Abstract
Total transcript amplification (TTA) from single eukaryotic cells for transcriptome analysis is established, but TTA from a single prokaryotic cell presents additional challenges with much less starting material, the lack of poly(A)-tails, and the fact that the messages can be polycistronic. Here, we describe a novel method for single-bacterium TTA using a model organism, Burkholderia thailandensis, exposed to a subinhibitory concentration of the antibacterial agent, glyphosate. Utilizing a B. thailandensis microarray to assess the TTA method showed low fold-change bias (less than twofold difference and Pearson correlation coefficient R ≈ 0.87–0.89) and drop-outs (4%–6% of 2842 detectable genes), compared with data obtained from the larger-scale nonamplified RNA samples. Further analysis of the microarray data suggests that B. thailandensis, when exposed to the aromatic amino acid biosynthesis inhibitor glyphosate, induces (or represses) genes to possibly recuperate and balance the intracellular amino acid pool. We validated our single-cell microarray data at the multi-cell and single-cell levels with lacZ and gfp reporter-gene fusions, respectively. Sanger sequencing of 192 clones generated from the TTA product of a single cell, with and without enrichment by elimination of rRNA and tRNA, detected only B. thailandensis sequences with no contamination. These data indicate that RNA-seq of TTA from a single cell is possible using this novel method.
Innovative methods in single-cell technology are needed to enhance the investigations of Bacteria and Archaea genomic material, particularly if we are to develop deeper insights into the functional and metafunctional genomics of these “prokaryotes.” Functional-genomics or transcriptomics of a single-cell can produce a wealth of information at resolutions that cannot be attained by analysis of multi-cell populations or communities. Such advances hinge on the development of innovative methods for single-cell isolation (Podar et al. 2009) and transcript amplification from a minute amount of starting material with low gene expression bias. Single eukaryotic cell mRNA amplification methods for transcriptome analysis, via microarray (Kurimoto et al. 2007; Scanlon et al. 2009) and mRNA sequencing (Tang et al. 2009), have recently been described. These existing methods of transcript amplification, pioneered for eukaryotic transcript amplification, involve multiple rounds of exponential (Kurimoto et al. 2006) and/or linear (Scanlon et al. 2009) amplification of cDNA. However, no study has described total transcript amplification (TTA) from a single bacterium, possibly due to the major challenges one may encounter when working with single-bacterium TTA. These challenges include (1) the low amount of RNA (∼0.1–2 pg/prokaryotic cell vs. ∼10–50 pg/eukaryotic cell); (2) the lack of poly(A)-tails for ease of tagging and mRNA amplification; and (3) the fact that the messages can be polycistronic, and full-length amplification is critical for detecting expression of all genes in an operon. Due to these characteristics of prokaryotic transcripts, our experience with TTA using existing linear and exponential amplification methods for single bacterial cells shows the methods are labor intensive and yield unusable data with extensive gene expression bias and low reproducibility. If these challenges could be resolved, one could envisage numerous applications that may provide a wealth of functional-genomic information that was not previously possible (Supplemental Fig. S1).
Here, we describe a novel method for TTA from a single prokaryotic cell. Using Burkholderia thailandensis as a model bacterium exposed to a subinhibitory concentration of the antibacterial agent glyphosate (GS) (Norris et al. 2009), we developed a novel method for TTA using ϕ29 polymerase multiple displacement amplification (MDA) of circularized cDNA. We used microarray to assess the reproducibility, level of gene expression bias, and gene presence that resulted from this novel method. This low bias and simple single-tube method is reproducible and is not labor intensive. The data yielded a less than twofold difference in fold-changes compared with the nonamplified samples. In a typical experiment, we could amplify and detect ∼94%–96% of the detectable transcripts (2842 genes) from a single cell by microarray. Exposure to GS up- or down-regulates many genes, resulting from GS inhibition of aromatic amino acid biosynthesis, to possibly compensate for amino acid imbalance. From the microarray data obtained through TTA of single cells exposed to GS versus no GS, we randomly picked five up-regulated genes, three down-regulated genes, and two control genes that showed no fold-change to validate our microarray data by reporter-gene fusions. We propose that this novel method can be applied to RNA-seq and will stimulate various important prokaryotic research areas that require single-cell level transcriptome analysis (Supplemental Fig. S1).
Results
Single-cell isolation and amplification method
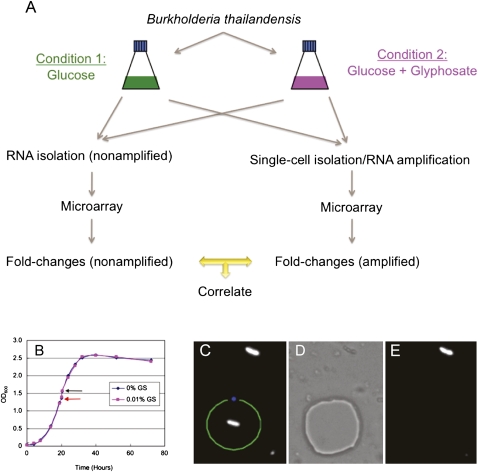
We utilized laser capture microdissection (Emmert-Buck et al. 1996) to isolate B. thailandensis single cells, followed by B. thailandensis microarray analysis to assess our single-cell TTA method. Although various single-cell isolation techniques have been described (Podar et al. 2009), we chose to use the Zeiss Laser Capture Microdissection (LCM) MicroBeam IV system (hereafter referred to as the Zeiss LCM) to isolate single B. thailandensis cells grown in 1× M9 minimal glucose media (MG) ± GS (Fig. 1). We have recently discovered that B. thailandensis is very sensitive to the herbicide GS (Norris et al. 2009) because bacteria, in the presence of GS, cannot synthesize aromatic amino acids (Fischer et al. 1986). At a subinhbitory GS concentration of 0.01% compared with no GS, there is no apparent difference in growth rate or final cell density, which renders GS exposure appropriate as a model for gene-expression analysis between these two growth conditions (Fig. 1B). Our approach (Fig. 1A) was to perform large-scale RNA preparation from each of the two cultures (nonamplified samples). Single cells were then isolated from both cultures (Fig. 1C–E) and subjected to our single-cell TTA method (amplified samples). Microarray experiments were then performed to obtain gene expression fold-change (between the two growth conditions, i.e., ± GS) for both the amplified and the nonamplified samples, which were then compared to assess the suitability of the TTA method (Fig. 1A). We expected that genes differentially expressed between the two conditions from the amplified RNA samples would correlate, with minimal fold-change bias, to data from the large-scale nonamplified RNA preparations from more than 109 bacteria.
Figure 1.
