a receptor) that undergoes either constitutive or ligand-mediated internalization, typically via a clathrin-mediated endocytic pathway. Currently, several aptamers have been isolated for their ability to bind with high affinity and specificity to various cell-surface targets (Table 1). These include aptamers to cancer biomarkers such as: prostate-specific membrane antigen (PSMA) [34]; nucleolin [53] and MUC1 [54]; aptamers to the receptor tyrosine kinase (RTK) Ret [55,56]; epidermal growth factor receptors (EGFRs) [57] and human epidermal growth factor receptor (HER)3 [58]; an aptamer to the serine/threonine kinase receptor transforming growth factor (TGF)-β type III receptor [59]; an RNA aptamer to the immune receptor CD4 [60] and aptamers to the HIV glycoprotein gp120 [22,61,62]. The PSMA and HIV gp120 RNA aptamers have been successfully utilized for delivery of siRNAs to PSMA or gp120-expressing cells, respectively [16,19–23,46]. Aptamers to several other cell-surface receptors are currently being evaluated for their ability to deliver therapeutic RNAs and small molecules to specific cell types.
Table 1.
Aptamers to cell-surface targets for intracellular delivery.
| DNA/RNA | Aptamers | Capable of directing internalization | Ref. |
|---|---|---|---|
| RNA | PSMA | Yes | [16,19,21,34,46–49,72–77] |
| RNA | CD4 | Yes | [18,45,60,67] |
| RNA | gp120 | Yes | [20,22] |
| RNA | Tenascin-C (TN-C) | Yes | [51,78] |
| RNA | 4-1BB | Uncertain | [31] |
| RNA | CTLA-4 | Uncertain | [79] |
| RNA | OX40 | Uncertain | [80] |
| RNA | EGFR | Yes | [57] |
| RNA | EGFRvIII | No | [81] |
| RNA | TGF-β type III receptor | Uncertain | [59] |
| RNA | HER3 | Uncertain | [58] |
| RNA | BACE | Uncertain | [82] |
| RNA | L-selectin | Uncertain | [83] |
| RNA | β-catenin | Uncertain | [84] |
| RNA | KRAS | Uncertain | [85] |
| RNA | Acetylcholine receptor (AChR) | Uncertain | [86] |
| RNA | Neurotensin-1 (NTS-1) | Uncertain | [87] |
| RNA | African trypanosomes (42 kDa protein) | Uncertain | [88] |
| RNA | RET receptor tyrosine kinase | Uncertain | [55] |
| DNA | Immunoglobulin heavy mu chain (IGHM) | Uncertain | [89] |
| DNA | Hemagglutinin | Uncertain | [90] |
| DNA | Mucin (MUC1) | Yes | [54,91] |
| DNA | Nucleolin (NCL) | Yes | [53,92–94] |
| DNA | Transferrin receptor (TfR) | Yes | [50] |
| DNA | PTK7 | Yes | [95–97] |
RNA and DNA aptamers selected to cell-surface targets. Several of these aptamers have been used as vehicles to deliver various cargos (siRNAs, toxins, chemotherapeutic agents and radionuclides) to cells expressing the aptamer target. BACE: β-secretase; EGFRvIII: Epidermal growth factor receptor variant III; gp120: Glycoprotein 120; HER: Human epidermal growth factor receptor; PSMA: Prostate-specific membrane antigen; PTK: Protein tyrosine kinase; TGF-β: Transforming growth factor-β.
Additional criteria for effective aptamer-mediated delivery of siRNAs require that the aptamer–siRNA conjugates be accessible to components of the RNAi machinery. First, the conjugates must be effectively delivered to the cytoplasm of target cells, where the RNAi machinery resides. Once in the cytoplasm, the conjugates must be recognized by components of the RNAi machinery (Dicer and RISC). Strategies to enable recognition of the conjugates by the RNAi machinery and to enhance endosomal release are discussed in the following sections.
RNA delivery using aptamers
Aptamer–siRNA conjugates/chimeras have been successfully generated as potential therapeutics for prostate cancer and HIV. The availability of cell-internalizing aptamers specific for the prostate cancer antigen PSMA [34], and the HIV glycoprotein gp120 [20,22], has made it possible to evaluate the use of RNA aptamers as delivery vehicles for siRNAs and, more importantly, establish design rules for optimal processing of the aptamer–siRNA conjugates by the RNAi machinery [16,19,20,22]. A description of the various aptamer–siRNA conjugates/chimeras targeting cancer or HIV is outlined below.
Cancer target: PSMA
Prostate-specific membrane antigen is expressed at high levels on the surface of primary and metastatic prostate cancers, but is not expressed on the surface of normal prostate epithelia [63,64]. Therefore, any molecule delivered via a PSMA aptamer is likely to be targeted specifically to cancer cells but not normal surrounding tissue. Two PSMA RNA aptamers (A9 and A10) identified by Lupold et al. [34] have been used by several groups to deliver siRNAs to prostate cancer cells in vitro [16,46] and in vivo [19,21,23,65].
In a study by Ellington et al., the PSMA aptamer A9 was noncovalently conjugated to 27mer Dicer substrates targeting housekeeping genes lamin A/C and GAPDH in proof-of-concept studies to validate the aptamer-delivery technology [16]. Biotinylated aptamer and 27mer Dicer substrates were noncovalently assembled on a streptavidin bridge, and the streptavidin–aptamer–siRNA particles were incubated with prostate cancer cell lines that either express (LNCaP) or lack (PC-3) PSMA on the cell surface (Figure 2a). An RNAi effect was observed only in PSMA-expressing cells exposed to the particles, demonstrating targeting specificity by the streptavidin–aptamer–Dicer substrate particle [16].
