Abstract
Centromeres are determined by poorly understood epigenetic mechanisms. Centromeres can be activated or inactivated without changing the underlying DNA sequences. However, virtually nothing is known about the epigenetic transition of a centromere from an active to an inactive state because of the lack of examples of the same centromere exhibiting alternative forms and being distinguishable from other centromeres. The centromere of the supernumerary B chromosome of maize provides such an opportunity because its functional core can be cytologically tracked, and an inactive version of the centromere is available. We developed a DNA fiber-based technique that can be used to assess the levels of cytosine methylation associated with repetitive DNA sequences. We report that DNA sequences in the normal B centromere exhibit hypomethylation. This methylation pattern is not affected by the genetic background or structural rearrangement of the B chromosome, but is slightly changed when the B chromosome is transferred to oat as an addition chromosome. In contrast, an inactive version of this same centromere exhibits hypermethylation, indicating that the inactive centromere was modified into a different epigenetic state at the DNA level.
The functional centromere is specified by the presence of a special histone H3 variant, known as CENH3 in plants. The establishment and maintenance of centromeres are not defined by the underlying DNA sequences but, rather, are determined by epigenetic mechanisms (Allshire and Karpen 2008). Centromeres can be inactivated (Earnshaw and Migeon 1985; Sullivan and Schwartz 1995; Han et al. 2006; Zhang et al. 2010), or they can also be activated as “neocentromeres” from non-centromeric regions (Williams et al. 1998; Nasuda et al. 2005; Marshall et al. 2008; Gong et al. 2009; Topp et al. 2009) or re-activated from a previously inactivated centromere (Han et al. 2009). Most strikingly, human neocentromeres were established at numerous locations associated with most chromosomes and are as fully functional as normal centromeres (Marshall et al. 2008). Despite the extensive and compelling evidence for the epigenetic regulation of centromere establishment and maintenance, there has been limited information as to what epigenetic modifications are important or specific to centromeric chromatin. The slow progress of this important research subject is largely due to the fact that centromeres in most multicellular eukaryotes are composed of megabase-sized arrays of satellite repeats (Henikoff et al. 2001; Jiang et al. 2003), which prevent DNA sequence-based fine mapping of epigenetic marks, such as DNA methylation and histone modifications.
Using an immunofluorescence assay on highly stretched meiotic pachytene chromosomes, we recently demonstrated a characteristic hypomethylation pattern associated with the satellite repeats in CENH3-associated chromatin (CEN chromatin) in both Arabidopsis thaliana and maize (Zea mays). In contrast, the same satellite repeats located in the pericentromeric regions were hypermethylated (Zhang et al. 2008; Koo and Jiang 2009). Thus, different members from the same satellite repeat family can either be hypomethylated or hypermethylated, depending on their association with CEN chromatin or with pericentromeric heterochromatin. In A. thaliana, the 178-bp centromeric repeats derived from CEN chromatin, which can be isolated by chromatin immunoprecipitation (ChIP) using anti-CENH3 antibodies, displayed a distinct distribution pattern of CG and CHG sites. This unique CG/CHG composition may provide the foundation for the differential methylation of these repeats from those located in pericentromeric regions (Zhang et al. 2008).
The supernumerary B chromosome of maize provides an excellent tool for centromere research. The functional core of the B centromere, which is associated with CENH3, has been well characterized and consists of ∼700 kb of complex repetitive DNA sequences (Jin et al. 2005). To further reveal the methylation status of DNA sequences associated with centromeres, we developed a DNA fiber-based technique to assess the levels of cytosine methylation associated with repetitive DNA sequences. This technique permitted us to precisely map the cytosine methylation associated with different repetitive DNA sequences located in the maize B centromere. We report that the DNA sequences associated with the functional B centromere exhibit hypomethylation. This distinct DNA methylation pattern has been largely maintained after the B chromosome was transferred into an oat genetic background and after it has undergone significant structural rearrangements. However, inactivation of the B centromere resulted in a drastic increase of DNA methylation of its underlying sequences. Thus, we demonstrate that the DNA sequences associated with active and inactive B centromeres are associated with distinct cytosine methylation patterns.
