Abstract
The filamentous fungus Aspergillus niger exhibits great diversity in its phenotype. It is found globally, both as marine and terrestrial strains, produces both organic acids and hydrolytic enzymes in high amounts, and some isolates exhibit pathogenicity. Although the genome of an industrial enzyme-producing A. niger strain (CBS 513.88) has already been sequenced, the versatility and diversity of this species compel additional exploration. We therefore undertook whole-genome sequencing of the acidogenic A. niger wild-type strain (ATCC 1015) and produced a genome sequence of very high quality. Only 15 gaps are present in the sequence, and half the telomeric regions have been elucidated. Moreover, sequence information from ATCC 1015 was used to improve the genome sequence of CBS 513.88. Chromosome-level comparisons uncovered several genome rearrangements, deletions, a clear case of strain-specific horizontal gene transfer, and identification of 0.8 Mb of novel sequence. Single nucleotide polymorphisms per kilobase (SNPs/kb) between the two strains were found to be exceptionally high (average: 7.8, maximum: 160 SNPs/kb). High variation within the species was confirmed with exo-metabolite profiling and phylogenetics. Detailed lists of alleles were generated, and genotypic differences were observed to accumulate in metabolic pathways essential to acid production and protein synthesis. A transcriptome analysis supported up-regulation of genes associated with biosynthesis of amino acids that are abundant in glucoamylase A, tRNA-synthases, and protein transporters in the protein producing CBS 513.88 strain. Our results and data sets from this integrative systems biology analysis resulted in a snapshot of fungal evolution and will support further optimization of cell factories based on filamentous fungi.
The saprotrophic filamentous fungus Aspergillus niger is found globally and exhibits a great diversity in its phenotype. A. niger has become one of the major workhorses in industrial biotechnology, being very efficient in producing both polysaccharide-degrading enzymes (particularly amylases, pectinases, and xylanases) or organic acids (mainly citric acid) in high amounts. It also has a long history of safe use (Schuster et al. 2002; van Dijck et al. 2003; van Dijck 2008). Commercial importance is illustrated by a world market for industrial enzymes of nearly US$ 5 billion in 2009, of which filamentous fungi account for roughly half of the production (Lubertozzi and Keasling 2008), and a global citric acid production of 9 × 106 metric tons in 2000 (Karaffa and Kubicek 2003).
In 2007, the genome sequence of A. niger strain CBS 513.88, used for industrial enzyme production, was published (Pel et al. 2007). This strain was derived from A. niger NRRL 3122, a strain developed for glucoamylase A production by classical mutagenesis and screening methods (van Lanen and Smith 1968). This work initiated a number of new genome-based investigations (Sun et al. 2007; Andersen et al. 2008a,b; Martens-Uzunova and Schaap 2008; Yuan et al. 2008a,b) but did not reveal differences between citric-acid-producing and enzyme-producing A. niger strains (Cullen 2007).
In this study, we present the nearly complete genome sequence of the citric-acid-producing A. niger wild-type strain ATCC 1015 and compare it to the genome sequence of the enzyme-producing strain CBS 513.88. The genetic diversity of these two A. niger strains was determined by applying systems biology tools as well as new bioinformatics methods to examine multi-level differences that distinguish the wild-type citric-acid-producing strain from the mutagenized glucoamylase A–producing strain.
Results
General genome statistics
The 34.85-Mb genome sequence of A. niger ATCC 1015 was generated using a shotgun approach and then further improved to a high-quality assembly of 24 finished contigs separated by 15 gaps (including eight from centromeric regions). Genome statistics are summarized in Table 1, with details in Supplemental Text 1. The full sequence and annotations are available from the Joint Genome Institute (JGI) Genome Portal (http://genome.jgi-psf.org/Aspni5) and from NCBI (accession number ACJE00000000).
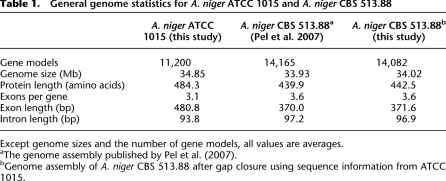
Table 1.
General genome statistics for A. niger ATCC 1015 and A. niger CBS 513.88
Except genome sizes and the number of gene models, all values are averages.
aThe genome assembly published by Pel et al. (2007).
bGenome assembly of A. niger CBS 513.88 after gap closure using sequence information from ATCC 1015.
The genome sequence of A. niger CBS 513.88 (Pel et al. 2007) was improved using the ATCC 1015 sequence to close 186 contig gaps and modify the gene models associated with these gaps (Table 1; Supplemental Table 1). The updated A. niger CBS 513.88 genome sequence is accessible through EMBL (accession numbers AM269948–AM270415).
We note a large difference (2882) in the number of called genes in the two strains (Table 1). A thorough analysis indicates an overprediction of genes in CBS 513.88 and an underprediction of genes in ATCC 1015, and 396/510 unique genes in CBS 513.88/ATCC 1015 (Supplemental Text 2; Supplemental Fig. 1; for details, see Supplemental Tables 2–9). For further validation of the absence/presence of individual proteins, we have performed gDNA hybridizations, which can be consulted for reference (Supplemental Table 16).
Unique genes in both strains suggest horizontal gene transfer to be a cause for the amylase hyper-producer phenotype
For the genes unique to the amylase-producing strain CBS 513.88 (Supplemental Table 6), the most notable genes are two alpha-amylases that are identical to the Aspergillus oryzae alpha-amylase, a possible cause for the amylase hyper-producer phenotype. We discuss this in detail below (Fig. 2). Furthermore, we find three possible polyketide synthases, which suggests a unique secondary metabolite profile for this strain.
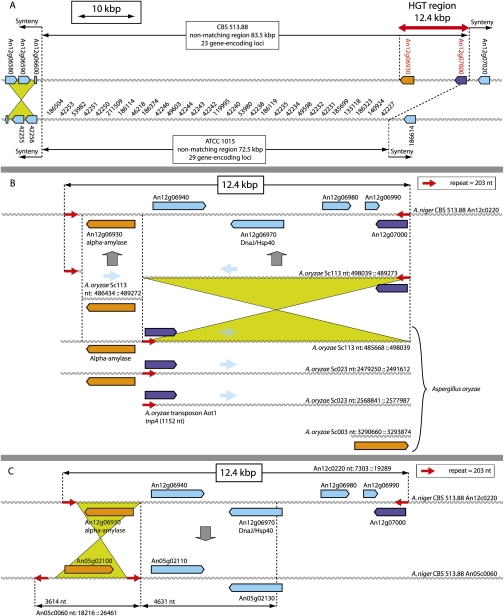
Figure 2.
Horizontal gene transfer of alpha-amylase genes from A. oryzae to A. niger CBS 513.88. (A) Unmatched region identified for the left arm of chromosome III spanning 65 kb for ATCC 1015 encoding 30 predicted genes and 85 kb for CBS 513.88 encoding 24 predicted genes. The unmatched region is flanked on one side by a small local inversion of three predicted genes (in green). (B) The 12.4-kb HGT region is part of an identical 12.7-kb region present in A. oryzae RIB40 supercontigs SC113 and SC023. In A. niger CBS 513.88, the transferred region is enclosed by a 203-bp inverted repeat (red arrow). An12g07000 (blue) is a putative transposase and identical to the A. oryzae transposon Aot1 tnpA gene. Similarly, the A. niger CBS 513.88 alpha-amylase encoding gene An12g06930 (orange) is identical to A. oryzae annotated genes AO090120000196 (SC113) and AO090023000944 (SC023). (C) Proposed duplication–recombination event between supercontig 12 and supercontig 05 of the 12-kb HGT region. Breakpoints are indicated with dotted lines. The fragment encoding alpha-amylase An05g02200 is identical to An12g06930 (orange). The breakpoints are flanked with additional copies of the 203-bp repeat region (red arrow). The region encoding genes An12g06940 to An12g06970 is identical to the region encoding An05g02210 to An05g02130. The downstream breakpoint occurred in the predicted gene coding region of An12g06970, thereby deleting the original start codon and upstream region.
Examining the genes found only in ATCC 1015 (Supplemental Table 6) does not point to any obvious cause for citric acid hyper-production, but four possible polyketide synthases and a putative NRPS are found to be unique for ATCC 1015, suggesting this strain has unique secondary metabolites as well. Multiple effects of this type are, indeed, seen for both strains in analyses below (Fig. 1; Supplemental Fig. 10; Table 2).