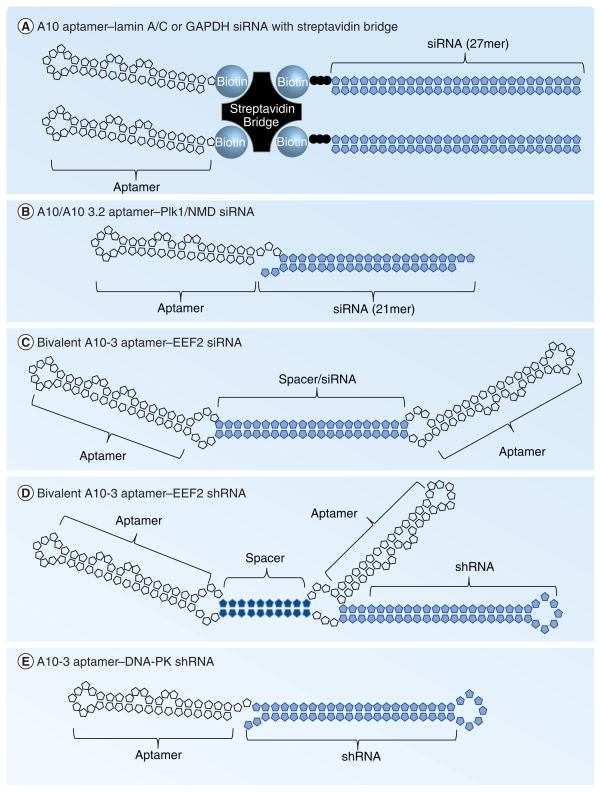
Figure 2. PSMA aptamer–siRNA conjugates.
(A) A10 and 27mer Dicer substrates against lamin A/C were biotinylated at the 3′-ends then assembled on a streptavidin bridge [16]. (B) PSMA A10 or A10-3.2 aptamers were linked to siRNAs against Plk1, Bcl-2 or components of the NMD pathway [19,21,23]. (C) PSMA A10-3 aptamer was dimerized using siRNA against EEF2 embedded within the linker sequence connecting two PSMA aptamers [46]. (D) Alternatively, the PSMA A10-3 aptamer was dimerized with a dsRNA linker and the EEF2 shRNA was appended to the 3′-end of one of the A10-3 aptamers [46]. (E) PSMA A10-3 aptamer was appended to an shRNA against DNA-PK [65], similar to the approach by Wullner et al. [46].
EEF: Eukaryotic elongation factor; PSMA: Prostate-specific membrane antigen; shRNA: Short hairpin RNA.
A covalent assembly approach was simultaneously described where the siRNA duplex was directly linked to the PSMA RNA aptamer A10 (Figure 2b) [19]. In this study, McNamara et al. covalently linked the passenger strands of siRNAs targeting prostate cancer survival genes Bcl-2 or polo-like kinase 1 (Plk1) to the 3′-end of the A10 PSMA aptamer. The guide strands of the siRNAs were then annealed to the respective passenger strands to create the functional siRNA duplex (Figure 2b). Specific silencing of Bcl-2 and Plk1 in prostate cancer cells expressing PSMA and efficient cell death was observed in vitro. The authors also went on to demonstrate efficacy of the PSMA aptamer-Plk1 siRNA chimera in vivo following intratumoral administration of the chimera in a xenograft model of human prostate cancer [19]. This work established aptamer-mediated siRNA delivery as a platform technology, although further optimization of the RNA conjugates was desired for therapeutic utility [21].
In subsequent work, Dassie et al. optimized the PSMA A10 aptamer–Plk1 siRNA chimera for systemic administration [21]. First, to enable future large-scale chemical synthesis of the RNA chimera, the aptamer portion of the chimera was reduced from 71 to 39 nucleotides (A10–3.2) while preserving specificity and high affinity (low nM) binding to PSMA. Next, steps were taken to improve the efficacy (silencing efficiency/specificity) of the chimera and to optimize its pharmacokinetic (PK) profile. Silencing efficiency and specificity of the siRNA portion of the chimera was improved by approximately 100-fold with the following modifications: swapping the passenger and guide strands of the siRNAs and engineering a two-nucleotide overhang at the 3′-end of the guide strand. These changes enhanced Dicer recognition and loading of the guide strand into RISC [21]. To extend the serum circulating time of the chimera, pyrimidines in the passenger strand were also 2′-fluoro modified for increased stability. A 20-kDa PEG group was added to the 5′-end of the passenger strand, thereby increasing the molecular weight and retarding renal clearance. Collectively, these modifications resulted in in vivo efficacy following systemic (intraperitoneal) administration of the chimera in immunocompromised mice bearing PSMA-expressing prostate tumors in their flanks. Importantly, regression after systemic administration of the optimized second-generation chimera was more pronounced and occurred at lower doses (~0.4 vs 1.4 mg/kg) compared with the first-generation chimera [19]. The next steps with this work are to evaluate the safety of the optimized chimera in larger animal models.
Due to the simple design and ease of production of the covalent assembly approach [19], several groups have since linked various silencing RNAs to the PSMA aptamer A10 (and to truncated versions of A10) for aptamer-targeted siRNA and shRNA delivery [21,23,46,65]. To enhance uptake of the PSMA aptamer–siRNA conjugate into PSMA-expressing cells, Wullner et al. designed a dimeric PSMA aptamer capable of inducing endocytosis of PSMA and enhancing uptake of an siRNA silencing eukaryotic elongation factor (EEF)2 into PSMA-expressing cells in vitro [46]. In their design, the siRNA sequence was either part of the spacer sequence linking the two PSMA A10–3 aptamers (Figure 2C) or was tethered to the 3′ end of one of the aptamer dimers as an shRNA (Figure 2D) [46]. When incubated with prostate cancer cells expressing PSMA, the dimeric chimeras promoted PSMA receptor internalization and uptake of the dimeric chimeras into cells, leading to enhanced silencing of the siRNA target gene EEF2 when compared with a monomeric A10–3 aptamer–EEF2 shRNA chimera.