Results
Development of a DNA fiber-based technique for mapping cytosine methylation
Methylation of repetitive DNA sequences is difficult to assess, especially when different members from the same repeat family are differentially methylated. Currently, the most common techniques for mapping DNA methylation, including genomic tiling arrays (Zhang et al. 2006; Zilberman et al. 2007) and shotgun bisulphite sequencing-based platforms (Cokus et al. 2008; Lister et al. 2008), cannot be used for evaluating methylation associated with highly repetitive DNA sequences. Therefore, we developed a DNA fiber-based technique to assess the levels of cytosine methylation associated with repetitive DNA sequences by combining an immunofluorescence assay with fluorescence in situ hybridization (FISH) on extended DNA fibers (see Methods).
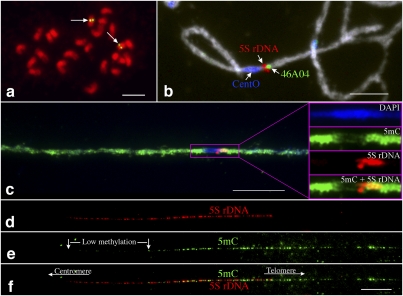
We first applied this technique on the 5S ribosomal DNA (5S rDNA) locus in rice (Oryza sativa) to prove its effectiveness in visualizing the differential methylation associated with a repetitive DNA element. Rice contains a single 5S rDNA locus that is located close to the centromere of chromosome 11 (Cen11) (Fig. 1a; Kamisugi et al. 1994). Rice centromeres contain a 155-bp centromere-specific satellite repeat CentO, ranging from ∼65 kb in Cen8 to ∼2 Mb in Cen 11 (Cheng et al. 2002). The entire CENH3 domain of Cen11 is embedded within the megabase-sized CentO array (Yan et al. 2008). The 5S rDNA locus is immediately adjacent to the CentO array in Cen11 (Fig. 1b; Supplemental Fig. S1).
Figure 1.
The hypermethylation–hypomethylation transition zone in the centromeric region of rice chromosome 11. (a) FISH mapping of the 5S rDNA on somatic metaphase chromosomes of rice. Arrows point to the single 5S rDNA locus (green) on rice chromosome 11. Bar, 5 μm. (b) Three-color FISH mapping of BAC clone 46A04 (green), 5S rDNA (red), and the CentO satellite repeat (blue) on meiotic pachytene chromosome 11 of rice. BAC 46A04 is the closest clone flanking the telomeric side of the 5S rDNA locus. Bar, 5 μm. (c) Immunodetection of 5mC on a super-stretched rice pachytene chromosome 11. The purple box includes the centromeric region, and the fluorescent signals in this region are exemplified and separated into different color channels. The 5S rDNA locus is located within the hypermethylation–hypomethylation transition zone. (d–f) Differential methylation along the 5S rDNA locus revealed by immunodetection of 5mC on DNA fibers. Approximately 41% of the 5S rDNA section toward the centromere is less methylated than the rest of the 5S rDNA array. Bars, 10 μm.
An immunofluorescence assay using an antibody against 5-methylcytosine (5mC) revealed that the Cen11 region occupied by the CentO array is hypomethylated compared to the pericentromeric regions (Yan et al. 2010). This methylation pattern is similar to the pattern at A. thaliana and maize centromeres (Zhang et al. 2008; Koo and Jiang 2009). The 5S rDNA locus was located in the hypomethylation–hypermethylation transition zone of Cen11 (Fig. 1c). We analyzed the 5mC signals associated with the 5S rDNA locus on extended DNA fibers. Significantly reduced 5mC signals were observed toward one end of each DNA fiber. The fiber section with a reduced level of 5mC accounted for 41.06% ± 4.86% (n = 32) of the length of the 5S rDNA fiber (Fig. 1d–f). The centromere–telomere orientation of this low-methylated 5S rDNA section as being proximal to the CentO array of Cen11 was confirmed by comapping with a closely flanking BAC clone 46A04 on DNA fibers (Supplemental Fig. S2). This differential methylation pattern was observed on all 32 DNA fibers analyzed. Thus, we were able to consistently visualize the differential methylation of the DNA sequences located in the centromeric region of rice chromosome 11.