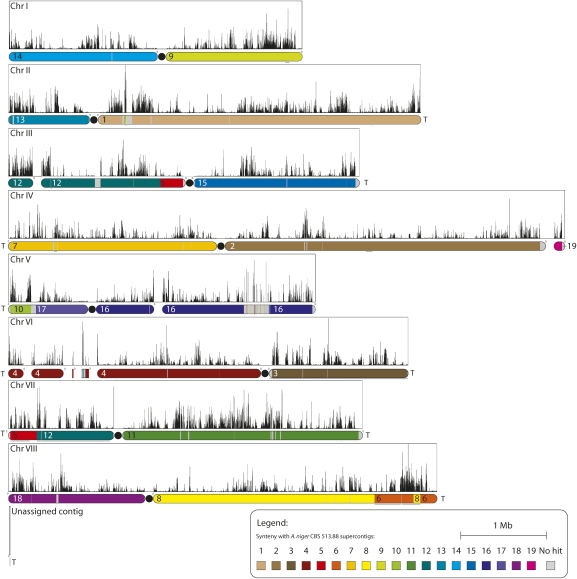
Figure 1.
Synteny map of the contigs of A. niger ATCC 1015 to the supercontigs of A. niger CBS 513.88. The coloring of the chromosomes shows syntenic regions in A. niger CBS 513.88. Arabic numerals show the number of the supercontig in A. niger CBS 513.88. Gray areas show regions not found in the CBS 513.88 genome sequence (Pel et al. 2007). (Filled black circles) Proposed locations of centromeric regions. Sequenced telomeres are marked with a T. Zeroes mark the first base of the contigs. A black line underneath a section of the chromosomes denotes inverted sequence. Black histograms show SNPs per kilobase (number of single nucleotide polymorphisms/kilobase) between the sequences of the two strains (y-axis: 0–160 SNPs/kb). Gaps between contigs and centromeres are not to scale. The alignment demonstrates almost complete synteny between the two strains, with the exception of a cross-over event between the left arms of chromosomes III and VII. An overview of the genes found in the gaps unique to ATCC 1015 can be found in Supplemental Table 2.
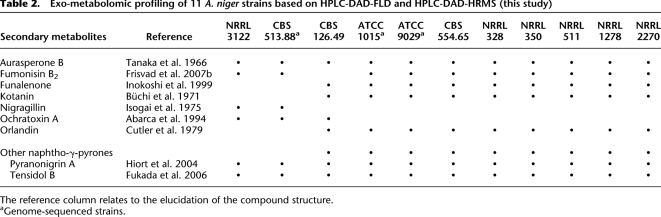
Table 2.
Exo-metabolomic profiling of 11 A. niger strains based on HPLC-DAD-FLD and HPLC-DAD-HRMS (this study)
The reference column relates to the elucidation of the compound structure.
aGenome-sequenced strains.
Synteny mapping shows 0.5 Mb of chromosomal rearrangements and a whole-arm inversion
The genomes of the two strains are largely syntenic (Fig. 1). For example, the 1429 protein-encoding genes of CBS 513.88 supercontig An01 that have predicted counterparts in ATCC 1015 are, with one exception, all mapped to chromosome 2b. A similar example is seen for all but one of 1486 protein-encoding genes on supercontig An02. Both exceptions encode putative Tan1-like transposases. Notwithstanding, a number of significant differences in genome configuration exist (Fig. 1). More than 0.5 Mb of additional genome sequence in ATCC 1015 resides in four large regions on chromosomes (Chr) II, III, and V (Fig. 1). The flanking sequences of three of these additional elements in ATCC 1015 are syntenic to continuous sequences in CBS 513.88 and are thus true differences in genome configuration (Supplemental Fig. 2; Supplemental Table 2; for details on ChrIII, see next section). The fourth region on the left arm of ChrV in ATCC 1015 could not be verified due to a contig gap for CBS 513.88. The contig gap in the right arm of ChrV in the genome sequence of ATCC 1015 is spanned by continuous sequence in CBS 513.88, containing 15 predicted genes. An overview of the genes found in the gaps unique to ATCC 1015 can be found in Supplemental Table 2.
Other striking differences include a large inversion in ChrVIII and the inversion and translocation of a large fragment between the left arms of ChrIII and ChrVII (Supplemental Fig. 3). Both were confirmed with PCR spanning the breakpoints (data not shown).
Finally, the presence of telomere sequences in genome data confirms an inversion of the complete right arm of ChrVI.
Two extra alpha-amylase-encoding genes likely obtained through HGT
An unmatched region identified for the left arm of ChrIII (Fig. 1) spans 72.5 kb of unique sequence in ATCC 1015. Remarkably, for CBS 513.88, a unique 85.3-kb sequence is found at this location (Fig. 2A). Gene annotation may be found in Supplemental Table 10. To confirm the actual absence of the region in the chromosome of ATCC 1015, we performed gDNA hybridizations, which did indeed support this finding (Supplemental Fig. 4).
Approximately 12 kb of this region shares >99.8% sequence identity at DNA level with genomic DNA from A. oryzae RIB40 (Fig. 2B). The complete 85-kb fragment including the 12-kb alpha-amylase region also appears to be involved in a recent duplication recombination event to ChrVII, (Fig. 2C). Thus, the CBS 513.88 genome harbors two additional alpha-amylase-encoding genes that are orthologs to the alpha-amylase-encoding genes (AO090023000944, AO090120000196) of A. oryzae RIB40. These findings suggest that CBS 513.88 and the parental strain NRRL 3122 (data not shown) acquired these duplicate alpha-amylase genes (An12g06930, An05g02100) through horizontal gene transfer (HGT). The occurrence of HGT in Aspergilli would further be supported by the presence of alpha-amylase-encoding genes in other black Aspergilli that display >99% nucleotide identity to those of A. oryzae RIB40 and the A. niger CBS 513.88 alpha-amylases (Korman et al. 1990; Shibuya et al. 1992). The origin of the remaining part of the unmatched region remains unclear. No significant similarity was observed at the DNA level, and only a few of the encoded proteins show similarity with other proteins present in the NR protein database.
There is also evidence that this HGT may have occurred by the action of transposases. The 12-kb HGT region is flanked by 202-bp inverted, perfect, terminal repeats (ITR) (Fig. 2A,B). Furthermore, this HGT region harbors another gene, An12g07000, that is identical to the A. oryzae tnpA transposase gene. Interestingly, in the A. oryzae RIB40 genome, the same 12-kb fragment has been duplicated twice and is present on multiple chromosomes, but only one perfect copy of the ITRs is retained (Fig. 2C).
Transposon presence is strain-specific
Transposon-like sequences were identified in both genomes and quantified (Supplemental Table 11). This comparison pinpoints a remarkable difference in the presence and amount of transposon-related sequences in ATCC 1015 and CBS 513.88 both for class I and for class II transposons: Only 16 sequences were identified in ATCC 1015, but 55 were detected in CBS 513.88. Whereas the ATCC 1015 genome contains no class I superfamily copia and a single copy of class II superfamily Fot1/pogo, these sequences are much more abundant in CBS 513.88, where 15 class I copia, and 13 class II Fot1/pogo are found.
SNP analysis reveals high mutation rates and hypervariable regions
We found 8 ± 16 SNPs/kb (average ± standard deviation) and a maximum of 163 SNPs/kb single-nucleotide polymorphisms (SNPs) between A. niger strains ATCC 1015 and CBS 513.88 (Supplemental Table 12). This value is much higher than the maximum of 9 SNPs/kb found by Cuomo et al. (2007) in a SNP analysis of two Fusarium graminearum strains. A comparison of the A. niger ATCC 1015 genome sequence to that of A. niger ATCC 9029 revealed markedly less variation between the two (2 ± 5 SNPs/kb) (Supplemental Fig. 5), indicating a large genomic variation in the A. niger group but little between ATCC 1015 and 9029. These polymorphisms are not uniformly distributed but cluster in hypervariable regions (Fig. 1). This is supported by a gDNA hybridization study, which confirms the presence of distinct hypervariable regions (Supplemental Fig. 4).
Gene comparisons reveal industrially relevant strain differences
To identify strain-defining systemic effects of the genome variation, results from the Imprint gene synteny analysis (see Supplemental Table 5; Methods) were used for additional genome-scale investigation.