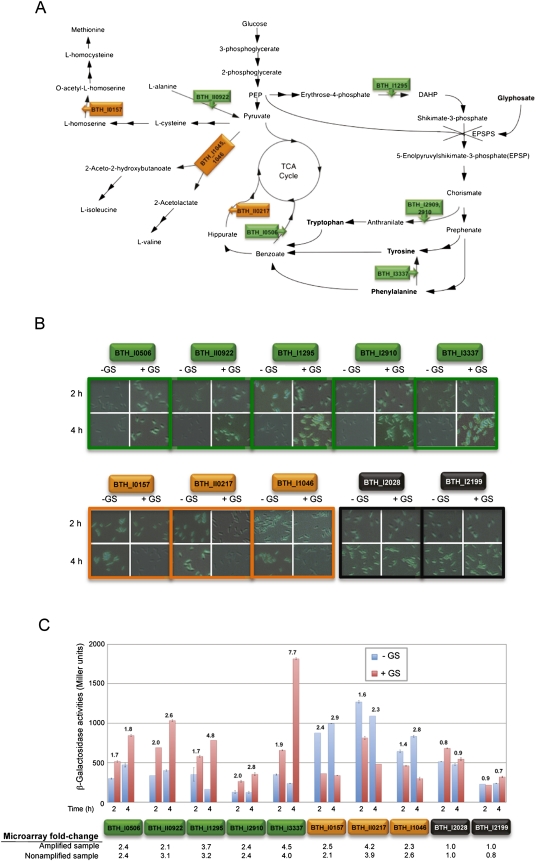
Single B. thailandensis cell isolation. (A) Experimental design for evaluating the single-cell transcript amplification method. B. thailandensis grown in two different conditions were used in large-scale (nonamplified) and single-cell level (amplified) microarray analysis. Fold-changes (between condition 1 and 2) of all genes detected from the nonamplified and amplified samples were then compared by correlation analysis. (B) Comparable growth curves of B. thailandensis in MG medium ± 0.01% GS (w/v) added at mid-log phase (red arrow) and harvested 30 min post-exposure (black arrow). (C) Fluorescent B. thailandensis cells were observed under 1000× magnification. The section of the membrane containing a single bacterium was drawn and cut by the focused laser (green line) and catapulted at a distance from the cell with unfocused low-intensity laser beam (blue spot), which aseptically catapulted and isolated the single cell into the lid of a 0.2-mL PCR tube containing the cell lysis buffer. (D) Bright-field mode showing the section of the membrane where the single bacterium had been. (E) Fluorescence mode confirming that the bacterium of interest has been transferred from the membrane slide to the PCR tube lid.
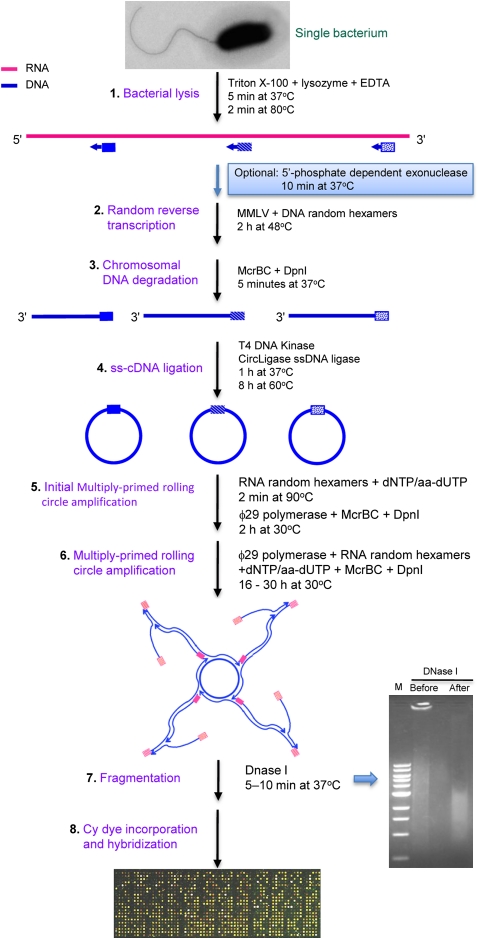
We devised a novel method for single-cell TTA, using ϕ29 polymerase for MDA of circularized cDNA (Fig. 2). Full details of the method can be found in the Methods section. Briefly, single B. thailandensis cells were isolated (Fig. 1C–E) and subjected to a lysis step with the nonionic detergent Triton X-100, EDTA, and lysozyme (Fig. 2). At this point, an optimal 5′-phosphate–dependent exonuclease step can be incorporated to remove rRNA and tRNA to enrich for mRNA, if the end product of the TTA method is to be used for deep sequencing (see Methods and below). Otherwise, cDNA synthesis was performed thereafter with DNA random hexamers and MMLV reverse transcriptase. McrBC and DpnI were added to remove bacterial chromosomal DNA and any potentially contaminating DNA, which are methylated and will be digested by these enzymes. The newly synthesized single-stranded (ss) cDNA (ss-cDNA) was 5′-end phosphorylated and ligated intramolecularly with CircLigase ssDNA ligase, which will not ligate ss-DNA <15 bp (e.g., excess random hexamers). The circularized ss-cDNA was then randomly primed with RNA hexamers and subjected to MDA with ϕ29 DNA polymerase. The use of thiophosphate-linked RNA random hexamers is critical to reduce falsely primed DNA by-product and primer dimers, while not interfering with the DNA synthesis (Takahashi et al. 2009). The thiophosphate-linked RNA random hexamers are also stable in the presence of ϕ29 DNA polymerase RNase activity (Lagunavicius et al. 2008). The ϕ29 DNA polymerase amplification was split into two steps (steps 5 and 6, Fig. 2), where the initial step was performed in a small total volume (∼10 μL) to increase the template and substrate concentrations. The second MDA step was performed by adding another 90 μL of a ϕ29 DNA polymerase and substrate mixture to increase the cDNA yield. As a precaution, we included McrBC and DpnI endonucleases again to prevent amplification of potentially contaminating DNA during the MDA steps of circularized cDNA. The highly polymerized double-stranded (ds) cDNA (ds-cDNA), labeled with 5-(3-aminoallyl)-2′-deoxyuridine-5′-triphosphate (aa-dUTP), was fragmented to ∼1–4 kb and incorporated with Cy dyes for microarray analysis (Fig. 2).
Figure 2.