A distinct methylation pattern associated with the centromere of the maize B chromosome
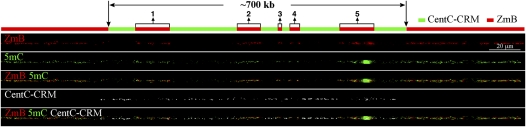
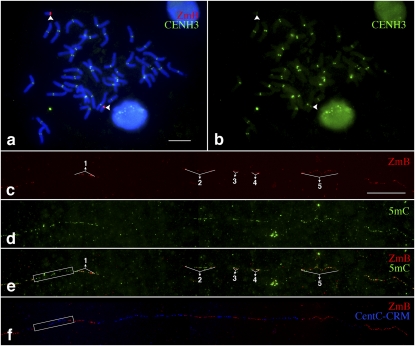
The maize B centromere contains a B-specific satellite repeat, ZmB, which allows the identification of the B centromere from the other maize centromeres (Alfenito and Birchler 1993). The functional B centromere, which is associated with CENH3, spans ∼700 kb of DNA consisting of five separate arrays of the ZmB repeat (Fig. 2; Jin et al. 2005). Each of the five ZmB arrays is flanked by intermingled CentC satellite repeats and the CRM retrotransposons (Fig. 2). Both CentC and CRM sequences are exclusively or highly enriched in the centromeres of both A and B chromosomes (Ananiev et al. 1998; Zhong et al. 2002; Jin et al. 2005; Lamb et al. 2005). Along with the 700-kb centromere core domain consisting of ZmB and CentC-CRM sequences, both sides of this domain are also flanked by megabase-sized arrays of ZmB repeats (Fig. 2). The B centromere can be readily identified on DNA fibers because of this unique composition of ZmB repeats (Jin et al. 2005).
Figure 2.
Mapping of DNA methylation in the centromere of maize B chromosome. The positions of the five ZmB arrays (1–5) in the functional centromere are labeled. Fiber-FISH of ZmB (red) and CentC-CRM (the white signals were digitally converted from blue fluorescence signals) and detection of 5mC (green) were performed on the same DNA fiber. (Top panel) A diagram of the DNA composition of the maize B centromere. The functional B centromere spans ∼700 kb and contains five distinct arrays (1–5) of the ZmB repeat. The association of this 700-kb region with CENH3 was previously demonstrated by Jin et al. (2005). Each ZmB repeat array is flanked by intermingled CentC and CRM repeats.
The fiber-based DNA methylation assay revealed bright 5mC signals associated with all five ZmB arrays in the B centromere (Fig. 2; Supplemental Fig. S3). In contrast, the intermingled CentC-CRM sequences flanking each of the five ZmB arrays were hypomethylated (Figs. 2, 3). We analyzed 15 independent DNA fibers, and no consistent 5mC signals were observed on the CentC-CRM sections in the B centromere. Sporadic fluorescent signals were detected on some DNA fibers. The positions of such signals on individual fibers were random, making it difficult to distinguish such signals from the background. Thus, such signals may represent either cytological artifacts or a low level of cytosine methylation that cannot be consistently detected by the present technique.
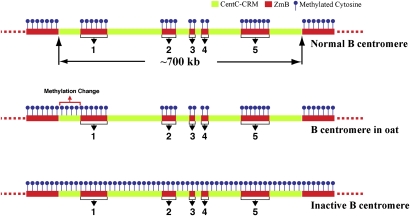
Figure 3.
Diagrams of DNA methylation patterns associated with maize B centromere. Methylation is only associated with the ZmB repeats, not with the Cent-CRM sequences in the normal B centromere. Methylation change was detected in a small region flanking the first ZmB array when the B chromosome is transferred to the genetic background of oats. In contrast, methylation was detected in the entire inactivated B centromere.
