Abstract
High-volume sequencing of DNA and RNA is now within reach of any research laboratory and is quickly becoming established as a key research tool. In many workflows, each of the short sequences (“reads”) resulting from a sequencing run are first “mapped” (aligned) to a reference sequence to infer the read from which the genomic location derived, a challenging task because of the high data volumes and often large genomes. Existing read mapping software excel in either speed (e.g., BWA, Bowtie, ELAND) or sensitivity (e.g., Novoalign), but not in both. In addition, performance often deteriorates in the presence of sequence variation, particularly so for short insertions and deletions (indels). Here, we present a read mapper, Stampy, which uses a hybrid mapping algorithm and a detailed statistical model to achieve both speed and sensitivity, particularly when reads include sequence variation. This results in a higher useable sequence yield and improved accuracy compared to that of existing software.
With ever increasing throughput of next-generation sequencing machines (Metzker 2010), time- and memory-efficient algorithms need no justification. However, sequence error rates are low, so why is sensitivity important? One answer is that reduced sensitivity in the presence of variation leads to undesired mapping biases, particularly for reads from regions of higher divergence and for reads containing indels. Similarly, improved sensitivity may enable analyses that are otherwise impossible, for example, to analyze samples that are divergent from available reference genomes or to help identify unknown splice donor and acceptor sites in mRNA-seq experiments (Wang et al. 2009). Finally, in any experiment, a fraction of reads will exhibit elevated error rates, and being able to reliably include data from these reads improves the power of downstream analyses and reduces the total cost of sequencing.
The read mapping algorithms underlying existing mappers largely come in two varieties. One category is hash-based, hashing either reads (MAQ [Li et al. 2008], ELAND [Cox 2007]) or the reference genome (Novoalign [www.novocraft.com], Mosaik [Quinlan et al. 2008]). A second category is based on the Burrows-Wheeler transform and associated data structures, which support fast retrieval of long inexact string matches. Mappers that use the Burrows-Wheeler transform (BWA [Li and Durbin 2009], SOAP2 [Li et al. 2009], Bowtie [Langmead et al. 2009]) are very fast but tend to be less sensitive than are the best hash-based mappers.
To achieve good sensitivity, Stampy also uses a hash table, representing the location of selected 15-mers in the reference genome. The hash table uses a novel data structure, which results in improved search times compared with those of standard implementations and in the efficient use of the available memory. The algorithm first identifies candidate mapping locations for each read using the hash. Specifically, the hash is searched for every overlapping 15-mer in the read, as well as their neighbors at one mismatch removed. For a 36-bp read, for example, this results in 1012 (22 × 46) search operations. The candidate mapping locations are filtered for sufficient sequence similarity to the read, and then an attempt is made to align the read to the reference at each qualifying location. A fast gapped aligner is used, which respects quality scores and considers short indels of up to 15 bp. Next, for a mate pair, the results of its alignment are considered. Finally, candidate reads or read pairs are realigned using a full probabilistic aligner that considers indels up to, by default, 30 bp. Full details of the algorithm are provided in the online Supplemental material.
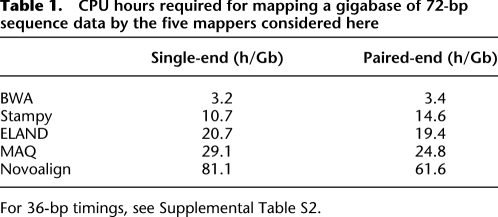
The resulting algorithm is sensitive and about as fast as MAQ (Supplemental Table S2). To achieve higher throughput, Stampy is recommended to be used in a hybrid mode, in which BWA is used to map the majority of reads that have a close representative in the reference. This results in a significant improvement in speed (Table 1; Supplemental Table S2), with no reduction in sensitivity. In fact, because of their fundamentally different algorithms, BWA and Stampy have somewhat complementary strengths; a particular strength of BWA, resulting from the use of the Burrows-Wheeler data structure, is in mapping highly repetitive reads that include sequence variation. Using BWA as a first stage allows Stampy to combine the advantages of BWA and its own algorithm, resulting in a further improvement in sensitivity and accuracy (data not shown).
Table 1.
CPU hours required for mapping a gigabase of 72-bp sequence data by the five mappers considered here
For 36-bp timings, see Supplemental Table S2.
To map against a mammalian-size genome, Stampy requires 2.7 Gb of memory shared between multiple instances running on a single node. An additional 3 Gb per instance is required to run BWA. Smaller genomes require proportionally less memory.