In a recent report from Gilboa and colleagues, the PSMA aptamer–siRNA chimera technology was applied to the field of cancer immunotherapy [23]. In order to successfully mount an immune response against tumors, unique antigens must be expressed by tumor cells to distinguish them from normal cells. In this study, the PSMA aptamer (A10) was used to deliver siRNAs against the nonsense-mediated mRNA decay (NMD) pathway. Normal cells use the NMD pathway to prevent expression of mRNAs containing premature termination codons. Therefore, inhibition of this pathway results in the generation of unique antigens that can be recognized by the immune system as foreign. First, siRNA duplexes against key components of the NMD pathway (Upf2, Smg1) were covalently linked to the 3′-end of the A10 PSMA aptamer as described by McNamara et al. (Figure 2b) [19]. The aptamer–siRNA chimeras demonstrated specific targeting to and silencing in PSMA-expressing mouse tumor cells in culture and in PSMA-expressing mouse tumors in vivo. Tumor volume was significantly reduced following systemic (intravenous) administration of the aptamer–siRNA chimeras compared with control nonsilencing or nontargeting chimeras. Importantly, in long-term studies with higher doses of the chimeras, most mice completely rejected the tumor cells. These studies highlight the power of targeting siRNAs against the NMD pathway to tumors and tumor metastases using RNA aptamers as a delivery vehicle in order to elicit a potent tumor-specific immune response.
While cancer immunotherapy is a powerful approach for eradicating cancer, many cancer patients are currently given multiple treatments (combination therapies), such as radiation and chemotherapy, to increase the success of killing their cancers. Towards this end, Lupold and colleagues conducted a high-throughput RNAi screen to identify novel siRNAs that sensitize PSMA+ prostate cancer cells to ionizing radiation [65]. After confirming radio-sensitization with several siRNAs (e.g., DNA-activated protein kinase [DNA-PK] or BRCA1), the group generated a PSMA aptamer–DNA-PK shRNA chimera and showed that in animals bearing PSMA+ tumors, pretreatment with the chimera 2–3 days before irradiation resulted in greater tumor regression than with radiation alone. Interestingly, instead of appending duplex siRNAs to the 3′-end of the A10–3 PSMA aptamer, Lupold and colleagues linked a shRNA against DNA-PK to the 3′-end of the A10–3 PSMA aptamer (Figure 2e). The shRNA is continuous with the aptamer strand (one molecule) and is thus fully 2′-fluoro modified for increased stability. In addition, because shRNAs, like miRNA precursors (pre-miRNA), are better substrates for Dicer, it is possible that this configuration may enhance processing of aptamer–shRNA chimeras by the RNAi machinery. A similar observation was made by Dassie et al. who compared processing of a PSMA–aptamer–Plk1 siRNA chimera that resembled a miRNA precursor to the standard aptamer–siRNA duplex design, and showed that the chimera resembling the pre-miRNA was several times more active than the aptamer–siRNA duplex chimera [21].
Cancer target: CD4
Work by Guo and co-workers has pioneered a unique method of noncovalently linking siRNAs to aptamers: packaging RNAs (pRNAs) [18,45]. pRNAs are derived from bacteriophage phi29 small RNAs that normally package DNA into procapsids and contain a 5′/3′ helical domain and an intermolecular interaction domain, which serves to form dimeric pRNA molecules. By substituting the 5′/3′ helical domains with an aptamer on one pRNA molecule and an siRNA on another pRNA molecule, an aptamer–siRNA conjugate was generated through dimerization of the intermolecular interaction domains of the two pRNAs (Figure 3) [18]. In proof-of-concept studies, an aptamer for CD4, a receptor predominantly expressed on T cells, was used to specifically deliver a 29mer Dicer substrate against survivin, a pro-survival gene that is often elevated in many types of cancer. This delivery mechanism resulted in reduced viability of CD4+ but not CD4− T lymphocytes in culture [18]. Future studies are necessary to evaluate the efficacy and safety of this approach in animal models of cancer.
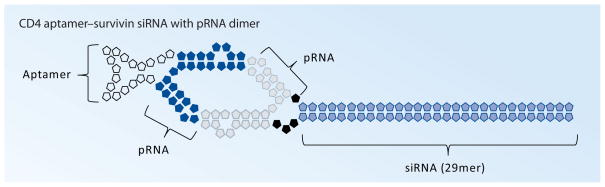
Figure 3. CD4 aptamer–pRNA–siRNA conjugates.
CD4 aptamer (left) and a 29mer Dicer substrate to survivin (right) were inserted into the 5′/3′ helical domain of pRNA. The two particles were joined by dimerization of the pRNAs [18].
pRNA: Packaging RNA.
HIV targets: gp120 & CD4
CD4 is expressed on the surface of a subset of T cells, and the interaction between CD4 and the HIV-1 cell-surface envelope glycoprotein, gp120, allows entry of the virus into T cells [24]. Thus, neutralizing aptamers against either receptor represent a potential anti-HIV therapy, and several groups have described RNA aptamers against both targets [24]. Rossi and colleagues were the first to report a dual-inhibitory aptamer–siRNA chimera for the treatment of HIV [20]. In this report, an inhibitory aptamer against gp120 tethered to a siRNA against tat/rev, two viral genes which drive replication of the virus, reduced HIV infectivity and replication in cultured T cells (Figure 4a). The tat/rev siRNA was covalently tethered to the gp120 aptamer, similar to the initial aptamer–siRNA chimera reported by McNamara et al. [19], except a 4-nucleotide linker was added between the 3′-end of the gp120 aptamer and the siRNA to increase flexibility of the chimera and processing by Dicer [20]. Interestingly, use of a 27-mer Dicer substrate siRNA linked to the 3′-end of the gp120 aptamer provided better silencing than a shorter 21-mer sequence (Figure 4a) [20].
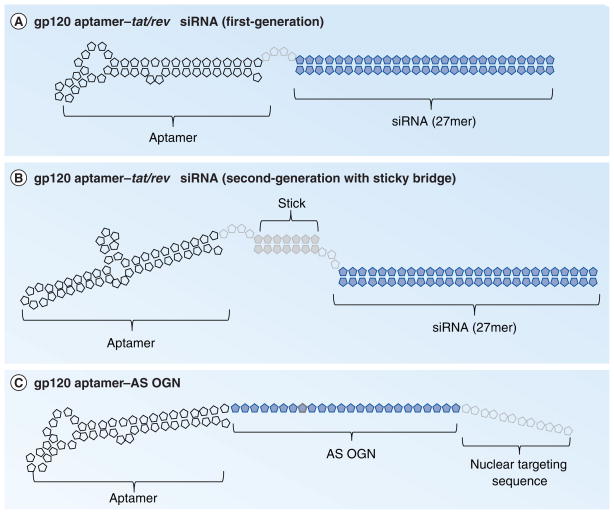
Figure 4. gp120 aptamer-RNA conjugates.