GO term over-representation analysis was performed on gene groups with distinct common properties (for Group overview, see Supplemental Table 5; for details, see Supplemental Table 13). Most interesting is the observation that 37 proteins involved in transcriptional regulation are nonfunctional in CBS 513.88 due to frameshifts or stop codons. This suggests a less stringent regulation of CBS 513.88 relative to ATCC 1015. A comparative shake-flask study of the two strains also suggests mutations in regulatory elements as nitrogen source utilization is impaired in the CBS 513.88 strain (data not shown). While the nitrogen catabolism regulator AreA was originally thought to be mutated in CBS 513.88 (Pel et al. 2007), a resequencing of areA showed the ORFs to be identical.
To detect potential differences in the metabolism of the two strains, we compared all genes encoding proteins that display differences at the amino acid level between the two strains on the metabolic network of A. niger (Andersen et al. (2008a) and mapped them to metabolic pathways (Supplemental Fig. 6). Mutations were found in the pathways for biosynthesis of proline, aspartate, asparagine, tryptophan, and histidine, which may be relevant to protein production. Also, mutations were found in the plasma membrane-bound ATPase, in the enzymes of the GABA shunt, of the TCA cycle, and in components of all steps of the electron transport chain, which could be relevant for the production of citric acid.
Phylogenetic analysis confirms high variability between genome sequences
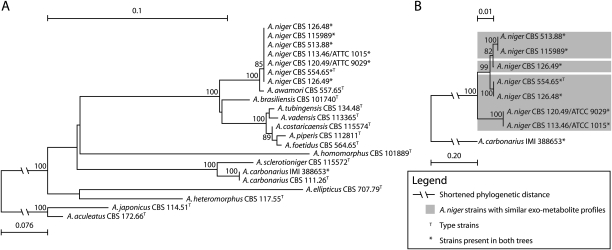
To place the comparison of A. niger ATCC 1015 and CBS 513.88 into the larger context of the Aspergillus section Nigri, variation in the two strains was analyzed by phylogenetic analysis of a part of the beta-tubulin sequence for a number of A. niger strains and other black Aspergilli (Fig. 3A). To further explore the genome variability across the A. niger species, we identified four 1-kb regions from ChrII, ChrIV, ChrVI, and ChrVIII that were identical in the ATCC 1015 and ATCC 9029 strains but had ∼20 SNPs/kb relative to the genome sequence of CBS 513.88. These regions were PCR-sequenced in seven A. niger strains including the CBS 513.88 progenitor, NRRL 3122, and the A. niger neotype strain, CBS 554.65 (Kozakiewicz et al. 1992) to form a phylogenetic tree with high variation (Fig. 3B). For previously sequenced strains, the resequencing results were identical to the previously determined genomic sequence, thereby excluding that the genomic variation is an artifact of low-fidelity sequencing.
Figure 3.
(A) Phylogenetic relationship of several black Aspergilli based on partial sequencing of beta-tubulin. The tree was rooted to Aspergillus aculeatus CBS 172.66. (B) Phylogenetic relationship of seven strains of A. niger based on sequencing of 1-kb variable regions from four chromosomes. The tree was rooted to the sequence obtained from Aspergillus carbonarius IMI 388653. Clades based on the exo-metabolomic groupings of Table 2 are shown. For both trees, bootstrap values above 80% of the 1000 performed reiterations are shown.
All A. niger strains have an identical sequence for beta-tubulin and form a strongly supported terminal clade together with Aspergillus awamori (Fig. 3A). The other taxonomically different species are phylogenetically distinct from this group, which is in accordance with the serial classification of Frisvad et al. (2007a). The latter clustering based on the selected regions (Fig. 3B) was able to separate the A. niger strains into three groups. Strain CBS 513.88 and its progenitor NRRL 3122 were identical for the regions, indicating limited SNP introduction due to classical strain improvement, and thus confirms high variation in the A. niger group.
Exo-metabolomic profiling describes three distinct groups of A. niger
To further compare the two sequenced strains on a system-wide, but nongenomic level, to each other and to other members of the A. niger species group, exo-metabolomic profiles of the two strains were compared to nine other strains, including the CBS 513.88 progenitor strain, NRRL 3122; the widely used laboratory strain ATCC 9029; and the A. niger neotype, CBS 554.65 (Table 2). This gave three distinct clades, which follow the clustering of Figure 3B, but does not conform to the phylogenetic distances calculated. Strain CBS 513.88 and its progenitor strain had very similar secondary metabolite profiles. These two strains differed from the other strains analyzed. Strain ATCC 1015 had a profile similar to seven other strains, although there were some quantitative differences. Strain CBS 126.49 had a unique profile.
In the case of ochratoxin A, it is interesting that in ATCC 1015 and ATCC 9029, a remnant of the PKS gene (An15g07920) of the putative ochratoxin cluster in A. niger CBS 513.88 (Pel et al. 2007) was identified, which could be related to a 21-kb deletion in both strains (Supplemental Fig. 7). We have noted that a putative NRPS is found in one of the unique chromosome regions found in ATCC 1015 (Fig. 1), which may account for some of the difference (Supplemental Table 2).
Transcriptome analysis of A. niger ATCC 1015 and CBS 513.88 growing on glucose
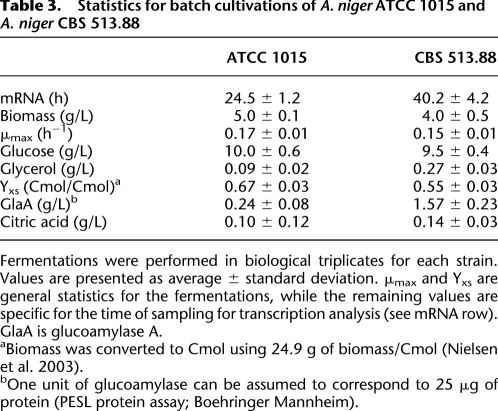
To evaluate the effect of the differences in genome sequence on the physiology of the two strains, batch cultivations in bioreactors and a comparative transcriptome analysis were performed. Strains ATCC 1015 and CBS 513.88 were grown under the same conditions in batch cultures in a glucose-based minimal medium. The GlaA-producing strain CBS 513.88 produced >1.2 g/L GlaA more than ATCC 1015, while producing ∼1 g/L biomass less. Other measured characteristics were similar for the two strains (Table 3). Statistical analysis showed 4784 significantly (adj. p < 0.05) differentially expressed genes, with an almost equal number of genes up-regulated in either strain (2431 in CBS 513.88 vs. 2353 for ATCC 1015).
Table 3.
Statistics for batch cultivations of A. niger ATCC 1015 and A. niger CBS 513.88
Fermentations were performed in biological triplicates for each strain. Values are presented as average ± standard deviation. μmax and Yxs are general statistics for the fermentations, while the remaining values are specific for the time of sampling for transcription analysis (see mRNA row). GlaA is glucoamylase A.
aBiomass was converted to Cmol using 24.9 g of biomass/Cmol (Nielsen et al. 2003).
bOne unit of glucoamylase can be assumed to correspond to 25 μg of protein (PESL protein assay; Boehringer Mannheim).
Examining the differential expression in the context of metabolic pathways (Supplemental Fig. 8), only the alternative oxidative pathway has uniformly higher expression in ATCC 1015, while a substantial subset of metabolism was up-regulated in CBS 513.88, including glycolysis and the TCA cycle. As the specific growth rates of the strains are similar, we suggest that this extra activity of central metabolism provides precursors for the higher productivity of glucoamylase. We also find increased expression of genes involved in amino acid metabolism, especially the entire biosynthetic pathways of threonine, serine, and tryptophan. Intriguingly, analysis of the amino acid composition of the glucoamylase A protein (Supplemental Fig. 9) revealed that GlaA is atypical in that it has a higher content of specifically tryptophan, threonine, and serine than 90% of the predicted genes of CBS 513.88. For all three amino acids, the content is almost twice as high as the average in the composition of total protein (Christias et al. 1975).
We initially thought that this was due to a frameshift mutation identified in the gene for the general amino acid–regulating transcription factor (cross-pathway control protein) CpcA (Supplemental Table 5), but resequencing of this gene showed this frameshift to be a sequencing error. The presence of two sense mutations was confirmed and is currently being investigated.
To further support that the increased production of GlaA is significant enough to affect amino acid biosynthesis, we used a genome-scale metabolic model of A. niger (Andersen et al. 2008a) to model the two strains under the growth conditions used (Table 3). The computed fluxes (Supplemental Table 14) show that for all three amino acids, the fluxes through the biosynthetic pathways must be at least twice as high in CBS 513.88 compared to ATCC 1015 to support the increased GlaA production, thus corresponding well with the transcriptome results.