Single-bacterium total transcript amplification strategy. This strategy is developed based on multiply primed rolling circle amplification of circularized cDNA using ϕ29 DNA polymerase. Blue boxes indicate DNA random hexamers, and pink boxes indicate RNA thiophosphate-linked random hexamers. The purpose and components of each step are indicated (for further details, see text). Abbreviations are as follows: aa-dUTP indicates 5-(3-aminoallyl)-2′-deoxyuridine-5′-triphosphate; dNTP, deoxyribonucleotide triphosphate; DpnI, restriction endonuclease from Diplococcus pneumoniae; EDTA, ethylenediaminetetraacetic acid; ϕ29 polymerase, DNA polymerase from a Bacillus subtilis phage Phi29; M, 1 kb DNA marker from New England BioLabs; McrBC, E. coli homing endonuclease; MMLV, Moloney murine leukemia virus reverse transcriptase; and ss-cDNA, single-stranded cDNA.
There are several advantages of this novel single-cell TTA method. In this study, the ϕ29 DNA polymerase provides rapid and efficient amplification from starting materials of a single cell with minimal fold-change bias (below). Several strategies were employed to eliminate template-independent amplification, including the use of thiophosphate-linked RNA random hexamers as previously described (Takahashi et al. 2009), elimination of exogenous DNA contamination with endonucleases (McrBC and DpnI), and minimization of the reaction volume (10 μL for initial amplification and 100 μL total). All manipulations were performed from beginning to end in a single tube in a PCR chamber. In addition, this simple method is not labor intensive and requires very few manipulation steps, thereby reducing potential manipulation-derived contaminants. The typical yield of ds-cDNA from a single cell is ∼25–30 μg, sufficient for microarray analysis (Fig. 3). If more ds-cDNA is required and further amplification is necessary for the technical microarray replicates below, additional ϕ29 DNA polymerase and substrate mixture can be added to the original amplification tube containing ∼25–30 μg of ds-cDNA. This will increase ds-cDNA yield to ∼75–90 μg.
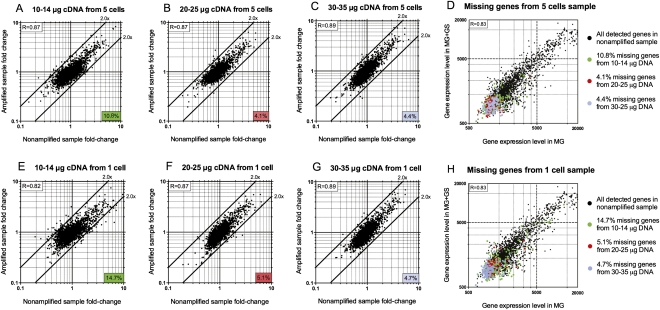
Figure 3.
Fold-change scatter plots of expressed genes obtained from nonamplified versus amplified samples. 10–14 μg (A), 20–25 μg (B), or 30–35 μg (C) of DNA amplified from five-cell samples were hybridized to different slides, and the fold-changes of detected genes were plotted against those obtained from the nonamplified sample. The number located at the right bottom corner of each plot indicates the percentage of missing genes (drop-outs) from each amplified sample compared with the nonamplified sample (2842 genes total). (D) Gene expression levels from the nonamplified sample (black dots) were compared between two growth conditions (MG ± 0.01% GS). Expression levels of genes that were missing in the five-cell amplified samples are colored green (as a result of using 10–14 μg of cDNA), red (using 20–25 μg of cDNA), or purple (using 30–35 μg of cDNA), and are overlaid on the same graph in D. Similar comparisons were conducted with different amounts of cDNA amplified from one-cell samples: 10–14 μg (E), 20–25 μg (F), or 30–35 μg (G). Missing genes or drop-outs from each sample were color-coded similarly and overlaid with the total number of genes detected in the nonamplified samples (H). The R value in the upper left corner of each plot represents the Pearson correlation coefficient. All microarray experiments in this figure were conducted without the optional mRNA enrichment step.
Microarray analysis of genes expressed at the single-cell level
Our final TTA method amplifies and detects ∼94%–96% of the total transcripts detectable by microarray (2842 genes), using from five cells down to the single-cell level. Although ∼10 μg of labeled ss-cDNA is sufficient for nonamplified samples in a typical microarray experiment, our amplified product is ds-cDNA. Therefore, to optimize single-cell microarray experiments for our amplified samples, we initially performed TTA on groups of five B. thailandensis cells and used different amounts of the amplified cDNA for each microarray slide (Fig. 3). When 10–14 μg of amplified cDNA from TTA of the five cells was used, ∼10.8% of the transcripts were not amplified to an amount detectable by microarray, while expression of these genes was detectable in the nonamplified samples (Fig. 3A). Increasing the amount of amplified cDNA used in microarray experiments to 20–25 μg reduced “drop-outs” to ∼4.1% (Fig. 3B). Further increasing the amount of amplified cDNA used to 30–35 μg did not significantly reduce the percentage of genes that was missing from the amplified samples (∼4.4%) (Fig. 3C). We next attempted amplification of total transcript from a single cell for microarray analysis, and the results were once again significantly improved at the higher concentration of cDNA used (Fig. 3E–G). To determine the reason for these missing genes (drop-outs) from the microarray data in the amplified samples, we examined their GC content, operon size, RNA secondary structure, or gene expression variation among the individual cells of the population (Supplementary Analysis). These analyses revealed these missing genes resulted from their low expression levels, which suggests the low abundance of their transcripts causes “drop-out” of these genes in the amplified samples (Fig. 3D,H). Although there were good Pearson correlations between the fold-change of the amplified and nonamplified samples in all these experiments (Fig. 3A–C, E–G), we recommend using 30 μg of amplified cDNA in a typical microarray experiment, as this amount of cDNA used yielded a minimal number of genes absent from the microarray data (∼4%–5% drop-outs) (Fig. 3C,G).