(A) First-generation HIV gp120 aptamer conjugated to 27mer Dicer substrates to tat/rev [20]. A 4-nucleotide linker connects the aptamer and siRNA. (B) Second-generation gp120 aptamer–tet/rev 27mer Dicer substrate conjugates were developed by a ‘sticky bridge’ approach by base-paring linker sequences engineered at the 3′-end of the aptamer to complementary sequences engineered at the 3′-end of the Dicer substrate sense strand [20]. (C) HIV gp120 (or CD4) aptamer was used to deliver AS OGN to carry out transcriptional gene silencing. A nuclear targeting sequence was appended to the 3′-end of the AS OGN to enable proper nuclear delivery.
AS OGN: Antisense oligonucleotide.
A subsequent study by the same group identified new anti-gp120 aptamers and described a ‘sticky bridge’ approach for appending siRNAs to the aptamer [22]. The sticky bridge approach consists of a GC-rich sequence that is extended from the 3′-end of the gp120 aptamer, and a complementary CG-rich sequence is added to the 3′-end of one of the siRNA strands (Figure 4b). This unique strategy theoretically allows any siRNA sequence to be appended to the 3′-end of the aptamer-stick without altering the conformation of the aptamer. Although the present study only reported delivery of a single siRNA to target cells, this approach could be extended to enable conjugation of multiple siRNAs onto a single aptamer [22].
Zhou et al., recently reported at the American Society of Gene and Cell Therapy (ASGCT) meeting, in vivo studies with a second-generation gp120 aptamer-rev/tat siRNA chimera, along with gp120 aptamer-delivered siRNAs against CD4 and transportin 3 [66]. Using the RAG-hu mice as a model for HIV infection, a reduction in infectivity was observed, particularly when the three siRNAs were used in a combination.
A different approach for targeting therapeutic RNAs to HIV via the gp120 aptamer was also recently presented by Morris and colleagues at the ASGCT meeting. The investigators used RNA aptamers to deliver RNA AS OGNs as a means to inhibit TGS. To apply this approach to HIV, Morris’s group created RNA chimeras using aptamers against either gp120 or CD4 tethered to a single-stranded antisense sequence (Figure 4C). A mir-29B targeting sequence added to the 3′-end of the OGN directed the chimera to the nucleus [Kevin Morris, Pers. Comm.]. Importantly, this is the first example of using an aptamer to deliver an AS OGN as well as application of the aptamer–chimera platform technology to TGS.
In another unpublished report, Lieberman and colleagues have described the use of RNA aptamers for CD4 to deliver anti-HIV siRNAs to CD4+ T cells [67], similar to the work with gp120 aptamer-directed Dicer substrates described above [20,22]. This aptamer–siRNA chimera was tested in human cervical explants for its ability to prevent HIV infection in a localized manner. The investigators speculate that their reagent can be applied topically to humans as a prophylactic against HIV infection. Interestingly, a topical route of administration of the aptamer–siRNA chimera may eliminate the need to optimize the reagent for systemic delivery (i.e., with stabilizing chemical modifications and PEGylation to retard renal clearance).
Challenges to aptamer-mediated RNA delivery
For therapeutic applications, the RNA aptamer–siRNA conjugates/chimeras must be fully optimized to reduce the potential for undesired immunostimulatory effects and to increase their serum stability and PK profiles [21]. Several attempts are currently being made to achieve these goals [21]. The isolation (e.g., improved selection methodologies) or synthesis (e.g., chemical synthesis of high-quality cGMP-grade long RNAs, chemical modifications of the nucleotides for increased stability or terminal modifications for increased stability and to bypass renal filtration) of RNA aptamers, which are optimally modified for in vivo applications, is likely to expand the potential applications of this technology. Similarly, the vast knowledge available from the RNAi field of chemical formulations for use of siRNAs in vivo can be easily translated to aptamers for the purpose of siRNA delivery.
A major challenge for using aptamers to deliver siRNAs to cells is cytoplasmic delivery. The aptamer–siRNA conjugates/chimeras must successfully be internalized (presumably by receptor-mediated internalization) and released into the cytoplasm where they can encounter the RNAi machinery. The current RNA conjugates/chimeras must escape from the early, sorting or late endosome prior to entering the lysosome where the presence of nucleases and low pH conditions are likely to result in siRNA degradation [68]. Currently, neither the uptake kinetics of the various RNA conjugates/chimeras described to date nor the mechanism by which these RNAs escape from the endosome are known. Molecules such as fusogenic lipids, peptides and pH-sensitive lipoplexes and polyplexes have been employed to promote the release of the siRNAs from the endosome [68]. However, none of these moieties have been tested to date in concert with aptamer–siRNA conjugates as a way to facilitate endosome escape.
Another limiting factor for the broad applicability of this delivery technology is the relative paucity of candidate aptamers for specific delivery. Only a handful of aptamers that bind to cell-surface receptors have been isolated, though several groups are developing cell-based methods to isolate aptamers against these complex, membrane-bound targets. A review by Shamah et al., nicely summarizes the various cell-based SELEX methodologies currently under investigation [69]. Cell-based selections for isolating aptamers with high affinity and specificity to cell-surface proteins have clear advantages over selections performed using purified recombinant proteins. For example, the complex glycosylation patterns and structural conformations of membrane proteins may not be replicated in recombinant protein preparations. Aptamers against TGF-β type III and Ret receptors have been identified using cell-based selections [55,59] and, given the role of these proteins in cancer and other diseases, these aptamers may turn out to be ideal candidates as siRNA delivery vehicles if they are internalized in a receptor-dependent manner.
Knowledge of RNA (tertiary) structure is also an important factor when generating aptamer–siRNA conjugates/chimeras. In particular, being able to predict how a chemically modified aptamer will fold when appended to a particular siRNA sequence can facilitate the design and characterization of these reagents.