For a broader analysis of trends revealed by the transcriptome profiles, a GO term over-representation analysis was conducted on the genes significantly up-regulated in either strain (Supplemental Table 15). The analysis confirmed that traits relevant for protein production—such as amino acid biosynthesis and tRNA aminoacylation—appear to be highly significant for CBS 513.88. For ATCC 1015, GO biological functions for electron transport (adj. p = 2.7 × 10−5), carbohydrate transport (adj. p = 6.8 × 10−3), and organic acid transport (adj. p = 0.035) were significant.
An examination of expression of individual genes showed that glaA had significantly increased expression in CBS 513.88 (adj. p = 84 × 10−6), but the fold change (3.2) was lower than the increase in enzyme activity (sixfold) (Table 3). Interestingly, all identified tRNA-aminoacyl synthases were found to be twofold to sixfold up-regulated in CBS 513.88.
Mapping of gene expression to the genome identifies secondary metabolite cluster activities and underpins the whole-arm inversion in chromosome VI
To examine differences in gene expression between the two strains relative to chromosome positions, the log2-ratios of the gene expression indices from the transcriptome analysis were mapped to the synteny diagrams of Figure 1 (Supplemental Fig. 10).
The analysis of this mapping identified a number of features. First, six regions not found in CBS 513.88 contain genes with uniformly higher expression in ATCC 1015, supporting the absence of these regions in CBS 513.88. Second, two active secondary metabolite clusters were identified on ChrVIII (contains a polyketide synthase [ID: 211885] that is unique for ATCC 1015) and ChrI (NRPS cluster, found in both strains [NRPS ID: 43555/An09g00520], but appears only to be active in CBS 513.88). Third, the inversion of the entire right arm of ChrVI (Fig. 1) is further supported. The telomeric position effect, which defines decreased expression in the vicinity of telomeres, has been described for Saccharomyces cerevisiae (Gottschling et al. 1990). Thus, if an arm has been inverted, reduced expression should be found at opposite ends in ATCC 1015 and CBS 513.88. This is indeed reflected in the log2-ratio on the right arm of ChrVI (Supplemental Fig. 10).
Discussion
In this study, we provide and compare the genomes of two strains of A. niger. These two strains have different phenotypes: one, the predecessor to efficient enzyme-producing strains having undergone some level of mutagenesis and selection, and the other a wild-type parent strain of high citric-acid-producing strains. This makes the comparison interesting both in terms of genomic research and industrial applications. We have supported the conclusions of our comparison with further experiments, allowing us to propose new hypotheses and conclusions within three main areas: (1) genetic diversity of the A. niger group, (2) horizontal gene transfer in fungi, and (3) fungal biotechnology (discussed separately below).
The diversity of the two strains was explored through a multi-level comparison (DNA, chromosome, gene, and protein) between both genome architectures and was supported by multiple types of experimental work. Interestingly, we observe a remarkable similarity at the level of chromosome, gene order, and gene identity, but also notable diversity was detected and further explored, namely, distinct regions with high levels of SNPs, a 0.8-Mb difference in genome size, several large insertion/deletions of up to 200 kb, different transposon populations, a major translocation/inversion, the inversion of an entire chromosomal arm, and a large set of unique genes in both species (see points 1–5 below):
About 400–500 unique genes were identified for each strain, most of which are evenly distributed over the chromosomes. This indicates strain evolution by a high frequency of loss and/or uptake of gene fragments. Interestingly, the opposite is seen in two strains of the pathogenic Aspergillus fumigatus, where 80% of the strain-specific genes (143 and 218, respectively) are found in a few, large isolate-specific genomic islands (Fedorova et al. 2008). Thus, this intra-species increased frequency of transfer/loss of genetic elements so far seems to be specific for the A. niger group. More genome sequences may prove it to be present in other Aspergilli as well.
The whole-arm inversion on ChrVI is a remarkable event. Over the characterized Aspergilli sequenced genomes, most described inversions were isolated as mutants, with a breakpoint in a gene, leading to a phenotype. Pel et al. (2007) reported high synteny between the centromeric parts of the arms of A. niger CBS 513.88 and Aspergillus nidulans FGSC A4. Even so, in this case, it is supported by the genome sequence, the presence of telomeric sequences, and a detectable telomeric positioning effect. From Figure 2 in Galagan et al. 2005, it is also clear that centromeric inversions must have occurred in the genealogy of the common ancestor of A. nidulans, A. oryzae, and A. fumigatus.
The large translocation/inversion event of ChrIII and ChrVII explains the discrepancies in chromosome size reported by Pel et al. (2007) between strain N400 (Verdoes et al. 1994) and calculated sizes of ChrIII and ChrVII for CBS 513.88. This fact and that the breakpoints are within two otherwise intact genes indicate that the event occurred after the branching of strains N400 and NRRL 3122, possibly in the mutagenesis of the predecessor of CBS 513.88.
The presence and functionality of transposons are qualitatively and quantitatively less complex in the genome of ATCC 1015 than in CBS 513.88 (Supplemental Table 11). Such a different distribution of transposons between strains has been observed before (Braumann et al. 2008) and was described to be inducible by mutagenesis and other stress-induced conditions (Strand and McDonald 1985). However, the number and scale of differences make a recent mutagenesis program unlikely to be the basis for the multiple HGT events. Furthermore, the detection of repeat sequences in a number of DNA regions was accompanied by strain-specific differences. Therefore, we suggest that the transposon populations have played a significant role in strain evolution.
Finally, the phylogenetic analysis (Fig. 3B) confirms the high genetic variation described for two strains to be general within the A. niger group and not the result of either CBS 513.88 or ATCC 1015 being atypical. This is again supported by the exo-metabolomic profiles of the 11 examined A. niger strains (Table 2). While the grouping of the exo-metabolites does not accurately reflect the cladograms in Figure 3, we see this as confirmation of the high genetic variation in the A. niger group, as the production of a given exo-metabolite may be changed by a single genomic event. Thus, we believe that the diversity of the two genome-sequenced strains is common for the A. niger group, which seems to have highly dynamic genomes.
The presence/absence of HGT in fungi has caused a lot of discussion (Galagan et al. 2005; Keeling and Palmer 2008; Khaldi and Wolfe 2008; Khaldi et al. 2008). In this study, we have shown HGT to be a most likely origin of two alpha-amylase genes in CBS 513.88 that might have been transferred to A. niger from another species, possibly A. oryzae. The HGT must have occurred before the separation of CBS 513.88 and ATCC 1015. The amylase genes are not found in the ATCC 1015 strain (also supported by gDNA hybridization), but they are in CBS 513.88 flanked by a transposon, which is found both in A. oryzae RIB40 and in a truncated form in ATCC 1015. With the evidence of HGT into Aspergillus clavatus from a Magnaporthe-related donor, presented by Khaldi et al. (2008), HGT is now identified in two separate cases with distantly related species and may thus be seen as a general phenomenon in filamentous fungi.
The initial reason for comparing the two strains has been to gain insight in the genetic basis for the two industrially relevant phenotypes. In citric-acid-producing ATCC 1015, we are intrigued to see higher expression of elements from the electron transport, carbohydrate transport, and organic acid transport. These factors are known to be involved in obtaining high citric acid yields. Higher expression levels even though citric acid production is not notably higher (see discussion below) suggests that these traits may have been contributing factors to the original selection of ATCC 1015 for citric acid production. For the GlaA-producing strain (CBS 513.88), we observe systemic changes: Mutations in regulatory genes (e.g., cpcA), higher expression of glaA itself, and up-regulation of all identified tRNA-synthases may all contribute to a more efficient enzyme producer. We also present the hypothesis that increased production of the amino acids serine, threonine, and tryptophan may be required for efficient GlaA production, as these are over-represented in GlaA.
While we have identified multiple possible contributors to the efficient amylase production of CBS 513.88, we have only seen a few factors associated with citric acid production. We propose multiple reasons for this: (1) CBS 513.88 was selected for increased GlaA production, while ATCC 1015 is a wild-type strain with only modest citric acid production (compared to current yields). (2) The medium used for the transcriptome analysis is not optimized for citric acid production, as this would not give the dispersed growth necessary for representative sampling (Karaffa and Kubicek 2003). (3) Protein synthesis is a complex process, with more factors involved than in the production and secretion of a simple organic acid. Since more than 6000 genes differ at the amino acid level between the two strains, it would be unlikely not to find many differences involved in protein production. For this reason, we have put our emphasis on genes found in multiple types of analyses, effects in entire pathways, and traits (GO) that are found to be statistically over-represented, which allows for robust conclusions.