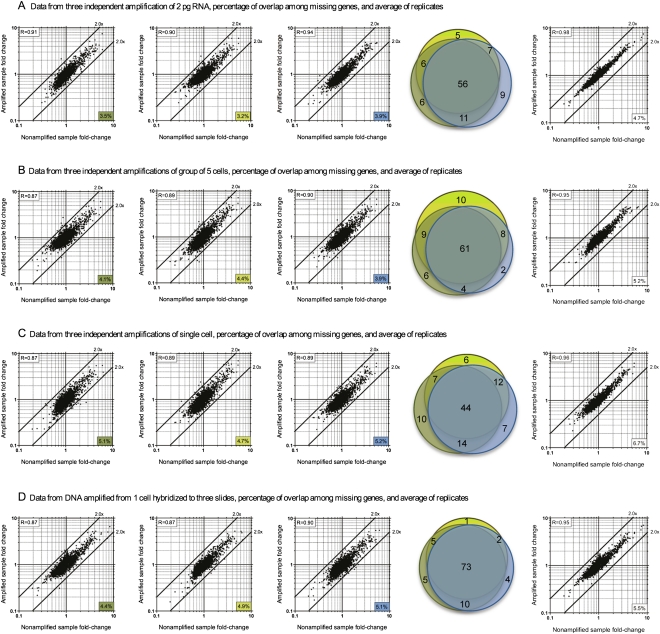
Having optimized the amount of amplified cDNA (30 μg) to use in microarray experiments, we performed biological (Fig. 4A–C) and technical replicates (Fig. 4D) to obtain fold-change data from 2 pg of diluted total RNA to an actual single cell. Initially, microarray experiments were performed in triplicate using three independent amplifications of 2 pg of purified and diluted RNA. We correlated the fold-change of the amplified to nonamplified sample (Figs. 1A, 4A). Each amplified replicate produces ∼3.2%–3.9% gene drop-outs, and the Venn diagram for the missing genes among these replicates indicated significant overlap among these low abundance transcripts (Fig. 4A). The averaged data from the triplicates showed excellent Pearson correlation of the fold-change to those of the nonamplified samples (R = 0.98) (Fig. 4A). A total of 4.7% drop-out genes in the averaged data indicated that we successfully amplified and detected ∼95% of the total transcript expressed relative to the nonamplified sample when starting with 2 pg of purified total RNA. Similar results were observed for three groups of five cells (Fig. 4B). When microarray data were obtained from three individual single cells as biological replicates, comparable fold-change correlations and percentages of missing genes were observed (first three graphs of Fig. 4C). The averaged data showed excellent correlations (R = 0.96), with 93.3% of the transcripts from single cells amplified and detected compared with the nonamplified sample (last graph of Fig. 4C). Technical replicates, where single-cell transcripts were amplified to sufficient level for three microarray slides, were relatively reproducible, confirming the consistency among different microarray experiments (Fig. 4D). An important point to note is that, in the averaged data, there was very little fold-change bias or skewing as a result of the amplification of transcripts from a single cell, and fold-change variations of all detectable genes were significantly less than a twofold difference (i.e., all dots lie well within the 2× difference lines in Fig. 4). The averaged data set from three TTA replicates of a single cell (Fig. 4C) indicated that reliable data could be generated by using this TTA method, and >93% of the transcripts from a single cell could be amplified and detected in microarray experiments.
Figure 4.
Microarray data fold-change comparison of nonamplified and amplified samples starting from 2 pg of diluted RNA (A), five cells (B), or one cell (C) as biological replicates; or a single cell hybridized to three different slides as technical replicates (D). The first three plots of each item are biological replicates (A–C) or technical replicates (D). The number in the bottom right corner of each plot indicates the percentage of genes that were missing in the amplified samples compared to the nonamplified samples. The Pearson correlation coefficient between the amplified and nonamplified fold-change data is shown at the upper left corner of each plot. The high correlation coefficient values (P < 0.0001) and the tight grouping of the dots within the twofold difference boundaries suggest a relatively low bias. The percentages of overlap among missing genes from each group are displayed as area-proportional Venn diagrams of three independent biological (A–C) or technical replicates (D). The color for each circle in the Venn diagram corresponds to the colored boxes in each scatter plot. The last plot of each item shows averaged data from the three biological (A–C) or technical replicates (D). All microarray experiments in this figure were performed without the optional mRNA enrichment step.
Correlation of the gene expression levels between amplified single cells versus nonamplified control and independent single cell amplifications was also analyzed (Supplemental Fig. S2). Expression levels from single cell amplified and nonamplified samples were poorly correlated (Supplemental Fig. S2A,B), indicating significant amplification bias, probably due to different amplification efficiencies among individual genes. However, correlations among transcripts amplified from independent single cells were significantly higher (Supplemental Fig. S2C,D), indicating more consistent amplification efficiencies for any particular gene in the two cells. Thus, any amplification bias of different genes due to differences in amplification efficiencies would “cancel out,” when comparing gene expression levels in single cells of two conditions to obtain fold-change. Much like real-time RT-PCR comparison to obtain reliable gene expression fold-change, amplification efficiencies of a particular gene in two different conditions must be consistent, while amplification efficiency variations between different genes are acceptable. Thus, single cell gene expression profiling using the method described here should be performed and compared between single cells in two different conditions (e.g., GS and no GS). However, this method is not recommended for obtaining gene fold-changes by comparing between amplified and nonamplified samples.
GS-dependent gene expression by single-cell microarray analysis and validation via reporter gene fusions
With the averaged microarray data generated from the TTA of a single bacterium performed in technical triplicates (Fig. 4D), we assembled a list of genes that were induced or repressed in the presence of GS (Supplemental Tables S1, S2). We chose five genes up-regulated in the presence of GS, three genes down-regulated by GS, and two control genes with no fold-change (Supplemental Table S3) to perform reporter-gene fusions with lacZ and gfp for microarray data confirmation. The genes selected for microarray validation are involved in amino acid metabolism (Fig. 5A). After 2 and 4 h exposure to GS, increased green fluorescence signals and β-galactosidase activities indicated that the expression of the five genes up-regulated in our microarray data did increase as a result of GS (Fig. 5B,C). As indicated by the fluorescence signals and β-galactosidase activities, expression levels of the three genes down-regulated in the microarray data also decreased as a result of GS, while expression levels of the two control genes showed only insignificant changes (Fig. 5B,C). Overall, gene fusion experiments and the fold-change from the nonamplified sample microarray data showed strong agreement to the single-cell amplified samples microarray data (Fig. 5). Only a few genes in the single-cell data with a fold-change of two or more were less than two in the nonamplified data (potential false positives) (Supplemental Tables S1 and S2). However, the majority of the genes expressed comparably in the amplified and nonamplified samples, suggesting a strong and reliable correlation between the fold-change data generated from single cells and large-scale nonamplified samples (Fig. 4D; Supplemental Tables S1, S2).
Figure 5.
Validation of microarray data via reporter-gene fusions. (A) Proposed amino acid metabolism pathways influenced by glyphosate (GS) in B. thailandensis. Two connecting arrows indicate two or more reaction steps. The target for GS is 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) (Fischer et al. 1986). In A, B, and C, green and orange boxes indicate genes induced and repressed, respectively, by GS. (B,C) gfp and lacZ reporters were fused to five GS-induced genes, three GS-repressed genes, and two GS-insensitive control genes (housekeeping gene controls in black boxes). (B) Cells were examined under 630 × magnification at 2 and 4 h post-exposure to GS. Differential interference contrast (DIC) and green fluorescence images were merged, and the representative fields are shown. (C) At the same time points, β-galactosidase activities for these fusion strains were determined in triplicate, and the Miller units were plotted with the SEM. The numbers above the bars in the histogram in C indicate fold-induction or fold-repression differences by GS, as measured by β-galactosidase assays. For comparison, the microarray data fold-change of the corresponding genes from the amplified sample and the nonamplified sample are also displayed below the graph. As a general trend, these gene-fusion data agree with the microarray data at both 2 and 4 h post-GS-exposure.