Finally, the proposed pharmaceutical application of this technology is likely to require improvements in chemical synthesis of longer RNAs (60–100 nucleotides long). RNAs up to 100 nucleotides in length are routinely synthesized/chemically modified using solid-phase phosphoramidite chemistry in an automated process used for small-scale synthesis of oligonucleotides [41,70]. However, large-scale, high-quality, cGMP-grade material of longer RNA sequences (aptamers/aptamers–siRNA chimeras) for therapeutic development still presents significant difficulties. Several potential solutions to this problem include the identification of shorter aptamer sequences through the use of shorter RNA libraries, extensive truncation of the RNAs down to the minimal functional sequence (15–30 nucleotides) and/or conjugation of siRNAs to aptamers via linking chemistries.
Future perspective
While the future of aptamer-mediated RNA delivery technology is limited by the number of cell-type specific aptamers available, novel selection methodologies for identifying cell-specific internalization-competent aptamers are currently being developed [71]. As discussed above, cell-based selection approaches hold the most promise for isolating aptamers that recognize the native conformation of proteins and glycosylation patterns, but such approaches still require an additional level of screening to find aptamers that are internalized and released into the cytoplasm of target cells. Work in our laboratory seeks to modify the existing cell-based SELEX methodology to identify internalization-competent aptamers. For the modified cell-based selections, complex RNA aptamer libraries are incubated with target cells in culture (e.g., cancer cells) at 37°C to allow intracellular uptake of the RNAs. Unbound RNAs or RNAs that bind to the cell surface but do not internalize can be removed by enzymatic digestion or stringent washes. Internalized RNAs can be recovered by TRIzol extraction. RT-PCR amplification of these RNAs followed by in vitro transcription is performed to generate the next round of selection. To increase specificity for a particular cell-type or a cell-specific antigen, a negative selection can be performed where the RNA library is precleared at each round of selection against a nontarget cell (e.g., normal cells or cells lacking the cell surface antigen). This approach may favor the identification of aptamers that internalize in specific cell-types but does not necessarily guarantee the isolation of aptamers that are efficiently released into the cytoplasm of target cells for siRNA delivery. Modifications to this approach that allow for the selective amplification of cytoplasmic-specific aptamers may enable the effective isolation of cytoplasm-targeting RNA sequences.
Currently we have isolated several aptamers capable of internalizing into specific cancer cell-lines and simultaneously delivering functional siRNAs against cancer-specific prosurvival genes [Thiel and Giangrande, Unpublished Data]. While still in their infancy, these cell-based approaches are likely to facilitate the identification of aptamers with increased stability, the ability to internalize into specific cell-types, and finally, the ability to escape endosomes and release their therapeutic RNA cargo into the cytoplasm of target cells.
Acknowledgments
We thank James O McNamara II for useful discussions and careful review of the manuscript.
Key Terms
- siRNA
Small-interfering RNA. Effector molecules of the RNAi pathway
- Aptamers
Synthetic structured single-stranded DNA or RNA oligonucleotide that binds to molecular targets with high affinity and specificity
- shRNA
Short hairpin RNA. Effector molecules of the RNAi pathway
- miRNA
Micro RNA. Endogenous effector molecules of the RNAi pathway
- Antisense oligonucleotide
Short nucleic acid polymer
- SELEX
Systematic evolution of ligands by exponential enrichment, an iterative process by which aptamers are discovered
Footnotes
For reprint orders, please contact reprints@future-science.com
Financial & competing interests disclosure
Kristina W Thiel acknowledges a Ladies Auxiliary to the Veterans of Foreign Wars Postdoctoral Fellowship. Paloma H Giangrande acknowledges support from the NIH (1RO1 CA138503-01A1, 1R21DE019953-01), and the Mary Kay Ash Foundation (MKACF 001-09). Paloma H Giangrande is a co-inventor on patent application PCT/US2007/012927_DELIVERY METHOD. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Potti A, Schilsky RL, Nevins JR. Refocusing the war on cancer: the critical role of personalized treatment. Sci Transl Med. 2010;2(28) doi: 10.1126/scitranslmed.3000643. 28cm13. [DOI] [PubMed] [Google Scholar]
- 2.Hamburg MA, Collins FS. The path to personalized medicine. N Engl J Med. 2010;363(4):301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 3.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457(7228):426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris KV. RNA-directed transcriptional gene silencing and activation in human cells. Oligonucleotides. 2009;19(4):299–306. doi: 10.1089/oli.2009.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akashi H, Matsumoto S, Taira K. Gene discovery by ribozyme and siRNA libraries. Nat Rev Mol Cell Biol. 2005;6(5):413–422. doi: 10.1038/nrm1646. [DOI] [PubMed] [Google Scholar]
- 6.Behlke MA. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18(4):305–320. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal S, Kandimalla ER. Antisense and siRNA as agonists of Toll-like receptors. Nat Biotech. 2004;22(12):1533–1537. doi: 10.1038/nbt1042. [DOI] [PubMed] [Google Scholar]
- 8.Sioud M. Does the understanding of immune activation by RNA predict the design of safe siRNAs? Front Biosci. 2008;13:4379–4392. doi: 10.2741/3011. [DOI] [PubMed] [Google Scholar]
- 9.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285(5433):1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 10▪▪.Song E, Zhu P, Lee S-K, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotech. 2005;23(6):709–717. doi: 10.1038/nbt1101. The first report of an antibody-based approach (antibody-protamine fusion protein) to specifically deliver siRNAs in vivo. [DOI] [PubMed] [Google Scholar]