In conclusion, our results establish a firm comparative genomics foundation on which to build and test hypotheses regarding enzyme production, organic acid production, and diversity within a “species group.”
Methods
ATCC 1015 genome assembly
The sequence reads were derived from four whole-genome shotgun (WGS) libraries: one with an insert size of 2–3 kb, two with an insert size of 6–8 kb, and one with an insert size of 35–40 kb. The reads were screened for vector using cross_match and then trimmed for vector sequence and quality (J Chapman, N Putnam, I Ho, and D Rokhsar, unpubl.). Reads shorter than 100 bases after trimming were excluded. The final data set included: 28,551 of 2–3-kb reads, containing 21.5 Mb of sequence; 160,479 of 6–8-kb reads, containing 123 Mb of sequence; and 38,651 of 35–40-kb reads, containing 23.8 Mb of sequence.
The data were assembled using release 2.7 of Jazz, a WGS assembler developed at the JGI (Aparicio et al. 2002; J Chapman, N Putnam, I Ho, and D Rokhsar, unpubl.). The genome size and sequence depth were initially estimated to be 36 Mb and 8.0, respectively. After removal of short (<1 kb) and redundant scaffolds (<5 kb with 80% or more of the length matching a scaffold >5 kb), the assembly included 43.7 Mb of scaffold sequence with 8.2 Mb (18.9%) of gaps in 350 scaffolds, with half of the scaffold sequence contained in the eight largest scaffolds of 1.81 Mb or longer. The sequence depth derived from the assembly was 7.88 ± 0.05. To estimate the completeness of the assembly, a set of 50,001 ESTs was BLAT-aligned to the unassembled trimmed reads, as well as the assembly itself. Forty-three thousand three hundred and twenty-three ESTs (86.6%) were >80% covered by the unassembled data; 43,978 (88.0%) were >50% covered; and 44,206 (88.4%) were >20% covered. By way of comparison, 48,798 ESTs (97.6%) showed hits to the assembly. Orientation of the chromosome arms was based on eight available telomere sequences, the supercontig orientation proposed by Pel et al. (2007), and research on linkage groups in A. niger (Bos et al. 1989; Debets et al. 1989, 1990a,b; Swart et al. 1992; Verdoes et al. 1994).
ATCC 1015 genome finishing
To perform genome improvement on A. niger ATCC 1015, initial read layouts from the whole-genome shotgun assembly were converted into the phred/phrap/consed pipeline (Gordon et al. 1998). Following manual inspection of the assembled sequences, finishing was performed by resequencing plasmid subclones and by walking on plasmid subclones or fosmids using custom primers. All finishing reactions were performed with 4:1 BigDye to dGTP BigDye terminator chemistry (Applied Biosystems). Repeats in the sequence were resolved by transposon-hopping 8-kb plasmid clones. Fosmid clones were shotgun sequenced and finished to fill large gaps, resolve large repeats, or to resolve chromosome duplications and extend into chromosome telomere regions.
The resulting 24 finished scaffolds were orientated where possible into chromosome structures using comparisons to Pel et al. (2007) and telomere positions. The improved genome consists of 34,853,277 bp with an estimated error rate of less than 1 error in 100,000 bp.
ATCC 1015 automatic annotation
Gene models in the genome of A. niger were predicted using Fgenesh (Salamov and Solovyev 2000), Fgenesh+ (Salamov and Solovyev 2000), and Genewise (Birney and Durbin 2000) integrated into the JGI annotation pipeline. Fgenesh was trained on a set of more than 2000 putative full-length transcripts derived from clustered A. niger ESTs and reliable homology-based gene models to show 81% sensitivity and 81% specificity of predictions on a test set. Homology-based gene predictors were seeded with BLASTX alignments of proteins from the NCBI nonredundant set of proteins. Thirty-one thousand five hundred and seventy-eight A. niger ESTs were sequenced using Sanger technology and were either directly mapped to genomic sequence when the ESTs included putative full-length (FL) genes or used to extend predicted gene models into FL genes by adding 5′ and/or 3′ UTRs. In addition, 386,515 ESTs with an average length of 104 nt were sequenced with 454 Life Sciences (Roche) GS20 sequencers and used in validation of predicted gene models. Since multiple gene models were generated for each locus, a single representative model was chosen based on homology and EST support and used for further analysis.
All predicted gene models were functionally annotated by sequence similarity to annotated genes from the NCBI nonredundant set and other specialized databases (such as KEGG) (Kanehisa et al. 2002, 2004) using BLAST and hardware accelerated double-affine Smith-Waterman alignments (http://www.timelogic.com). Functional and structural domains were predicted in protein sequences using the InterPro software (Zdobnov and Apweiler 2001). All genes were also annotated according to Gene Ontology (Ashburner et al. 2000; Harris et al. 2004), eukaryotic orthologous groups (KOGs) (Koonin et al. 2004), and KEGG metabolic pathways (Kanehisa et al. 2004).
ATCC 1015 sequence availability
A. niger assemblies, annotations, and analyzes are available through the interactive JGI Genome Portal at http://genome.jgi-psf.org/Aspni5/Aspni51.home.html. Genome assemblies together with predicted gene models and annotations were also deposited at NCBI under the project accession number ACJE00000000.
Strains
The following A. niger strains were used for experiments (deposition numbers in different collections are given as well): CBS 513.88 = FGSC A1513 = IBT 29270, ATCC 1015 = IBT 28639 = NCTC 3858a = NRRL 1278 = NRRL 350 = NRRL 511 = NRRL 328 = Thom 167 = CBS 113.46 = ATCC 10582 = IMI 031821 = LSBH Ac4 = Thom 3528.7, NRRL 3 = IBT 28539 = MUCL 30480 = DSM 2466 = CECT 2088 = VTT D-85240 = NRRL 566 = WB3 = ATCC 9029 = N400 = CBS 120.49 = IMI 041876, NRRL 326 = IBT 27876 = WB 326 = CBS 554.65 = ATCC 16888 = IHEM 3415 = IMI 050566 = Thom 2766 = JCM 10254 = IFO 33023 (ex tannin-gallic acid fermentation; A. niger neotype) (Kozakiewicz et al. 1992) NRRL 328 = IBT 27878 = NRRL 350 = IBT 27877 = CBS 113.46, NRRL 337 = CBS 126.48 = ATCC 10254 = DSM 734 = IFO 6428 = IMI 015954 = WB 337, NRRL 363 = IBT 3617 = IBT 5764 = CBS 126.49 = ATCC 10698 = IFO 6648, NRRL 511 = IBT 27875, NRRL 1278 = IBT 27872, NRRL 2270 = IBT 26391 = ATCC 11414 = VTT D-77050 = IMI 075353 = A60 = S.M. martin A-1-233 = Wisconsin 72-4, derived from ATCC 1015, and NRRL 3122 = IBT 23538 = ATCC 22343 = CBS 115989 (this strain is a wild-type progenitor of CBS 513.88). Strain histories for ATCC 1015, ATCC 9029, and NRRL 3122 are reviewed by Baker (2006).
Exo-metabolite profiling
A. niger strains were inoculated on Czapek yeast autolysate (CYA), yeast extract sucrose (YES) agar, and CYA with 5% NaCl agar (CYAS agar). For medium composition, see Frisvad and Samson (2004). All strains were three-point inoculated on these media and incubated at 25°C in darkness for a week, after which five plugs (6 mm diameter) along a diameter of a fungal colony were cut out and extracted (Smedsgaard 1997). The extracts were analyzed by HPLC-DAD fluorescence (Frisvad and Thrane 1987; Smedsgaard 1997) and by HPLC-HR-MS (Nielsen and Smedsgaard 2003; Frisvad et al. 2007b). The secondary metabolites were identified by comparison with authentic standards (fumonisin B2, ochratoxin A, nigragillin, kotanin, and orlandin) and by UV spectra and high-resolution mass spectra using electrospray ionization.
CBS 513.88 gap closure
The near-complete ATCC 1015 genome sequence was used to verify contig order and orientation and to estimate gap sequence length between contigs in CBS 513.88. Gap-flanking PCR primers (20-mers) giving rise to PCR products with a minimal overlap of 100 bp were automatically designed with primer-3 (Rozen and Skaletsky 2000) using custom scripts. PCR products of expected size were purified using a QIAquick PCR Purification Kit (QIAGEN) and sequenced by Baseclear. The updated sequence for A. niger CBS 513.88 is available from EMBL (http://www.ebi.ac.uk/embl/) under accession nos. AM269948–AM270415.