Among the up-regulated genes in particular, three genes targeted for validation were responsible for aromatic amino acid (AAA) biosynthesis (BTH_I1295, BTH_I2909, and BTH_I3337) and two genes were responsible for shuffling benzoate and alanine to possibly replenish central pathway intermediates (BTH_I0506 and BTH_II0922) (Fig. 5A). Of the three genes down-regulated due to GS, two targeted genes convert pyruvate in the synthesis of other amino acids, potentially balancing other amino acid species (e.g., Met, Ile, and Val), and one gene was involved in conversion of TCA cycle intermediates (Fig. 5A). It seems that GS not only affects the EPSPS target (Fig. 5A) and reduces AAA synthesis but also has an overall influence on the amino acid pool. Reducing AAA signals a concurrent reduction in other amino acids (Met, Ile, Val, and Ala) to balance the cellular amino acid population.
Potential application to next-generation sequencing
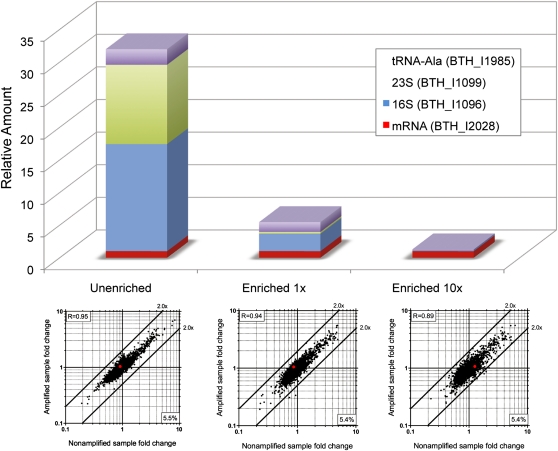
We next evaluated if this TTA method can be used for transcriptome analysis by deep sequencing. RNA-seq methods for prokaryotes require mRNA enrichment to eliminate rRNA and tRNA, which can be upward of 90%–95% of the total RNA in the cell, to reduce the number of reads and costs. Additionally, transcriptome profiling by deep sequencing is less tolerant of potential contaminants or nonspecific amplification. Therefore, we analyzed our TTA products using real-time RT-PCR and Sanger sequencing to assess the potential use of this method for transcriptome analysis by deep sequencing. An mRNA enrichment step was added to our amplification method to amplify transcripts from single cells (Fig. 2). By real-time RT-PCR, we then analyzed amounts of rRNAs (23S and 16S) and a tRNA (Ala) relative to the mRNA amount of one gene (BTH_I2028) before and after enrichment in amplified single cell samples grown in MG medium (Fig. 6). The amounts of rRNAs and one tRNA species were significantly reduced after enrichment, relative to the mRNA transcript of BTH_I2028 (Fig. 6). A microarray analysis was then performed on the enriched or unenriched samples amplified from single cells grown in MG medium (same as the real-time experiment above) and MG + GS medium. Data from the enriched sample showed low fold-change bias resulting from the enrichment, with an optimized amount of 5′-phosphate–dependent exonuclease (1× = 1 × 10−5 units of enzymes). However, a higher level of amplification bias was observed when more exonuclease (10× = 1 × 10−4 units of enzymes) was used, possibly due to nonspecific digestion of mRNA (Fig. 6).
Figure 6.
Evaluation of mRNA enrichment in amplified single cell samples. Top panel presents mRNA amount of a gene (BTH_I2028) relative to genes of 16S rRNA, 23S rRNA, and a tRNA-Ala detected by real-time RT-PCR. Unenriched represents amplified sample without the mRNA enrichment step. Enriched (1× or 10×) means treated with 1 × 10−5 U (as described in Methods) or 1 × 10−4 U of the Terminator 5′-Phosphate–Dependent Exonuclease, respectively. The relative transcriptional levels of rRNAs and tRNA are significantly higher than the BTH_I2028 gene (mRNA) in the unenriched sample but are greatly reduced in the enriched samples. Microarray analysis was performed for amplified samples (enriched and unenriched); fold-changes were compared to nonamplified samples as shown in the bottom plots. Fold-change correlation for the BTH_I2028 gene is indicated by the red dots. The number at the bottom right corner represents the percentage of transcripts that were missing in the single cell. Enrichment with 10× the amount of 5′-phosphate–dependent exonuclease resulted in a slightly higher fold-change bias as indicated by the Pearson correlation coefficient shown at the upper left corner for each plot.
Single-cell cDNA libraries were constructed to analyze the purity of our enriched and amplified ds-cDNA to determine the potential for RNA-seq application (Supplemental Fig. S3). Sanger sequencing reads of 96 independent clones from each of the unenriched and enriched cDNA libraries were analyzed, and ∼96% (188 out of 192) matched to B. thailandensis sequences (Supplemental Table S4). Approximately 4% did not match to any sequences in GenBank. This is typical in RNA-seq experiments, where ∼5% of sequences cannot be assigned (i.e., no match) (Yoder-Himes et al. 2009). This demonstrated the effectiveness of the strategies to eliminate template-independent amplification and exogenous DNA contamination. The significant reduction of rRNA sequences in the enriched sample was confirmed by both real-time RT-PCR and sequencing data, suggesting successful depletion of rRNAs in the enrichment step. Taken together, these results showed promise to apply our TTA method for transcriptome profiling by deep sequencing, which may allow detection of low abundance transcripts that were drop-outs in our less sensitive microarray analysis approach.
Discussion
There are currently no published methods for single prokaryotic cell TTA. However such a method is desperately needed if we are to expand multiple areas in prokaryote functional genomics (Supplemental Fig. S1). The model used in this study, B. thailandensis exposed to GS, was appropriate and sufficient for assessment of our TTA method because we could compare the transcriptomic fold-change data obtained from single cells to data obtained from the larger-scale nonamplified RNA samples. Since there were excellent microarray data correlations of fold-change between the amplified and nonamplified samples in this study and the differences were significantly less than twofold when three replicates were averaged (Fig. 4), we are confident that the fold-changes generated from TTA of single cells between two different conditions are reliable (Supplemental Tables S1, S2). In summary, we have developed a single-tube TTA method that is simple, requiring very few steps. This method yielded reproducible data, low fold-change bias, and a high number of genes efficiently amplified from a single prokaryotic cell (94%–96% presence or low transcript drop-out as detectable by microarray).