- 11.Kumar P, Wu H, McBride JL, et al. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448(7149):39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 12.Morrissey DV, Lockridge JA, Shaw L, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotech. 2005;23(8):1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 13.Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2004;12(5):461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 14.Zimmermann TS, Lee ACH, Akinc A, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441(7089):111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 15.Kim S-S, Garg H, Joshi A, Manjunath N. Strategies for targeted nonviral delivery of siRNAs in vivo. Trends Mol Med. 2009;15(11):491–500. doi: 10.1016/j.molmed.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16▪.Chu TC, Twu KY, Ellington AD, Levy M. Aptamer mediated siRNA delivery. Nucl Acids Res. 2006;34(10):e73. doi: 10.1093/nar/gkl388. In this proof-of-concept work, a streptavidin bridge was used to assemble a prostate-specific membrane antigen (PSMA) aptamer–siRNA conjugate to accomplish specific delivery to PSMA+ prostate cancer cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peer D, Zhu P, Carman CV, Lieberman J, Shimaoka M. Selective gene silencing in activated leukocytes by targeting siRNAs to the integrin lymphocyte function-associated antigen-1. Proc Natl Acad Sci USA. 2007;104(10):4095–4100. doi: 10.1073/pnas.0608491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18▪.Guo S, Tschammer N, Mohammed S, Guo P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum Gene Ther. 2005;16(9):1097–1110. doi: 10.1089/hum.2005.16.1097. Packaging RNAs (pRNA) were used to form aptamer–siRNA conjugates for targeted siRNA delivery to CD4+ T lymphocytes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19▪.McNamara JO, II, Andrechek ER, Wang Y, et al. Cell type-specific delivery of siRNAs with aptamer–siRNA chimeras. Nat Biotechnol. 2006;24(8):1005–1015. doi: 10.1038/nbt1223. This in vivo study established aptamer–siRNA chimeras as a platform technology to specifically deliver siRNAs against pro-survival genes to PSMA+ human prostate cancer tumors. [DOI] [PubMed] [Google Scholar]
- 20▪▪.Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer–siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16(8):1481–1489. doi: 10.1038/mt.2008.92. An inhibitory aptamer was combined with a silencing siRNA to deliver a two-hit punch to HIV-infected T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21▪▪.Dassie JP, Liu X-Y, Thomas GS, et al. Systemic administration of optimized aptamer–siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotech. 2009;27(9):839–846. doi: 10.1038/nbt.1560. Optimization of the first-generation PSMA aptamer–siRNA chimera achieved systemic delivery in an in vivo model of prostate cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22▪.Zhou J, Swiderski P, Li H, et al. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucl Acids Res. 2009;37(9):3094–3109. doi: 10.1093/nar/gkp185. New anti-gp120 inhibitory aptamers were identified and a unique ‘sticky bridge’ strategy was introduced to non-covalently conjugate siRNAs to the gp120 aptamer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23▪▪.Pastor F, Kolonias D, Giangrande PH, Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature. 2010;465(7295):227–230. doi: 10.1038/nature08999. This groundbreaking tumor immunotherapy study used aptamers to deliver siRNAs against components of the nonsense-mediated mRNA decay pathway, which generated novel antigens on the surface of PSMA+ tumor cells and resulted in an immune response to the tumor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J, Rossi J. Aptamer-targeted cell-specific RNA interference. Silence. 2010;1(1):4. doi: 10.1186/1758-907X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soutschek J, Akinc A, Bramlage B, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 26.Nishina K, Unno T, Uno Y, et al. Efficient in vivo delivery of siRNA to the liver by conjugation of α-tocopherol. Mol Ther. 2008;16(4):734–740. doi: 10.1038/mt.2008.14. [DOI] [PubMed] [Google Scholar]
- 27.Muratovska A, Eccles MR. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett. 2004;558(1–3):63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- 28.Chiu YL, Ali A, Chu CY, Cao H, Rana TM. Visualizing a correlation between siRNA localization, cellular uptake, and RNAi in living cells. Chem Biol. 2004;11(8):1165–1175. doi: 10.1016/j.chembiol.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 30.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 31.McNamara JO, Kolonias D, Pastor F, et al. Multivalent 4–1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J Clin Invest. 2008;118(1):376–386. doi: 10.1172/JCI33365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burmeister PE, Lewis SD, Silva RF, et al. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem Biol. 2005;12(1):25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 33.Rusconi CP, Scardino E, Layzer J, et al. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419(6902):90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- 34▪.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62(14):4029–4033. PSMA A9 and A10 aptamers characterized in this study were the initial aptamers used, in subsequent studies, to deliver siRNAs to target cells. [PubMed] [Google Scholar]
- 35.Ruckman J, Green LS, Beeson J, et al. 2′-fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165): Inhibition of receptor binding and VEGF-induced vascular permeability through interacions requiring the exon 7-encoded domain. J Biol Chem. 1998;273(32):20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 36.Biesecker G, Dihel L, Enney K, Bendele RA. Derivation of RNA aptamer inhibitors of human complement C5. Immunopharmacology. 1999;42(1–3):219–230. doi: 10.1016/s0162-3109(99)00020-x. [DOI] [PubMed] [Google Scholar]
- 37.Healy JM, Lewis SD, Kurz M, et al. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm Res. 2004;21(12):2234–2246. doi: 10.1007/s11095-004-7676-4. [DOI] [PubMed] [Google Scholar]
- 38.Beielman L, Karpeisky A, Matulic-Adamic J, Haeberli P, Sweedler D, Usman N. Synthesis of 2′-modified nucleotides and their incorporation into hammerhead ribozymes. Nucl Acids Res. 1995;23(21):4434–4442. doi: 10.1093/nar/23.21.4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adler A, Forster N, Homann M, Goringer HU. Post-SELEX chemical optimization of a trypanosome-specific RNA aptamer. Comb Chem High Throughput Screen. 2008;11(1):16–23. doi: 10.2174/138620708783398331. [DOI] [PubMed] [Google Scholar]