Sequencing of variable regions
Regions were amplified using standard PCR techniques with the four following primer pairs (the name describes chromosome number and direction): (1) Chr02-Fwd: 5′-GGACACTGCTTGATGTGATG-3′, Chr02-Rev: 5′-GAGAGACGTACGAAAGGTTG-3′; (2) Chr04-Fwd: 5′-CGATCTGCGACCAGGA-3′, Chr04-Rev: 5′-CATAACGGATTCGTCGCTG-3′; (3) Chr06-Fwd: 5′-CTTGAAGGCGTTGAGGTC-3′, Chr06-Rev: 5′-GCGAGTATGTGGCTAACATC-3′; (4) Chr08-Fwd: 5′-GGTATGTCACATTCCRTCCA-3′, Chr08-Rev: 5′-GCTTGCAGTGAGCAAGGA-3′. The sequences on ChrII, ChrVI, and ChrVIII overlap with predicted genes. The PCR products were purified using a QIAquick PCR Purification Kit (QIAGEN) and sequenced by Agencourt Bioscience Corporation (GenBank accession number GU296708–GU296739).
Sequencing of beta-tubulin
Amplification of part of the beta-tubulin gene was performed using the primers Bt2a (GGTAACCAAATCGGTGCTGCTTTC) and Bt2b (ACCCTCAGTGTAGTGACCCTTGGC) (Glass and Donaldson 1995). Both strands of the PCR fragments were sequenced with the ABI Prism Big DyeTM Terminator v.3.0 Ready Reaction Cycle sequencing kit. Samples were analyzed on an ABI PRISM 3700 Genetic Analyzer, and contigs were assembled using the forward and reverse sequences with the program SeqMan from the LaserGene package (GenBank accession numbers GU296686–GU296707).
Calculation of phylogenetic tree
Sequences were aligned using ClustalX and trimmed to the first common base at both ends before making the final alignment. For the tree of Figure 3B, four trimmed sequences were joined for each strain before making the final alignment. Tree calculations were performed using TreeCon (van de Peer and de Wachter 1994). Distances were estimated using the Jukes and Cantor algorithm taking insertions and deletions into account. Topology was inferred using neighbor joining with bootstrap values based on 1000 reiterations.
Comparison of protein-encoding loci
The loci were compared using megablast of CBS 513.88 cDNA sequences Version 2008 with standard setting followed by parsing of the output for local pairwise sumscore of HSPs.
Fermentation procedure
Growth media
The A. niger batch cultivation medium was 20 g/L glucose monohydrate, 7.3 g/L (NH4)2SO4, 1.5 g/L KH2PO4, 1.0 g/L MgSO4·7H2O, 1 g/L NaCl, 0.1 g/L CaCl2·2H2O, 0.05 mL/L antifoam 204 (Sigma), and 1 mL/L trace element solution. The trace element solution composition was 7.2 g/L ZnSO4·7H2O, 0.3 g/L NiCl2·6H2O, 6.9 g/L FeSO4·7H2O, 3.5 g/L MnCl2·4H2O, and 1.3 g/L CuSO4·5H2O. The A. niger complex medium was 2 g/L yeast extract, 3 g/L tryptone, 10 g/L glucose monohydrate, 20 g/L agar, 0.52 g/L KCl, 0.52 g/L MgSO4·7H2O, 1.52 g/L KH2PO4, and 1 mL/L of trace elements solution. The trace elements solution was 0.4 g/L CuSO4·5H2O, 0.04 g/L Na2B4O7·10H2O, 0.8 g/L FeSO4·7H2O, 0.8 g/L MnSO4·H2O, 0.8 g/L Na2MoO4·2H2O, and 8 g/L ZnSO4·7H2O.
Spore propagation
The bioreactors were inoculated with spores of A. niger CBS 513.88 or ATCC 1015 strains previously propagated on complex media plates and incubated for 8 d at 30°C. The same stock of spores was used to inoculate all the plates. Spores were harvested by adding 10 mL of Tween 80 0.01%, centrifuged at 4000 rpm for 10 min, and washed three times with a suitable amount of 0.9% NaCl. The fermentors were inoculated with a spore suspension to obtain a final concentration of 5.7 × 109 spores/L.
Batch cultivations
A. niger batch cultivations were carried out in 5-L reactors with a working volume of 4.5 L. The bioreactors were equipped with two Rushton four-blade disc turbines, and pH and temperature control. Inlet air was controlled with a mass flowmeter. The concentrations of oxygen and carbon dioxide in the exhaust gas were monitored with a gas analyzer (1311 Fast response Triple gas, Innova combined with multiplexer controller for Gas Analysis MUX100, B. Braun Biotech International). The temperature was maintained at 30°C, and the pH was controlled by automatic addition of 2 M NaOH. The pH was initially set to 3.0 to prevent spore aggregation; when spores started to germinate, the pH was gradually increased to 4.5. Similarly, the stirring speed was initially set to 200 rpm and the aeration rate to 0.05 vvm (volume of gas per volume of liquid per minute) to prevent loss of hydrophobic spores from the medium to the head space of the reactor. After germination, these parameters were progressively increased to 600 rpm and 0.89 vvm and kept steady throughout all the rest of the fermentation.
Sampling
For quantification of cell mass and extracellular metabolites, the fermentation broth was withdrawn from the reactor, filtered, and washed. The filter cakes were used for cell weight determination. The filtrate was filtered once more and frozen at −20°C for later HPLC analysis. For gene expression analysis, mycelium was harvested in the exponential phase of growth at half the maximum biomass density by filtration through sterile Miracloth (Calbiochem) and washed with a suitable amount of 0.9% NaCl solution. The mycelium was quickly dried by squeezing and subsequently frozen in liquid nitrogen. Samples were stored at −80°C until used for RNA extraction.
Cell mass determination
Cell dry weight was determined using paper filters (Whatmann cat. no. 1001070). The filters were pre-dried in an oven for 24 h at 100°C, cooled down in a desiccator for 2 h, and subsequently weighed. A known volume of cell culture was filtered, and the residue was washed with 0.9% NaCl and dried on the filter for 24 h in an oven at 100°C. The filter was weighed again, and the cell mass concentration was calculated. Dry biomass was converted to Cmol using 24.9 g biomass/Cmol (Nielsen et al. 2003).
Quantification of sugars and extracellular metabolites
The concentrations of sugar and organic acids in the filtrates were determined using HPLC on an Aminex HPX-87H ion-exclusion column (Bio-Rad). The column was eluted at 60°C with 5 mM H2SO4 at a flow rate of 0.6 mL/min. Metabolites were detected with a refractive index detector and an UV detector.
Determination of glucoamylase activity
The activity of glucoamylase in culture filtrates was quantified spectrophotometrically using p-nitrophenyl β-maltoside as the substrate for glucoamylase (McCleary et al. 1991). Ninety microliters of 200 mM sodium acetate (pH 4.4) was added to 10 μL of enzyme solution (standards or fermentation broth), mixed, and shaken for 30 sec in a microtiter plate, then incubated for 5 min at 40°C. Ten microliters of this mixture was mixed with 10 μL of substrate containing p-nitrophenyl β-maltoside plus thermostable β-glucosidase (Megazyme). This preparation was shaken for 30 sec and incubated for 10 min at 40°C. The reaction was stopped by the addition of 150 μL of trizma base solution 2%. The sample preparation was shaken again for 30 sec and measured at 405 nm in a spectrophotometer. The absorbance measurement was related to enzyme units per milliliter according to the calibration curve. One unit of enzyme is the amount of enzyme that liberates 1 μmol of glucose per minute at pH 4.8 and 60°C. One unit can be assumed to correspond to 25 μg of protein (PESL protein assay; Boehringer Mannheim).
Transcriptome analysis
Extraction of total RNA, preparation of biotin-labeled cRNA, and microarray processing
Total RNA was isolated from 70–90 mg of frozen mycelium. Twenty micrograms of fragmented biotin-labeled cRNA was prepared from ∼1 μg of total RNA. Fifteen micrograms of fragmented cRNA was hybridized to the 3AspergDTU GeneChip, washed, stained, and scanned. The scanned probe array images (.DAT files) were converted into .CEL files using the Affymetrix GeneChip Operating Software. All steps were performed as in Andersen et al. (2008b).
Analysis of transcription data
Statistical analysis of the Affymetrix CEL-files was performed as described in Andersen et al. (2008b). Note that this method uses the medianpolish algorithm (Irizarry et al. 2003), which should make calculations of gene expression indexes relatively robust to differences in the genome sequences. A cutoff value of adjusted p < 0.05 was set to assess statistical significance. Normalized and raw data values are deposited with GEO as series GSE10983.