We envisage that the method described will be used for transcriptome analysis by deep sequencing (Croucher et al. 2009; Passalacqua et al. 2009) of the amplified ds-cDNA, with an optional step to eliminate rRNAs and tRNAs for enrichment of mRNA (Figs. 2, 6). Deep sequencing may increase the dynamic range of detectable genes expressed to include low or highly expressed genes that may not be detectable or differentiated by microarray (Wang et al. 2009), and this may decrease or eliminate drop-outs. The limitation of this method is that antisense transcripts are not detectable, because the final product of our TTA method is ds-cDNA, and thus it is not strand specific (Vivancos et al. 2010). Our method yielded ds-cDNA, which could potentially be used in tiling arrays, possibly identifying untranslated regions and novel operons (Toledo-Arana et al. 2009). Hence, this method, beyond microarray, has the potential to detect global gene expression of a single cell, novel operons, and novel coding and noncoding regions (Sorek and Cossart 2010) in both cultured and noncultured prokaryotes (Supplemental Fig. S1).
Finally, we envisage that our method can be applied to single eukaryotic cell transcript amplification. This could be performed by substituting the DNA random hexamers to a poly(T) oligo during cDNA synthesis, and an mRNA enrichment step is not necessary. The remaining steps of the protocol are otherwise identical.
Methods
Bacterial strains, media, and culturing conditions
Escherichia coli EPMax10B-lacIq-pir (Norris et al. 2010) was routinely used as a cloning strain. The B. thailandensis wild-type strain, E264 (Brett et al. 1998), and its derivatives were cultured in LB or 1× M9 minimal medium (Sambrook and Russell 2001) supplemented with 20 mM glucose (MG). For the B. thailandensis microarray and reporter-gene fusion studies (Fig. 5B,C), B. thailandensis wild-type strain E264 and derivatives were grown in MG medium + 1% (w/v) Brij-58 and exposed to a final concentration of 0.01% (w/v) GS. Brij-58 (1% w/v) was added to all cultures to prevent bacterial clumping during growth.
Molecular reagents
T4 polynucleotide kinase, dNTPs, MMLV (Moloney murine leukemia virus) reverse transcriptase, and endonucleases (McrBC and DpnI) were purchased from New England Biolabs. Ready-Lyse lysozyme, Terminator 5′-Phosphate–Dependent Exonuclease, CircLigase ssDNA ligase, and ϕ29 DNA polymerase were purchased from Epicentre Biotechnologies. Inorganic pyrophosphatase was purchased from Roche Applied Science. TRIzol reagent, RNaseOut reagent, RNase-free DNA random hexamers, and Live/Dead BacLight cell stain were purchased from Invitrogen. The RNeasy Mini Kit and RNAprotect reagent were purchased from Qiagen. DNA oligonucleotide primers and random RNA hexamers with five thiophosphate-linkages (6R5S) (Takahashi et al. 2009) were synthesized through Integrated DNA Technology (IDT). DNase I, aminoallyl-dUTP (aa-dUTP), and all the other chemicals used in this study were purchased from Sigma. Cy3 and Cy5 dyes were purchased from Amersham Biosciences.
Single bacterium level transcript preparation
Large-scale total RNA was extracted from B. thailandensis wild-type cells (Fig. 1A) using TRIzol reagent and purified with the RNeasy Mini Kit by following the manufacturer's total RNA cleanup protocol. Two picograms of RNA, representing the single bacterium level of RNA, was prepared by serially diluting the purified total RNA to a final concentration of 10 pg/μL in DEPC water containing 1 U/μL of RNaseOut. Then, 0.2 μL (2 pg) of the final dilution was directly added to 2 μL of the lysis buffer (100 mM Tris-Cl at pH 8.0, 200 mM KCl, 0.2 mM EDTA, 0.1% Triton X-100, 2 mM DTT, 0.04 U/μL RNaseOut, 2 × 10−7 U/μL Ready-Lyse lysozyme).
Single bacterium isolation
For single cell isolation, B. thailandensis cells were treated with RNAprotect reagent, and stained with Live/Dead BacLight fluorescent dyes. Stained cells were then smeared onto a PALM membrane Frameslide (Carl Zeiss), and observed under 1000× magnification on the Zeiss LCM system. Sections of membrane containing one or five fluorescent bacteria were cut by the focused laser and catapulted with unfocused low-intensity laser beam into 2 μL of the lysis buffer contained within a 0.2-mL PCR tube lid. The cDNA synthesis and amplification of the single cell total transcript were then performed as below.
Single bacterium transcript amplification
The B. thailandensis cells or diluted 2 pg total RNA was incubated in the lysis buffer for 5 min at 37°C, then heated for 2 min to 80°C. When necessary (e.g., RNA-seq is used rather than microarray), an optional mRNA enrichment step should be performed here. A 0.5 μL of the enrichment mixture (1 × 10−5 U Terminator 5′-Phosphate–Dependent Exonuclease and 5 nmol MgCl2) is added and incubated for 10 min at 37°C. A 2 μL aliquot of RT mixture was then added, consisting of 4 U RNaseOut, dNTPs (0.25 nmol each), preheated DNA random hexamers (0.2 pg for one cell, 1 pg for five cells, or 2 pg for 2 pg diluted total RNA), 10 nmol MgCl2, and 2 U MMLV. The reverse transcription was carried out for 2 h at 48°C. A 0.24 μL aliquot of endonucleases mixture (4 nmol GTP, 1 U McrBC, and 2 U DpnI) was then added to each tube, followed by incubation for 15 min at 37°C to degrade the chromosomal DNA. ss-DNA phosphorylation and ligation were accomplished by adding a 0.7 μL aliquot of the ss-DNA ligation mixture (0.25 nmol ATP, 12.5 nmol MnCl2, 1 U T4 polynucleotide kinase, 10 U CircLigase ss-DNA Ligase) to each tube followed by incubation for 1 h at 37°C and then for 8 h at 60°C for ligation. The circularized ss-DNA (∼5 μL) was then used as template in a two-step multiply primed rolling circle amplification (MPRCA) reaction as follows. First, a 25 mM dNTP/aa-dUTP stock was prepared following an established protocol, where a 1:2 dTTP:aa-dUTP ratio was used to efficiently label DNA with high GC% (http://pfgrc.jcvi.org/index.php/microarray/protocols.html). A 4.5 μL mixture (1× ϕ29 reaction buffer, 20 nmol DTT, 0.5 μL dNTPs/aa-dUTP stock, 15 pmol of RNA random hexamers 6R5S, and 4 nmol GTP) was then added, followed by heating for 2 min to 90°C then immediate cooling on ice. McrBC (2 U, 0.2 μL) was then added to each tube and incubated for 5 min at 37°C. Fifty units of ϕ29 polymerase (0.5 μL) was added, followed by incubation for 2 h at 30°C to initiate the MPRCA reaction in a small total volume (∼10 μL). After the initial MPRCA, a larger volume (90 μL) of the MPRCA mixture (1× ϕ29 reaction buffer, 360 nmol DTT, 4.5 μL dNTPs/aa-dUTP, 150 pmol RNA random hexamers, 100 U ϕ29 polymerase, 90 nmol GTP, 5 U McrBC, 10 U DpnI, and 20 U pyrophosphatase) was added, and the reaction was carried out for another 32 h at 30°C. After this step, the cDNA yield is typically 25–30 μg. For the higher yields (75–90 μg) required to perform microarray technical replicates, another 200 μL of the MPRCA mixture (1× ϕ29 reaction buffer, 800 nmol DTT, 10 μL dNTPs/aa-dUTP, 300 pmol RNA random hexamers, 200 U ϕ29 polymerase, 200 nmol GTP, 10 U McrBC, 20 U DpnI, and 40 U pyrophosphatase) was added to the reaction. The reaction mixture (300 μL total) was then aliquoted into three 0.2-mL PCR tubes and incubated for another 16 h at 30°C. After amplification, the reaction was stopped by heat-inactivation at 65°C for 10 min. The newly synthesized double-stranded cDNA was then randomly fragmented to 1–4 kb in length with DNase I and extracted with phenol/chloroform (1:1). Fragmented cDNA was precipitated with 0.1 volume of 3 M sodium acetate (pH 5.2) and 3 volumes of isopropanol and then incubated at −80°C for 1 h. The DNA pellet was then collected by centrifugation at 20,000g for 10 min, washed with 70% ethanol, and vacuum dried.