- 40.Floege J, Ostendorf T, Janssen U, et al. Novel approach to specific growth factor inhibition in vivo: antagonism of platelet-derived growth factor in glomerulonephritis by aptamers. Am J Pathol. 1999;154(1):169–179. doi: 10.1016/S0002-9440(10)65263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9(7):537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keefe AD, Cload ST. SELEX with modified nucleotides. Curr Opin Chem Biol. 2008;12(4):448–456. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 43.Thiel KW, Giangrande PH. Therapeutic applications of DNA and RNA aptamers. Oligonucleotides. 2009;19(3):209–222. doi: 10.1089/oli.2009.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 45.Khaled A, Guo S, Li F, Guo P. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett. 2005;5(9):1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46▪.Wullner U, Neef I, Eller A, Kleines M, Tur MK, Barth S. Cell-specific induction of apoptosis by rationally designed bivalent aptamer–siRNA transcripts silencing eukaryotic elongation factor 2. Curr Cancer Drug Targets. 2008;8(7):554–565. doi: 10.2174/156800908786241078. Bivalent PSMA aptamers were used to deliver siRNAs to PSMA+ cells with increased efficiency compared to monovalent counterparts. [DOI] [PubMed] [Google Scholar]
- 47.Farokhzad OC, Cheng J, Teply BA, et al. Targeted nanoparticle–aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci USA. 2006;103(16):6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Radovic-Moreno Aleksandar F, Alexis F, et al. Co-delivery of hydrophobic and hydrophilic drugs from nanoparticle–aptamer bioconjugates. ChemMedChem. 2007;2(9):1268–1271. doi: 10.1002/cmdc.200700121. [DOI] [PubMed] [Google Scholar]
- 49.Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug–PLGA–PEG nanoparticles. Proc Natl Acad Sci USA. 2008;105(45):17356–17361. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen CH, Dellamaggiore KR, Ouellette CP, et al. Aptamer-based endocytosis of a lysosomal enzyme. Proc Natl Acad Sci USA. 2008;105(41):15908–15913. doi: 10.1073/pnas.0808360105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hicke BJ, Stephens AW, Gould T, et al. Tumor targeting by an aptamer. J Nucl Med. 2006;47(4):668–678. [PubMed] [Google Scholar]
- 52.Borbas KE, Ferreira CSM, Perkins A, Bruce JI, Missailidis S. Design and synthesis of mono- and multimeric targeted radiopharmaceuticals based on novel cyclen ligands coupled to anti-MUC1 aptamers for the diagnostic imaging and targeted radiotherapy of cancer. Bioconjug Chem. 2007;18(4):1205–1212. doi: 10.1021/bc0700741. [DOI] [PubMed] [Google Scholar]
- 53.Bates PJ, Kahlon JB, Thomas SD, Trent JO, Miller DM. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J Biol Chem. 1999;274(37):26369–26377. doi: 10.1074/jbc.274.37.26369. [DOI] [PubMed] [Google Scholar]
- 54.Ferreira CSM, Matthews CS, Missailidis S. DNA aptamers that bind to MUC1 tumour marker: design and characterization of MUC1-binding single-stranded DNA aptamers. Tumour Biol. 2006;27(6):289–301. doi: 10.1159/000096085. [DOI] [PubMed] [Google Scholar]
- 55▪.Cerchia L, Duconge F, Pestourie C, et al. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS Biol. 2005;3(4):e123. doi: 10.1371/journal.pbio.0030123. This cell-based study modified the systematic evolution of ligands by exponential enrichment methodology to identify aptamers against particular cell-surface receptors. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Pestourie C, Cerchia L, Gombert K, et al. Comparison of different strategies to select aptamers against a transmembrane protein target. Oligonucleotides. 2006;16(4):323–335. doi: 10.1089/oli.2006.16.323. [DOI] [PubMed] [Google Scholar]
- 57.Li N, Larson T, Nguyen HH, Sokolov KV, Ellington AD. Directed evolution of gold nanoparticle delivery to cells. Chem Commun (Camb) 2010;46(3):392–394. doi: 10.1039/b920865h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen CB, Chernis GA, Hoang VQ, Landgraf R. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc Natl Acad Sci USA. 2003;100(16):9226–9231. doi: 10.1073/pnas.1332660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohuchi SP, Ohtsu T, Nakamura Y. Selection of RNA aptamers against recombinant transforming growth factor-β type III receptor displayed on cell surface. Biochimie. 2006;88(7):897–904. doi: 10.1016/j.biochi.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 60.Kraus E, James W, Barclay AN. Cutting edge: novel RNA ligands able to bind CD4 antigen and inhibit CD4+ T lymphocyte function. J Immunol. 1998;160(11):5209–5212. [PubMed] [Google Scholar]
- 61.Dey AK, Khati M, Tang M, Wyatt R, Lea SM, James W. An aptamer that neutralizes R5 strains of human immunodeficiency virus type 1 blocks gp120-CCR5 interaction. J Virol. 2005;79(21):13806–13810. doi: 10.1128/JVI.79.21.13806-13810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62▪.Khati M, Schuman M, Ibrahim J, Sattentau Q, Gordon S, James W. Neutralization of infectivity of diverse R5 clinical isolates of human immunodeficiency virus type 1 by gp120-binding 2′F-RNA aptamers. J Virol. 2003;77(23):12692–12698. doi: 10.1128/JVI.77.23.12692-12698.2003. The inhibitory aptamer against gp120 was used in the first-generation chimera to deliver siRNAs against rev/tat. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Israeli RS, Powell CT, Corr JG, Fair WR, Heston WDW. Expression of the prostate-specific membrane antigen. Cancer Res. 1994;54(7):1807–1811. [PubMed] [Google Scholar]
- 64.Su SL, Huang I-P, Fair WR, Powell CT, Heston WDW. Alternatively spliced variants of prostate-specific membrane antigen RNA: ratio of expression as a potential measurement of progression. Cancer Res. 1995;55(7):1441–1443. [PubMed] [Google Scholar]
- 65.Ni X, Zhang Y, DeWeese TL, Lupold SE. Prostate tumor radiosensitization through PSMA-aptamer-targeted siRNA knock-down of DNA repair pathways. Presented at: 13th Annual Meeting American Society of Gene & Cell Therapy; Washington, DC, USA. 19–22 May 2010. [Google Scholar]
- 66.Zhou J, Neff PC, Liu X, et al. Systemic administration of dendrimer- and aptamer–siRNA conjugates efficiently suppresses HIV-1 infection. Presented at: 13th Annual Meeting American Society of Gene & Cell Therapy; Washington, DC, USA. 19–22 May 2010. [Google Scholar]
- 67.Dove A. An apt approach. Nat Med. 2010;16(3):258–260. doi: 10.1038/nm0310-258. [DOI] [PubMed] [Google Scholar]