GO-term enrichment analysis
A. niger ATCC 1015 ORF lists were examined for GO-term enrichment using R-2.5.1 (http://www.R-project.org) with BioConductor (Gentleman et al. 2004) and the topGO-package v. 1.2.1 using the elim algorithm to remove local dependencies between GO terms (Alexa et al. 2006). GO-term assignments were based on automatic annotation of the A. niger ATCC 1015 gene models. Where nothing else is noted, p < 0.05 is used as the cutoff for significance.
Genomic DNA hybridization analysis
Genomic DNA was prepared with the GNOME kit (Bio101). This DNA was fragmented in a Hydroshear (Genemachines) as per the manufacturer's protocol for 20 cycles with speed code 4, to yield DNA fragments of ∼1.5 kb. This fragmented DNA was further digested to a size of 50–200 nt by partial DNase I digestion (Invitrogen) and labeled with biotin using biotin ddUTP and Terminal deoxynucleotide transferase of the “Bioarray Terminal Labelling kit” (Enzo). Thirteen micrograms of fragmented and labeled DNA was hybridized to the dsmM_ANIGERa_coll511030F GeneChip, washed, stained, and scanned. The images were converted into data files, probe set values, and signal log ratios using the Affymetrix GCOS software. The obtained data set is given in Supplemental Table 16. The dsmM_ANIGERa_coll511030F library and platform information is deposited at GEO under number GPL6758.
Genome-scale modeling
Flux calculation was performed as described in Andersen et al. (2008a) using the A. niger iMA871 model. Specifically, fluxes were calculated for ATCC 1015 and CBS 513.88 using the growth conditions of the experiments described in Table 3 and using the concentrations of glucose, glycerol, glucoamylase, and citric acid from Table 3. The calculated fluxes may be found in Supplemental Table 14.
Synteny analysis
The finished version of the A. niger ATCC 1015 genome sequence was divided into fragments of 1 kb and compared to the nucleotide sequence of A. niger CBS 513.88 using BLASTN (McGinnis and Madden 2004). A cutoff value of 1 × 10−75 was used. The BLAST results were parsed using a custom-made Perl script, giving for each 1-kb fragment the location of the hit and the number of SNPs, insertions, and deletions. The same analysis was performed for a comparison of the A. niger ATCC 1015 and A. niger ATCC 9029 genome sequences.
Transcription mapping
The nucleotide sequence of the gene models of the A. niger ATCC 1015 genome sequence v1.0 was mapped to the 24 contigs of the finished sequence by BLASTN. A cutoff value of 1 × 10−100 was used. An adapted version of the Imprint algorithm (see below) was used to find the coordinates of each gene. Log2-ratios of the average expression ratios in each of the two strains were mapped to the coordinate of the first base of the gene finding a hit in the target genome.
Imprint algorithm
The object of the algorithm is, for every coding sequence of A. niger ATCC 1015, to present the corresponding genomic sequence from A. niger CBS 513.88 (the Imprint), so that an unbiased comparison of two sets of gene callers can be conducted.
The CDSs of A. niger ATCC 1015 were compared to the nucleotide sequence of A. niger CBS 513.88 using BLASTN (McGinnis and Madden 2004). The BLASTN output was parsed to extract—for each gene—the alignment to the A. niger CBS 513.88 sequence. This provided for each gene, the part of the CDS having a hit in the CBS 513.88 sequence, and the corresponding sequence from CBS 513.88 sequence (the Imprint). The CDS and the Imprint were compared to produce a comprehensive list of insertions, deletions, silent mutations, sense mutations, frameshift mutations, and nonsense mutations for each CDS pair. Detail on the analysis and manual curation of the results can be found in Supplemental Text 3. The final summary is found in Supplemental Table 5. All genes were assigned to one of the categories of Supplemental Table 5. Genes were only assigned to categories E–F if none of the requirements of categories G–L were fulfilled. Manual and automatic annotation was added for 9400 genes, including all genes with >0.5% difference in amino acid sequence.
Acknowledgments
This study was partially funded by the Danish Research Agency for Technology and Production (to M.R.A., S.L.M., J.B.N., and M.L.N.) and the National Council of Research Conacyt-Mexico (to M.P.S.). Genome sequencing was performed under the auspices of the U.S. Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396. Preparation of genomic DNA and some analysis of the genome were supported by the DOE Office of the Biomass Program.
Footnotes
[Supplemental material is available for this article. The A. niger ATCC 1015 whole genome sequence has been submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank/) under accession no. ACJE00000000. The sequence data from the phylogeny study have been submitted to GenBank under accession nos. GU296686–GU296739. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under series accession no. GSE10983. The dsmM_ANIGERa_coll511030F library and platform information have been submitted to GEO under accession no. GPL6758.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.112169.110.
References
- Abarca ML, Bragulat MR, Castella G, Cabanes FJ 1994. Ochratoxin A production by strains of Aspergillus niger var. niger. Appl Environ Microbiol 60: 2650–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A, Rahnenführer J, Lengauer T 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Andersen MR, Nielsen M, Nielsen J 2008a. Metabolic model integration of the bibliome, genome, metabolome and reactome of Aspergillus niger. Mol Syst Biol 4: 178 doi: 10.1038/msb.2008.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen MR, Vongsangnak W, Panagiotou G, Salazar M, Lehmann L, Nielsen J 2008b. A tri-species Aspergillus micro array: Comparative transcriptomics of three Aspergillus species. Proc Natl Acad Sci 105: 4387–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SE 2006. Aspergillus niger genomics: Past, present and into the future. Med Mycol Suppl 1 44: S17–S21 [DOI] [PubMed] [Google Scholar]
- Birney E, Durbin R 2000. Using GeneWise in the Drosophila annotation experiment. Genome Res 10: 547–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos CJ, Debets AJ, Kobus G, Slakhorst SM, Swart K 1989. Adenine and pyrimidine genes of Aspergillus niger and evidence for a seventh linkage group. Curr Genet 16: 307–310 [DOI] [PubMed] [Google Scholar]
- Braumann I, van den Berg M, Kempken F 2008. Strain-specific retrotransposon-mediated recombination in commercially used Aspergillus niger strain. Mol Genet Genomics 280: 319–325 [DOI] [PubMed] [Google Scholar]
- Büchi G, Klaubert D, Shank R, Weinreb S, Wogan GN 1971. Structure and synthesis of kotanin and desmethylkotanin, metabolites of Aspergillus glaucus. J Org Chem 36: 1143–1147 [DOI] [PubMed] [Google Scholar]
- Christias C, Couvaraki C, Georgopoulos SG, Vomvoyanni V 1975. Protein content and amino acid composition of certain fungi evaluated for microbial protein production. Appl Microbiol 29: 250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen D 2007. The genome of an industrial workhorse. Nat Biotechnol 25: 189–190 [DOI] [PubMed] [Google Scholar]
- Cuomo CA, Güldener U, Xu JR, Trail F, Turgeon BG, Di Pietro A, Walton JD, Ma LJ, Baker SE, Rep M, et al. 2007. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317: 1400–1402 [DOI] [PubMed] [Google Scholar]
- Cutler HG, Crumley FG, Cox RH, Hernandez O, Cole RJ, Dorner JW 1979. Orlandin: a nontoxic fungal metabolite with plant growth inhibiting properties. J Agric Food Chem 27: 592–595 [DOI] [PubMed] [Google Scholar]
- Debets A, Swart K, Bos C 1989. Mitotic mapping in linkage group V of Aspergillus niger based on selection of auxotrophic recombinants by Novozym enrichment. Can J Microbiol 35: 982–988 [DOI] [PubMed] [Google Scholar]
- Debets A, Holub E, Swart K, van den Broek H, Bos C 1990a. An electrophoretic karyotype of Aspergillus niger. Mol Gen Genet 224: 264–268 [DOI] [PubMed] [Google Scholar]
- Debets A, Swart K, Bos C 1990b. Genetic analysis of Aspergillus niger: Isolation of chlorate resistance mutants, their use in mitotic mapping and evidence for an eighth linkage group. Mol Gen Genet 221: 453–458 [DOI] [PubMed] [Google Scholar]