Two-color microarray and data analysis
For microarray analysis of the nonamplified samples, cDNA was synthesized from 10 μg of total RNA following established protocols (http://pfgrc.jcvi.org/index.php/microarray/protocols.html). Typically, 8–12 μg of nonamplified cDNA or 25–30 μg of amplified cDNA was used for each condition in microarray hybridization. Both the nonamplified and amplified cDNA were labeled with Cy3 (no-GS condition) and Cy5 (GS condition) dyes and then hybridized to the B. thailandensis 70-mer triplicate arrays (GEO accession number GPL7113) following the established protocols (http://pfgrc.jcvi.org/index.php/microarray/protocols.html). Microarray slides were scanned with a GenePix 4000B microarray scanner using GenePix Pro software 5.1. Individual TIFF images from each channel were processed with Spotfinder software 3.2.1 (available at http://www.tm4.org) to quantify the gene expression levels. Raw microarray data were processed and normalized by low intensity filtering, total intensity normalization, LOWESS normalization, standard deviation regularization, and in-slide replicate analysis using MIDAS software 2.21 (available at http://www.tm4.org). Finally, normalized gene expression data were used to generate data tables using MEV software 4.5.1 (available at http://www.tm4.org).
Fold-change data for all detected genes were obtained from triplicates of the nonamplified or amplified samples. GS-induced genes (fold-change ≥ 2, P ≤ 0.05) are summarized in Supplemental Table S1, and GS-repressed genes (fold-change ≥ 2, P ≤ 0.05) are summarized in Supplemental Table S2.
Scatter plots and Venn diagrams
GraphPad Prism 5 software was used generate all scatter plots and calculate the correlation coefficients in Figures 3, 4, and 6 and Supplemental Figure S2. The area-proportional Venn diagrams were drawn based on images generated using a free online software (http://bioinforx.com/free/bxarrays/venndiagram.php).
Gene assignment and pathway designation
Gene description assignment for some genes was assisted by reference to the Burkholderia Genome database (http://www.burkholderia.com). Genes involved in the GS pathway (Fig. 5A) were assigned according to the Kyoto Encyclopedia of Genes and Genomes (KEGG).
Microarray data validation via gene fusion studies
Eight genes predicted to be involved in the GS pathway (Fig. 5A) and two controls genes were chosen for validation of the microarray data. Two promoter-less reporter-genes, gfp and lacZ, were integrated downstream from each target gene via two-step λ red protein-mediated recombineering (data to be published elsewhere). Briefly, the gfp-pheS-gat fragment was amplified from pUCP28T-gfp-pheS-gat (laboratory plasmid) using oligos containing 40–45 bp homologous sequences to the downstream region of the target gene. The PCR product was then integrated downstream from the target gene after the stop codon to make a transcriptional fusion in the B. thailandensis chromosome via λ red protein-mediated recombineering. Positive integration was selected on 1× M9 minimal medium supplemented with 0.04% (w/v) GS and screened by PCR. Next, the second λ red protein-mediated recombineering was accomplished by introducing the lacZ fragment (with the 5′-end homologous to the downstream region of the gfp gene, and the 3′-end homologous to the downstream region of the gat gene) to replace the pheS-gat fragment. The final gfp-lacZ fusion strains were obtained via pheS counter-selection on cPhe-containing media, as previously described (Barrett et al. 2008), and confirmed by PCR using oligos which anneal to the target genes and the lacZ gene.
These newly engineered fusion strains were first grown in LB medium overnight. Cells were harvested by centrifugation, washed twice with 1× M9 minimal medium, and resuspended in the same medium. Resuspended cells were diluted 100× into MG medium + 1% (w/v) Brij, and two identical cultures of each fusion strain were grown to mid-log phase, at which point GS was added to one of the cultures to a final concentration of 0.01% (w/v). At 2 and 4 h post-exposure to GS, samples of both cultures (with and without GS) were taken for fluorescence microscopy and β-galactosidase assay. To immobilize bacteria for fluorescence microscopy, cells were mixed with warm (42°C) molten agarose to a final concentration of 0.1% (w/v) and were immediately mounted between glass slides and coverslips to solidify the agar. Fluorescence was observed under the 38HE filter set on a Zeiss Axio Observer D1 microscope, and images were recorded with an AxioCam MRc 5 camera. Differential interference contrast (DIC) and green fluorescence images were merged at the time of capture using Zeiss AxioVision software. Multiple images were captured for each sample, and representative fields are shown in Figure 5B. β-Galactosidase assay (Sambrook and Russell 2001) was performed in triplicate on these fusion strains, and average activities are shown in Figure 5C with SEM. For comparison of the fold-change data from the microarray and β-galactosidase assay, the GS-induced gene fold-changes were calculated as the gene expression levels in the presence of GS divided by those in the absence of GS; the GS-repressed or GS-insensitive gene fold-changes were calculated based on gene expression levels in the absences of GS, divided by gene expression levels in the presence of GS.
Microarray data accession number
Microarray data are available in the NCBI GEO repository under accession number GSE23419.