The DNA methylation pattern of the B centromere is maintained in a rearranged B chromosome
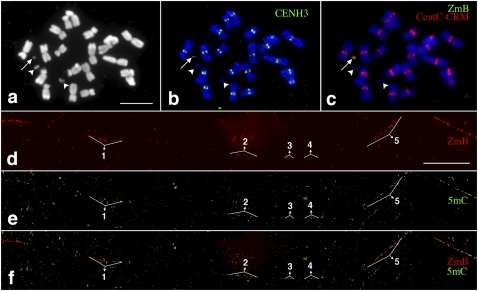
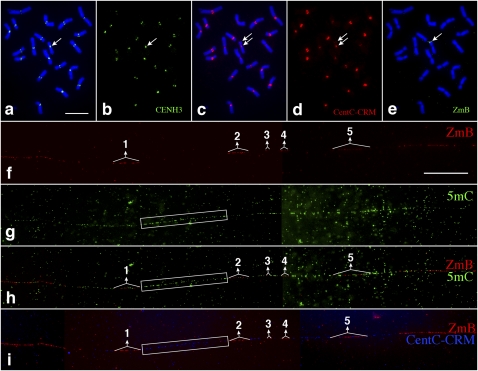
We analyzed the DNA methylation associated with the centromere of a mini-B chromosome (Fig. 4a–c). This mini-B chromosome was derived from the complex A-B translocation chromosome B-9-Dp9 (Han et al. 2007) and has undergone extensive rearrangements from breakage–fusion–bridge (BFB) cycles. The size of the mini-B chromosome was reduced to ∼7% (7.43% ± 1.47%, n = 6) of the normal B chromosome, but it retained a fully functioning centromere (Fig. 4b) and showed faithful segregation and transmission in somatic cells. Fiber-FISH analysis indicated that the centromere of the mini-B chromosome was intact and contained all five distinct ZmB arrays (Fig. 4d). Fiber-based 5mC mapping revealed that the mini-B centromere maintained an identical DNA methylation pattern as the normal B centromere (Fig. 4e,f).
Figure 4.
Mapping of DNA methylation in a significantly rearranged minichromosome derived from the maize B chromosome. (a) A metaphase cell derived from a maize line containing a mini-B chromosome (arrow). Two arrowheads point to the two satellites associated with maize chromosome 6. Bar, 5 μm. (b) Immunofluorescence assay using anti-CENH3 antibody (green). The arrow points to the CENH3 signal associated with the minichromosome. (c) FISH mapping of CentC-CRM (red) and ZmB (green) repeats. The arrow points to the signals associated with the minichromosome. (d) Fiber-FISH signals from the ZmB repeat located in the mini-B chromosome. Bar, 20 μm. (e) Detection of 5mC on the same DNA fiber as d. (f) A merged image of the ZmB and 5mC signals. The ZmB signals were all associated with 5mC signals on the DNA fiber. The five ZmB repeat arrays associated with the CENH3 domain are labeled in d–f.
The DNA methylation pattern of the B centromere is changed in an oat–maize B chromosome addition line
The maize B chromosome has been transferred to the genetic background of oat (Avena sativa) using wide crosses between oat and maize (Fig. 5a,b; Kynast et al. 2001). In oat–maize chromosomal addition lines, the centromeres of maize chromosomes incorporate the CENH3 from oat (Jin et al. 2004). Fiber-based 5mC mapping revealed that the B centromere in the oat background displayed a similar DNA methylation pattern as in the normal B centromere in maize, as all five ZmB arrays were hypermethylated (Fig. 5c–e). However, we observed unambiguous 5mC signals associated with the CentC-CRM sequences immediately flanking the first ZmB array on nine of the 12 DNA fibers analyzed (Fig. 5e,f). In contrast, 5mC signals were not observed in the same region of the B chromosome in the maize background (Figs. 2, 3). These results show that the DNA methylation of this flanking region was modified after the B chromosome was transferred into the oat background.
Figure 5.
Mapping of DNA methylation in the centromere of a maize B chromosome in an oat genetic background. (a) A metaphase cell, derived from the oat–maize B chromosome addition line, was analyzed with an immunofluorescence assay using a maize anti-CENH3 antibody combined with FISH using a ZmB probe. The arrowheads point to the two B chromosomes. Bar, 10 μm. (b) Digitally separated CENH3 signals from the immunofluorescence assay. Arrowheads point to the CENH3 signals from the B chromosomes. (c) Fiber-FISH signals from the ZmB repeat located in the B chromosome in an oat genetic background. Bar, 20 μm. (d) Detection of 5mC on the same DNA fiber as c. (e) A merged image of the ZmB and 5mC signals. The region immediately flanking the first ZmB array is boxed and shows a similar methylation level as the ZmB arrays. (f) Fiber-FISH signals derived from ZmB (red) and a mixed probe with both CentC and CRM sequences (blue). The box, which covers the same region as in e, includes CentC-CRM sequences.
An inactivated B centromere showed a dramatically changed DNA methylation pattern
Han et al. (2006) isolated a maize A-B translocation chromosome, 9-Bic-1. This translocation chromosome retains the majority of maize chromosome 9 and also contains a fragment from the B chromosome including a complete but inactivated B centromere (Fig. 6a–d; Han et al. 2006). A fiber-based DNA methylation assay revealed nearly uniform 5mC signals throughout the inactivated B centromere (Fig. 6f–i). The five ZmB arrays and their flanking CentC-CRM sequences in the inactivated B centromeres were equally methylated (Figs. 3, 6). Such a uniform methylation pattern was observed on all 15 different fibers analyzed. These results demonstrated that cytosine methylation is now associated with the CentC-CRM arrays after the inactivation of the B centromere.