- 68.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123(8):1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 69.Shamah SM, Healy JM, Cload ST. Complex target SELEX. Acc Chem Res. 2008;41(1):130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 70.Caruthers M. Gene synthesis machines: DNA chemistry and its uses. Science. 1985;230(4723):281–285. doi: 10.1126/science.3863253. [DOI] [PubMed] [Google Scholar]
- 71.Thiel KW, Thiel WH, Liu X, et al. Delivery of chemo-sensitizing siRNAs to HER2-positive breast cancer cells using RNA aptamers. Presented at: 13th Annual Meeting American Society of Gene & Cell Therapy; Washington, DC, USA. 19–22 May 2010. [Google Scholar]
- 72.Chu TC, Marks JW, III, Lavery LA, et al. Aptamer: toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006;66(12):5989–5992. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- 73.Bagalkot V, Farokhzad OC, Langer R, Jon S. An aptamer–doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew Chem Int Ed Engl. 2006;45(48):8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 74.Cheng J, Teply BA, Sherifi I, et al. Formulation of functionalized PLGA–PEG nanoparticles for in vivo targeted drug delivery. Biomaterials. 2007;28(5):869–876. doi: 10.1016/j.biomaterials.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Farokhzad OC, Jon S, Khademhosseini A, Tran T-NT, LaVan DA, Langer R. Nanoparticle-Aptamer Bioconjugates. Cancer Research. 2004;64(21):7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 76.Gu F, Zhang L, Teply BA, et al. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. Proc Natl Acad Sci USA. 2008;105(7):2586–2591. doi: 10.1073/pnas.0711714105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jayaprakash S, Wang X, Heston W, Kozikowski A. Design and synthesis of a PSMA inhibitor–doxorubicin conjugate for targeted prostate cancer therapy. ChemMedChem. 2006;1(3):299–302. doi: 10.1002/cmdc.200500044. [DOI] [PubMed] [Google Scholar]
- 78.Hicke BJ, Marion C, Chang Y-F, et al. Tenascin-C aptamers are generated using tumor cells and purified protein. J Biol Chem. 2001;276(52):48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- 79.Santulli-Marotto S, Nair SK, Rusconi C, Sullenger B, Gilboa E. Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 2003;63(21):7483–7489. [PubMed] [Google Scholar]
- 80.Dollins CM, Nair S, Boczkowski D, et al. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem Biol. 2008;15(7):675–682. doi: 10.1016/j.chembiol.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Y, Kuan C-T, Mi J, et al. Aptamers selected against the unglycosylated EGFRvIII ectodomain and delivered intracellularly reduce membrane-bound EGFRvIII and induce apoptosis. Biol Chem. 2008;390(2):137–144. doi: 10.1515/BC.2009.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rentmeister A, Bill A, Wahle T, Walter J, Famulok M. RNA aptamers selectively modulate protein recruitment to the cytoplasmic domain of β-secretase BACE1 in vitro. RNA. 2006;12(9):1650–1660. doi: 10.1261/rna.126306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Connell D, Koenig A, Jennings S, et al. Calcium-dependent oligonucleotide antagonists specific for L-selectin. Proc Natl Acad Sci USA. 1996;93(12):5883–5887. doi: 10.1073/pnas.93.12.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee HK, Choi YS, Park YA, Jeong S. Modulation of oncogenic transcription and alternative splicing by β-catenin and an RNA aptamer in colon cancer cells. Cancer Res. 2006;66(21):10560–10566. doi: 10.1158/0008-5472.CAN-06-2526. [DOI] [PubMed] [Google Scholar]
- 85.Tanaka Y, Akagi K, Nakamura Y, Kozu T. RNA aptamers targeting the carboxyl terminus of KRAS oncoprotein generated by an improved SELEX with isothermal RNA amplification. Oligonucleotides. 2007;17(1):12–21. doi: 10.1089/oli.2006.0035R1. [DOI] [PubMed] [Google Scholar]
- 86.Ulrich H, Ippolito JE, Pagan O, et al. In vitro selection of RNA molecules that displace cocaine from the membrane-bound nicotinic acetylcholine receptor. Proc Natl Acad Sci USA. 1998;95(24):14051–14056. doi: 10.1073/pnas.95.24.14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Daniels DA, Sohal AK, Rees S, Grisshammer R. Generation of RNA aptamers to the G-protein-coupled receptor for neurotensin, NTS-1. Anal Biochem. 2002;305(2):214–226. doi: 10.1006/abio.2002.5663. [DOI] [PubMed] [Google Scholar]
- 88.Ulrich H, Magdesian MH, Alves MJM, Colli W. In vitro selection of RNA aptamers that bind to cell adhesion receptors of Trypanosoma cruzi and inhibit cell invasion. J Biol Chem. 2002;277(23):20756–20762. doi: 10.1074/jbc.M111859200. [DOI] [PubMed] [Google Scholar]
- 89.Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy μ chain in Burkitt’s lymphoma cells. Mol Cell Proteomics. 2007;6(12):2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 90.Jeon SH, Kayhan B, Ben-Yedidia T, Arnon R. A DNA aptamer prevents influenza infection by blocking the receptor binding region of the viral hemagglutinin. J Biol Chem. 2004;279(46):48410–48419. doi: 10.1074/jbc.M409059200. [DOI] [PubMed] [Google Scholar]
- 91.Ferreira CSM, Cheung MC, Missailidis S, Bisland S, Gariepy J. Phototoxic aptamers selectively enter and kill epithelial cancer cells. Nucl Acids Res. 2009;37(3):866–876. doi: 10.1093/nar/gkn967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008;68(7):2358–2365. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]
- 93.Girvan AC, Teng Y, Casson LK, et al. AGRO100 inhibits activation of nuclear factor-κB (NF-κB) by forming a complex with NF-κB essential modulator (NEMO) and nucleolin. Mol Cancer Ther. 2006;5(7):1790–1799. doi: 10.1158/1535-7163.MCT-05-0361. [DOI] [PubMed] [Google Scholar]
- 94.Cao Z, Tong R, Mishra A, et al. Reversible cell-specific drug delivery with aptamer-functionalized liposomes. Angew Chem Int Ed Engl. 2009;48(35):6494–6498. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- 95.Shangguan D, Cao ZC, Li Y, Tan W. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clin Chem. 2007;53(6):1153–1155. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- 96.Xiao Z, Shangguan D, Cao Z, Fang X, Tan W. Cell-specific internalization study of an aptamer from whole cell selection. Chemistry. 2008;14(6):1769–1775. doi: 10.1002/chem.200701330. [DOI] [PubMed] [Google Scholar]
- 97.Shangguan D, Cao Z, Meng L, et al. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J Proteome Res. 2008;7(5):2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]