Real-time RT-PCR
Primers and probes for each target were designed using Integrated DNA Technologies Primer Quest software (http://www.idtdna.com) and are shown in Supplemental Table S5. Amplified ds-cDNA from isolated single cells grown in MG medium were used as template for real-time RT-PCR, which was performed as previously described (Son et al. 2007). Real-time PCR was conducted in eight replicates for each target. To control for variations between runs, all PCRs were performed at the same time in one 96-well plate.
We followed the previously reported data analysis method (Peirson et al. 2003) to provide more accurate quantitative real-time PCR data. Real-time RT-PCR data were averaged over eight replicates for each target, and fold-changes were calculated using DART-PCR (Peirson et al. 2003). Accordingly, the average efficiencies of each target are within 4.1% differences (<5%) and permit accurate analysis. The expression level of mRNA BTH_I2028 was taken as 1 for each amplified sample tested, and the amounts of tRNA and rRNAs were normalized relative to this value.
cDNA library construction for sequencing
For construction of the single-cell cDNA library, aliquots of the same preparation of amplified ds-cDNA samples for real-time RT-PCR above were used. Amplified ds-DNA (10 μg) from unenriched or enriched (1 × 10−5 U exonuclease) samples were randomly fragmented with DNase I, blunt-ended with T4 DNA polymerase and 1 mM dNTPs, and ran on a 1% agarose gel. Fragments in two size ranges (0.1–1.5 kb and 1.5–4.0 kb) were extracted from the agarose gel and cloned into the SmaI site of pUC19 vector for library construction (Supplemental Fig. S3). Sanger sequencing reads of the inserts were generated at a local sequencing facility and identified using a homology search with the BLASTn algorithm (http://www.ncbi.nlm.nih.gov).
Competing interest statement
A provisional patent application (no. 61383699, submitted September 16, 2010) has been filed for the single bacterium TTA method described in this paper. T.T.H., Y.K., and M.H.N. are named as the inventors on this patent.
Acknowledgments
This project was supported by Award no. AI065359 from the National Institute of Allergy and Infectious Diseases and by the Center of Biomedical Research Excellence grant P20RR018727 from the National Center for Research Resources of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official view of the funding agencies.
Authors' contributions: Y.K. performed the single-cell TTA, microarray analysis, real-time RT-PCR study, and cDNA library construction. M.H.N. performed the fluorescence microscopy experiments and contributed to data analysis. J.Z-S. contributed to data analysis. W.C.N. generated the B. thailandensis microarray slides, provided technical advice and performed critical reading of the manuscript. S.P.D. secured funding for the Carl Zeiss LCM system used in this study, assisted with the single cell isolation training, and critical editing of this manuscript. T.T.H. conceived of the method and supervised the experiments. T.T.H. and Y.K. wrote the manuscript.
Footnotes
[Supplemental material is available for this article. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE23419. The sequencing data have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. JG731867–JG732058.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.116103.110.
References
- Barrett AR, Kang Y, Inamasu KS, Son MS, Vukovich JM, Hoang TT 2008. Genetic tools for allelic replacement in Burkholderia species. Appl Environ Microbiol 74: 4498–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett PJ, DeShazer D, Woods DE 1998. Burkholderia thailandensis sp. nov., description of Burkholderia pseudomallei-like species. Int J Syst Bacteriol 48: 317–320 [DOI] [PubMed] [Google Scholar]
- Croucher NJ, Fookes MC, Perkins TT, Turner DJ, Marguerat SB, Keane T, Quail MA, He M, Assefa S, Bahler J, et al. 2009. A simple method for directional transcriptome sequencing using Illumina technology. Nucleic Acids Res 37: e148 doi: 10.1093/nar/gkp811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA 1996. Laser capture microdissection. Science 274: 998–1001 [DOI] [PubMed] [Google Scholar]
- Fischer R, Berry A, Gaines CG, Jensen RA 1986. Comparative action of glyphosate as a trigger of energy drain in eubacteria. J Bacteriol 168: 1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Ono Y, Uno KD, Yamada RG, Ueda HR, Saitou M 2006. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res 34: e42 doi: 10.1093/nar/gkl1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Saitou M 2007. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc 2: 739–752 [DOI] [PubMed] [Google Scholar]
- Lagunavicius A, Kiveryte Z, Zimbaite-Ruskuliene V, Radzvilavicius T, Janulaitis A 2008. Duality of polynucleotide substrates for Phi29 DNA polymerase: 3′→5′ RNase activity of the enzyme. RNA 14: 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris MH, Kang Y, Lu D, Wilcox BA, Hoang TT 2009. Glyphosate resistance as a novel select-agent-compliant, non-antibiotic selectable marker in chromosomal mutagenesis of the essential genes asd and dapB of Burkholderia pseudomallei. Appl Environ Microbiol 75: 6062–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris MH, Kang Y, Wilcox B, Hoang TT 2010. Stable site-specific fluorescent tagging constructs optimized for Burkholderia species. Appl Environ Microbiol 76: 7635–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passalacqua KD, Varadarajan A, Ondov BD, Okou DT, Zwick ME, Bergman NH 2009. Structure and complexity of a bacterial transcriptome. J Bacteriol 191: 3203–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirson SN, Butler JN, Foster RG 2003. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res 31: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M, Keller M, Hugenholtz P 2009. Single cell whole genome amplification of uncultivated organisms. In Uncultivated microorganisms (ed. Epstein SS), Vol. 10, pp. 83–99 Springer-Verlag, Berlin/Heidelberg/New York [Google Scholar]
- Sambrook J, Russell DW 2001. Molecular cloning: A laboratory manual.2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Scanlon MJ, Ohtsu K, Timmermans MC, Schnable PS 2009. Laser microdissection-mediated isolation and in vitro transcriptional amplification of plant RNA. Curr Protoc Mol Biol Chapter 25: Unit25.A.3 doi: 10.1002/0471142727.mb25a03s87 [DOI] [PubMed] [Google Scholar]
- Son MS, Matthews WJJ, Kang Y, Nguyen DT, Hoang TT 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun 75: 5313–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Cossart P 2010. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat Rev Genet 11: 9–16 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Yamamoto K, Ohtani T, Sugiyama S 2009. Cell-free cloning using multiply-primed rolling circle amplification with modified RNA primers. Biotechniques 47: 609–615 [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. 2009. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6: 377–382 [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, et al. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459: 950–956 [DOI] [PubMed] [Google Scholar]
- Vivancos AP, Guell M, Dohm JC, Serrano L, Himmelbauer H 2010. Strand-specific deep sequencing of the transcriptome. Genome Res 20: 989–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder-Himes DR, Chain PS, Zhu Y, Wurtzel O, Rubin EM, Tiedje JM, Sorek R 2009. Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc Natl Acad Sci 106: 3976–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]