Figure 6.
Mapping of DNA methylation in an inactivated B centromere. (a) Immunofluorescence assay using a maize anti-CENH3 antibody on somatic metaphase chromosomes of maize line 9-Bic-1. The arrow points to the 9-Bic-1 chromosome containing a normal centromere from maize chromosome 9 and an inactivated centromere of the maize B chromosome. CENH3 signals, indicated by an arrow, were only observed in the functional centromere derived from chromosome 9. Bar, 10 μm. (b) Digitally separated CENH3 signals. The arrow points to the single CENH3 signal associated with the dicentric chromosome. (c) FISH of the same metaphase cell using a mixed probe containing both CentC satellite and CRM sequences. The two arrows point to the normal centromere and the inactivated B centromere, respectively. (d) Digitally separated FISH signals. CentC-CRM signals were associated with both the normal and the inactivated B centromeres (arrows). (e) FISH of the same metaphase cell using a ZmB probe. The arrow points to the distinct signal associated with the inactivated B centromere. (f) Fiber-FISH signals from the ZmB repeat in the inactivated B centromere. Bar, 25 μm. (g) Detection of 5mC on the same DNA fiber as f. (h) A merged image of the ZmB and 5mC signals. The 5mC signals were distributed throughout the DNA fiber. (i) Fiber-FISH signals derived from ZmB (red) and a mixed probe with both CentC and CRM sequences (blue). The five ZmB repeat arrays associated with the CENH3 domain are labeled. The boxes cover the centromeric region flanked by the ZmB repeat arrays #1 and #2. This region contains mainly CentC-CRM sequences and is extensively methylated.
Discussion
It has been well documented that CEN chromatin is composed of alternating blocks of nucleosomes that contain either H3 or CENH3 (Blower et al. 2002; Chueh et al. 2005; Yan et al. 2008). Four of the 12 rice centromeres have been well characterized. The sizes of the functional cores of these four centromeres (Cen4, Cen5, Cen7, and Cen8) range from 420 kb to 820 kb and consist of five to seven CENH3 nucleosome blocks. Each CENH3 nucleosome block is flanked by H3 nucleosome blocks (Yan et al. 2008). The size (∼700 kb) and structure (alternating ZmB and CentC-CRM sequence blocks) of the maize B centromere is highly similar to the four rice centromeres. Although the functional B centromere spans five ZmB repeat arrays, ZmB repeats were clearly not as enriched as the CentC repeats in ChIP experiments using anti-CENH3 antibodies (Jin et al. 2005). Thus, the majority of the ZmB repeats in the CEN chromatin are likely associated with H3 nucleosomes, while the flanking CentC-CRM sequences are associated with CENH3 nucleosomes.
CEN chromatin is embedded in pericentromeric heterochromatin (Sullivan and Karpen 2004; Cam et al. 2005). In Schizosaccharomyces pombe, conditional deletion of a centromere resulted in the formation of neocentromeres that were recurrently located adjacent to telomeric heterochromatin (Ishii et al. 2008). In addition, synthetic heterochromatin on plasmid-based minichromosomes in S. pombe can promote de novo CENH3 and kinetochore assembly (Kagansky et al. 2009). These results suggest an intrinsic association of heterochromatin with the establishment and maintenance of CENH3 chromatin. However, the exact role of heterochromatin in centromere determination remains to be determined. It is well known that DNA sequences associated with heterochromatin are hypermethylated in higher eukaryotes. Heterochromatin may provide insulation for the CENH3 loading zone. Loading of newly synthesized CENH3 occurs in different cell cycle windows from that of H3 (Lermontova et al. 2006; Jansen et al. 2007). Hypomethylation of DNA associated with CEN chromatin may be more favorable to differential decondensation than the flanking H3 nucleosome blocks, which may aid the loading of newly synthesized CENH3. Thus, differential methylation of the ZmB and CentC-CRM blocks in the B centromere might be favorable for precise CENH3 loading and propagation in specific regions.