- Fedorova ND, Khaldi N, Joardar VS, Maiti R, Amedeo P, Anderson MJ, Crabtree J, Silva JC, Badger JH, Albarraq A, et al. 2008. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet 4: e1000046 doi: 10.1371/journal.pgen.1000046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad JC, Samson R 2004. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of the food and air-borne terverticillate Penicillia and their mycotoxins. Stud Mycol 49: 1–52 [Google Scholar]
- Frisvad JC, Thrane U 1987. Standardized high performance liquid chromatography of 182 mycotoxins and other fungal metabolites based on alkylphenone indices and UV VIS spectra (diode array detection). J Chromatogr 404: 195–214 [DOI] [PubMed] [Google Scholar]
- Frisvad JC, Larsen TO, de Vries R, Meijer M, Houbraken J, Cabañes FJ, Ehrlich K, Samson RA 2007a. Secondary metabolite profiling, growth profiles and other tools for species recognition and important Aspergillus mycotoxins. Stud Mycol 59: 31–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad JC, Smedsgaard J, Samson R, Larsen T, Thrane U 2007b. Fumonisin B2 production by Aspergillus niger. J Agric Food Chem 55: 9727–9732 [DOI] [PubMed] [Google Scholar]
- Fukuda T, Hasegawa Y, Hagimori K, Yamaguchi Y, Masuma R, Tomoda H, Omura S 2006. Tensidols, new potentiators of antifungal miconazole activity, produced by Aspergillus niger FKI-2342. J Antibiot (Tokyo) 59: 480–485 [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Baştürkmen M, Spevak CC, Clutterbuck J, et al. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 5: R80 doi: 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61: 1323–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P 1998. Consed: A graphical tool for sequence finishing. Genome Res 8: 195–202 [DOI] [PubMed] [Google Scholar]
- Gottschling D, Aparicio O, Billington B, Zakian V 1990. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 1: D258–D261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiort J, Maksimenka K, Reichert M, Perović-Ottstadt S, Lin WH, Wray V, Steube K, Schaumann K, Weber H, Proksch P, et al. 2004. New natural products from the sponge-derived fungus Aspergillus niger. J Nat Prod 67: 1532–1543 [DOI] [PubMed] [Google Scholar]
- Inokoshi J, Shiomi K, Masuma R, Tanaka H, Yamada H, Omura S 1999. Funalenone, a novel collagenase inhibitor produced by Aspergillus niger. J Antibiot (Tokyo) 52: 1095–1100 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Isogai A, Horii T, Suzuki A, Murakoshi S, Ikeda K, Sato S, Tamura S 1975. Isolation and identification of nigragillin as an insecticidal metabolite produced by Aspergillus niger. Agric Biol Chem 39: 739–740 [Google Scholar]
- Kanehisa M, Guto S, Kawashima S, Nakaya A 2002. The KEGG databases at GenomeNet. Nucleic Acids Res 30: 42–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res 32: D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaffa L, Kubicek CP 2003. Aspergillus niger citric acid accumulation: do we understand this well working black box? Appl Microbiol Biotechnol 61: 189–196 [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD 2008. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet 9: 605–617 [DOI] [PubMed] [Google Scholar]
- Khaldi N, Wolfe KH 2008. Elusive origins of the extra genes in Aspergillus oryzae. PLoS ONE 3: e3036 doi: 10.1371/journal.pone.0003036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaldi N, Collemare J, Lebrun MH, Wolfe KH 2008. Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biol 9: R18 doi: 10.1186/gb-2008-9-1-r18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, et al. 2004. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol 5: R7 doi: 10.1186/gb-2004-5-2-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korman DR, Bayliss FT, Barnett CC, Carmona CL, Kodama KH, Royer TJ, Thompson SA, Ward M, Wilson LJ, Berka RM 1990. Cloning, characterization, and expression of two alpha-amylase genes from Aspergillus niger var. awamori. Curr Genet 17: 203–212 [DOI] [PubMed] [Google Scholar]
- Kozakiewicz Z, Frisvad JC, Hawksworth DL, Pitt JI, Samson RA, Stolk AC 1992. Proposals for nomina specifica conservanda and rijicienda in Aspergillus and Penicillium (Fungi). Taxon 41: 109–113 [Google Scholar]
- Lubertozzi D, Keasling JD 2008. Developing Aspergillus as a host for heterologous expression. Biotechnol Adv 27: 53–75 [DOI] [PubMed] [Google Scholar]
- Martens-Uzunova ES, Schaap PJ 2008. An evolutionary conserved D-galacturonic acid metabolic pathway operates across filamentous fungi capable of pectin degradation. Fungal Genet Biol 45: 1449–1457 [DOI] [PubMed] [Google Scholar]
- McCleary BV, Bouhet F, Driguez H 1991. Measurement of amyloglucosidase using p-nitrophenyl b-maltoside as substrate. Biotechnol Tech 5: 255–258 [Google Scholar]
- McGinnis S, Madden T 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32: W20–W25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KF, Smedsgaard J 2003. Fungal metabolite screening: database of 474 mycotoxins and fungal metabolites for dereplication by standardized liquid chromatography-UV-mass spectrometry methodology. J Chromatogr A 1002: 111–136 [DOI] [PubMed] [Google Scholar]
- Nielsen J, Villadsen J, Liden G 2003. Bioreaction engineering principles, 2nd ed Kluwer Academic/Plenum, New York [Google Scholar]
- Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, et al. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25: 221–231 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H 2000. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics methods and protocols: Methods in molecular biology (ed. Krawetz S, Misener S), pp. 365–386 Humana, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res 10: 516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster E, Dunn-Coleman N, Frisvad JC, van Dijck PWM 2002. On the safety of Aspergillus niger—a review. Appl Microbiol Biotechnol 59: 426–435 [DOI] [PubMed] [Google Scholar]
- Shibuya I, Tamura G, Ishikawa T, Hara S 1992. Cloning the alpha-amylase cDNA of Aspergillus shirousamii and its expression in Saccharomyces cerevisiae. Biosci Biotechnol Biochem 56: 174–179 [DOI] [PubMed] [Google Scholar]
- Smedsgaard J 1997. Micro-scale extraction procedure for standardised screening of fungal metabolite production in cultures. J Chromatogr A 760: 264–270 [DOI] [PubMed] [Google Scholar]
- Strand DJ, McDonald JF 1985. Copia is transcriptionally responsive to environmental stress. Nucleic Acids Res 13: 4401–4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Lu X, Rinas U, Zeng A 2007. Metabolic peculiarities of Aspergillus niger disclosed by comparative metabolic genomics. Genome Biol 8: R182 doi: 10.1186/gb-2007-8-9-r182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart K, Debets AJ, Kobus G, Bos CJ 1992. Arginine and proline genes of Aspergillus niger. Antonie van Leeuwenhoek 61: 259–264 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Wang PL, Yamada O, Tamura L 1966. Yellow pigments of Aspergillus niger and Asp. awamori. Part. I. Isolation of aurasperone A and related pigments. Agric Biol Chem 30: 107–113 [Google Scholar]
- van de Peer Y, de Wachter R 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10: 569–570 [DOI] [PubMed] [Google Scholar]
- van Dijck PWM 2008. The importance of Aspergilli and regulatory aspects of Aspergillus nomenclature. In Aspergillus in the genomic era (ed. Varga J, Samso RA), pp. 249–257 Wageningen Academic, Wageningen, The Netherlands [Google Scholar]
- van Dijck PWM, Selten GCM, Hempenius RA 2003. On the safety of a new generation of DSM Aspergillus niger production strains. Regul Toxicol Pharmacol 38: 27–35 [DOI] [PubMed] [Google Scholar]
- van Lanen JM, Smith MB 1968. Process of producing glucamylase and an alcohol product. US patent 3,418,211. Hiram Walker & Sons, Inc, Peoria, IL [Google Scholar]
- Verdoes JC, Calil MR, Punt PJ, Debets F, Swart K, Stouthamer AH, van den Hondel CA 1994. The complete karyotype of Aspergillus niger: The use of introduced electrophoretic mobility variation of chromosomes for gene assignment studies. Mol Gen Genet 244: 75–80 [DOI] [PubMed] [Google Scholar]
- Yuan X, Roubos J, van den Hondel C, Ram A 2008a. Identification of InuR, a new Zn(II)2Cys6 transcriptional activator involved in the regulation of inulinolytic genes in Aspergillus niger. Mol Genet Genomics 279: 11–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan XL, van der Kaaij RM, van den Hondel CA, Punt PJ, van der Maarel MJ, Dijkhuizen L, Ram AF 2008b. Aspergillus niger genome-wide analysis reveals a large number of novel α-glucan acting enzymes with unexpected expression profiles. Mol Genet Genomics 279: 545–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848 [DOI] [PubMed] [Google Scholar]