It is interesting to note that the inactivation of the B centromere resulted in a change of methylation of the CentC-CRM sequences. Both CentC and CRM elements are highly repetitive DNA elements, which typically are heavily methylated in plant genomes except for those associated with functional centromeres (Zhang et al. 2008). We propose that propagation of CENH3 nucleosomes may prevent the methylation of the included DNA sequences and maintain them in a hypomethylated state. The positioning of nucleosomes influences DNA methylation in both plant and animal genomes (Chodavarapu et al. 2010). CENH3 nucleosomes are substantially more conformationally rigid than H3 nucleosomes (Black et al. 2007; Sekulic et al. 2010). Micrococcal nuclease digestion generates a smeared pattern for CEN chromatin, but a nucleosome ladder pattern for pericentromeric chromatin in yeasts (Polizzi and Clarke 1991; Baum et al. 2006). Thus, the unique nucleosome structure of CEN chromatin may prevent methylation of the associated DNA sequences. Consequently, the loss of CENH3 leads to the underlying CentC-CRM sequences to be unprotected from DNA methyltransferases and to become heavily methylated after the B centromere inactivation.
Methods
Plant materials and chromosome preparations
A maize B73 line containing a B chromosome was used for cytological analysis of intact B chromosomes. A maize line containing the mini-B chromosome was derived from breakage–fusion–bridge (BFB) cycles of the B-9-Dp9 translocation chromosome (Han et al. 2007). The maize line containing an inactivated B centromere was developed at the University of Missouri. The oat–maize B chromosome addition line was provided by Howard Rines and Ronald Phillips of the University of Minnesota. Rice variety Nipponbare was used for chromosomal and DNA fiber mapping of Cen11. Preparations of mitotic and meiotic chromosomes and super-stretched pachytene chromosomes followed published protocols (Cheng et al. 2001; Koo and Jiang 2009). All preparations were stored at −80°C until they were used.
FISH, fiber-FISH, and immunodetection of CENH3 and 5mC on chromosomes
The FISH and fiber-FISH procedures were conducted following previously published protocols (Jiang et al. 1995; Fransz et al. 1996). DNA probes of the CentO repeat, 5S rDNA, and BAC 46A04 were labeled with digoxigenin-11-dUTP, biotin-16-dUTP, and/or DNP-11-dUTP, depending on whether two or three probes were used in a FISH experiment. After post-hybridization washes, the probes were detected with Alexafluor 488 streptavidin for biotin-labeled probes, and rhodamine-conjugated anti-digoxigenin for dig-labeled probe. The DNP-labeled probe was detected with rabbit anti-DNP, followed by amplification with a chicken anti-rabbit Alexafluor 647 antibody. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) in Vectashield antifade solution (Vector Laboratories). The combination of FISH and immunodetection of CENH3 followed published protocols (Jin et al. 2004).
To detect 5mC on chromosomes, the slides were denatured in 70% formamide in 2× SSC (0.3 M NaCl, 30 mM trisodium citrate at pH 7.0) for 2.5 min at 80°C, washed for 5 min in ice-cold 70% ethanol, and incubated in 0.5% bovine serum albumin (BSA) in PBST (0.14 M NaCl, 8 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl, 0.5% Tween 20) for 30 min at 37°C. The slides were subsequently incubated with mouse antiserum raised against 5mC (1:250) (Eurogentec) in TNB (0.1 M Tris-HCl at pH 7.5, 0.15 M NaCl, 0.5% blocking reagent). Mouse antibodies were detected using rabbit anti-mouse Alexafluor 488 antibody (Invitrogen).
Combining FISH and immunodetection of 5mC on extended DNA fibers
DNA fibers were prepared according to published protocols (Jackson et al. 1998). For detection of 5mC on DNA fibers, slides were pretreated with 100 μg/mL DNase-free RNase A (Sigma) in 2× SSC for 30 min at 37°C and washed two times for 5 min each in 2× SSC. The slides were treated with 1 μg/mL pepsin (Roche) in 10 mM HCl for 10 min at 37°C and placed in ddH2O for 1 min to stop the reaction. The slides were washed two times in 2× SSC for 5 min each and dehydrated in 70% and 95% ethanol.
In most experiments we combined 5mC detection with FISH using a single DNA probe. In these experiments the FISH probe mixture (50% formamide, 10% dextran sulfate, 50 ng of labeled DNA in 2× SSC) was applied to the slide, covered with a coverslide (22 × 22 mm), and sealed with rubber cement. The slide was heated for denaturation for 3 min in an 80°C oven and then incubated for 14–18 h at 37°C. After hybridization the slide was washed in 2× SSC for 5 min at room temperature, 30% formamide in 2× SSC for 10 min at 42°C, 2× SSC for 5 min at room temperature, and 1× PBS (0.14 M NaCl, 8 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl at pH 7.4) for 10 min at room temperature. The slide was treated with 0.5% BSA in PBST for 30 min to reduce nonspecific binding of the 5mC antibodies. The slide was then incubated with the anti-5mC antibody diluted 1:200 in the blocking solution for 20–24 h at room temperature.
The 5mC antibody and biotin-labeled probes were detected with red and green, respectively, using three successive layers of antibodies— Layer 1: rabbit anti-mouse IgG (Jackson ImmunoResearch) 1:100 + avidin-FITC (Vector Labs) 1:100 in TNB; Layer 2: chicken anti-rabbit Alexafluor 594 (Invitrogen) 1:200 + biotinylated anti-avidin from goat (Vector Labs) 1:200 in TNB; and Layer 3: avidin-FITC 1:100 in TNB. All antibody incubations were done at 37°C; the first layer was incubated for 1 h and the last two for 45 min each. Antibody washes consisted of three successive washes, 5 min each at room temperature using TNT (0.1 M Tris-HCl, 0.15 M NaCl, 0.05% Tween 20 at pH 7.5). A final wash in 1× PBS was performed, and the slides were drained and mounted in Vectashield without counterstaining dyes. After recording the fluorescence signals, the slides were washed in 4T (4× SSC/0.05% Tween 20) buffer for 1 h at 37°C and dehydrated in an ethanol series. The slides were reprobed with new FISH probes to detect additional sequences on the same fibers.
The fluorescence signals were captured using a Hamamatsu CCD camera. The images were processed with Meta Imaging Series 7.5 software using an Olympus BX51 epifluorescence microscope equipped with FITC-Cy3-Cy5-DAPI four-way filter sets (Olympus). The final contrast of the images was processed using Adobe Photoshop CS3 software.
Acknowledgments
We thank R. Phillips and H. Rines (University of Minnesota) for providing the oat–maize B chromosome addition line. This research was supported by grants DBI-0421671 and DBI-0922703 from the National Science Foundation.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.116202.110.
References
- Alfenito MR, Birchler JA 1993. Molecular characterization of a maize B chromosome centric sequence. Genetics 135: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Karpen GH 2008. Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat Rev Genet 9: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev EV, Phillips RL, Rines HW 1998. Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc Natl Acad Sci 95: 13073–13078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M, Sanyal K, Mishra PK, Thaler N, Carbon J 2006. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc Natl Acad Sci 103: 14877–14882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Brock MA, Bédard S, Woods VL Jr, Cleveland DW 2007. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci 104: 5008–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH 2002. Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2: 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SIS 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Cheng ZK, Stupar RM, Gu MH, Jiang JM 2001. A tandemly repeated DNA sequence is associated with both knob-like heterochromatin and a highly decondensed structure in the meiotic pachytene chromosomes of rice. Chromosoma 110: 24–31 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Dong F, Langdon T, Ouyang S, Buell CB, Gu MH, Blattner FR, Jiang J 2002. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14: 1691–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. 2010. Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueh AC, Wong LH, Wong N, Choo KH 2005. Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere. Hum Mol Genet 14: 85–93 [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Migeon BR 1985. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma 92: 290–296 [DOI] [PubMed] [Google Scholar]
- Fransz PF, Alonso-Blanco C, Liharska TB, Peeters AJM, Zabel P, de Jong JH 1996. High-resolution physical mapping in Arabidopsis thaliana and tomato by fluorescence in situ hybridization to extended DNA fibres. Plant J 9: 421–430 [DOI] [PubMed] [Google Scholar]
- Gong ZY, Yu HX, Huang J, Yi CD, Gu MH 2009. Unstable transmission of rice chromosomes without functional centromeric repeats in asexual propagation. Chromosome Res 17: 863–872 [DOI] [PubMed] [Google Scholar]
- Han FP, Lamb JC, Birchler JA 2006. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci 103: 3238–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han FP, Gao Z, Yu WC, Birchler JA 2007. Minichromosome analysis of chromosome pairing, disjunction, and sister chromatid cohesion in maize. Plant Cell 19: 3853–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han FP, Gao Z, Birchler JA 2009. Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell 21: 1929–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS 2001. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293: 1098–1102 [DOI] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K 2008. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321: 1088–1091 [DOI] [PubMed] [Google Scholar]
- Jackson SA, Wang ML, Goodman HM, Jiang JM 1998. Application of fiber-FISH in physical mapping of Arabidopsis thaliana. Genome 41: 566–572 [PubMed] [Google Scholar]
- Jansen LET, Black BE, Foltz DR, Cleveland DW 2007. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JM, Gill BS, Wang GL, Ronald PC, Ward DC 1995. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci 92: 4487–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JM, Birchler JA, Parrott WA, Dawe RK 2003. A molecular view of plant centromeres. Trends Plant Sci 8: 570–575 [DOI] [PubMed] [Google Scholar]
- Jin W, Melo JR, Nagaki K, Talbert PB, Henikoff S, Dawe RK, Jiang J 2004. Maize centromeres: Organization and functional adaptation in the genetic background of oat. Plant Cell 16: 571–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin WW, Lamb JC, Vega JM, Dawe RK, Birchler JA, Jiang JM 2005. Molecular and functional dissection of the maize B centromere. Plant Cell 17: 1412–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC 2009. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science 324: 1716–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisugi Y, Nakayama S, Nakajima R, Ohtsubo H, Ohtsubo E, Fukui K 1994. Physical mapping of the 5S ribosomal RNA genes on rice chromosome 11. Mol Gen Genet 245: 133–138 [DOI] [PubMed] [Google Scholar]
- Koo DH, Jiang JM 2009. Super-stretched pachytene chromosomes for fluorescence in situ hybridization mapping and immunodetection of DNA methylation. Plant J 59: 509–516 [DOI] [PubMed] [Google Scholar]
- Kynast RG, Riera-Lizarazu O, Vales MI, Okagaki RJ, Maquieira SB, Chen G, Ananiev EV, Odland WE, Russell CD, Stec AO, et al. 2001. A complete set of maize individual chromosome additions to the oat genome. Plant Physiol 125: 1216–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JC, Kato A, Birchler JA 2005. Sequences associated with A chromosome centromeres are present throughout the maize B chromosome. Chromosoma 113: 337–349 [DOI] [PubMed] [Google Scholar]
- Lermontova I, Schubert V, Fuchs J, Klatte S, Macas J, Schubert I 2006. Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18: 2443–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KHA 2008. Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 82: 261–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR 2005. Stable barley chromosomes without centromeric repeats. Proc Natl Acad Sci 102: 9842–9847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizzi C, Clarke L 1991. The chromatin structure of centromeres from fission yeast: Differentiation of the central core that correlates with function. J Cell Biol 112: 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, Black BE 2010. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature 467: 347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH 2004. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S 1995. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet 4: 2189–2197 [DOI] [PubMed] [Google Scholar]
- Topp CN, Okagaki RJ, Melo JR, Kynast RG, Phillips RL, Dawe RK 2009. Identification of a maize neocentromere in an oat–maize addition line. Cytogenet Genome Res 124: 228–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BC, Murphy TD, Goldberg ML, Karpen GH 1998. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat Genet 18: 30–37 [DOI] [PubMed] [Google Scholar]
- Yan H, Talbert PB, Lee HR, Jett J, Henikoff S, Chen F, Jiang J 2008. Intergenic locations of rice centromeric chromatin. PLoS Biol 6: e286 doi: 10.1371/journal.pbio.0060286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Kikuchi S, Neumann P, Zhang W, Wu Y, Chen F, Jiang J 2010. Genome-wide mapping of cytosine methylation revealed dynamic DNA methylation patterns associated with genes and centromeres in rice. Plant J 63: 353–365 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zhang WL, Lee HR, Koo DH, Jiang JM 2008. Epigenetic modification of centromeric chromatin: Hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell 20: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WL, Friebe B, Gill BS, Jiang JM 2010. Centromere inactivation and epigenetic modifications of a plant chromosome with three functional centromeres. Chromosoma 119: 553–563 [DOI] [PubMed] [Google Scholar]
- Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, Nagaki K, Birchler JA, Jiang J, Dawe RK 2002. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14: 2825–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S 2007. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39: 61–69 [DOI] [PubMed] [Google Scholar]