Downstream analyses depend on accurate estimates of the reliability of read mapping. Stampy uses an approximate Bayesian model to estimate the “mapping quality,” the probability that a read, or read pair, is mapped incorrectly. The use of probabilistic models, rather than thresholds, is the main reason for Stampy's improved sensitivity and also allows a consistent treatment of read pairs spanning large indels and structural variation. The model considers three scenarios: (1) that the correct candidate locus was not considered due to an excess of errors or variants in the read; (2) that the best-matching location is incorrect despite the correct locus having been considered, either because an exact repeat was chosen or because read errors cause a near-repeat to match better; and (3) that the original sequence is not represented in the reference. The likelihood of a read pair (r1,r2) mapping to loci (x0,y0) is modeled as
where we have ignored strandedness for simplicity; here Pr is the alignment likelihood and includes priors on read errors, single nucleotide polymorphisms (SNPs), and indel polymorphisms; Pd models the insert size distribution and includes a prior on the occurrence of large indels and other structural variants leading to anomalous inferred insert sizes; and Pu is the uniform distribution over the genome. The posterior probability that an incorrect locus was inferred is
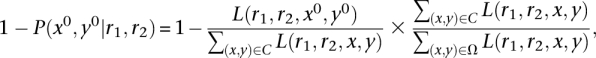 |
where C is the set of candidate positions, and Ω denotes all pairs of genomic coordinates. The last factor cannot be calculated efficiently but is likely to be close to 1 or 0, depending on whether the true locus is in C. We may therefore replace it by the probability that the true candidate was not considered, which we estimate from the nucleotide quality scores. Finally, to identify cases where a read sequence is not represented in the reference, a likelihood ratio test assesses whether the inferred sequence similarity is sufficiently unlikely to have occurred randomly, assuming a random reference sequence. The resulting P-value is used to cap the posterior. In addition, if P is too high, the mapping is not reported. Although the random sequence model is rather approximate, this process removes a large majority of erroneous maps, with a small effect on sensitivity (data now shown). For details, see the online Supplemental material.
Results
To test the performance of Stampy, we first simulated reads from the human genome reference and inserted errors following the empirical error distribution in sequence data taken from the 1000 Genomes Project (The 1000 Genomes Project Consortium 2010). To these we added mutations according to a variety of schemes and annotated the reads with their place of origin and mutational content. These data sets were used to compare the performance of Stampy with four of the more widely used read mappers that generate mapping quality scores: BWA, MAQ, ELAND, and Novoalign (see Supplemental material, section 2).
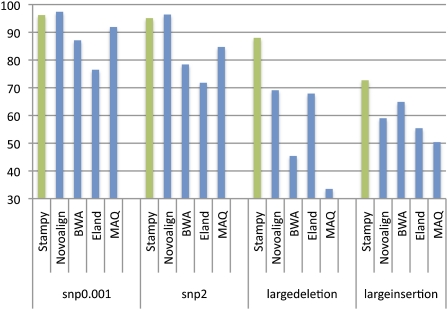
The bulk of sequence reads of a human sample contains an approximate 0.1% fraction of SNP variants. Depending on the read length and paired-end status, on this data set the recall rate of Stampy ranges from 82%–96%, similar to Novoalign and MAQ (87%–97% and 84%–92%) and somewhat better than BWA and ELAND (70%–87% and 70%–77%) (Fig. 1; Supplemental Table S3). To investigate the balance between sensitivity and specificity, we computed receiver operating characteristic (ROC) curves by thresholding on the reported mapping quality, which show a broadly similar picture (Supplemental Fig. S2).
Figure 1.
Recall rates for four sets of 2 million simulated 72-bp paired-end reads, mapped back to the human reference by five read mapping algorithms. Reads included errors following an empirical distribution, as well as additional simulated polymorphisms: 0.1% single nucleotide variants (snp0.001), two single nucleotide variants per read (snp2), and a single large deletion or insertion per read pair (largedeletion and largeinsertion). For details of the simulation procedure, see Supplemental material.
We also assessed mapping quality calibration, which SNP and indel callers depend on to produce well-calibrated likelihoods and posterior scores and which is only partially addressed by ROC curves. ELAND and Novoalign are systematically over- and under-confident, while on this data set Stampy, BWA, and MAQ produce well-calibrated mapping qualities for both long and short reads and both single and paired-end reads (Supplemental Fig. S5).
We next looked at indel mutations. The increasing read length that Illumina sequencing machines are capable of producing means that a nonnegligible fraction of reads (e.g., 2% of 72-bp paired-end reads in human) is expected to overlap with such mutations. Besides their intrinsic interest, correctly dealing with indels is important to avoid spurious SNP calls because of incorrect alignments. We generated a set of reads, each of which containing a single insertion or deletion of up to 30 bp, and computed recall rates, ROC curves, and mapping quality calibration as before.
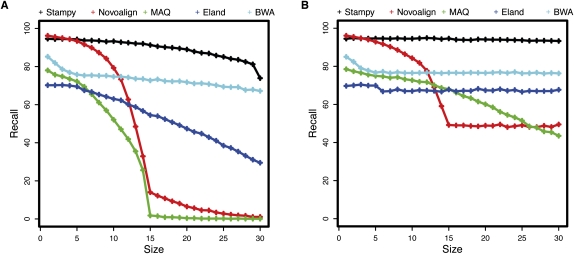
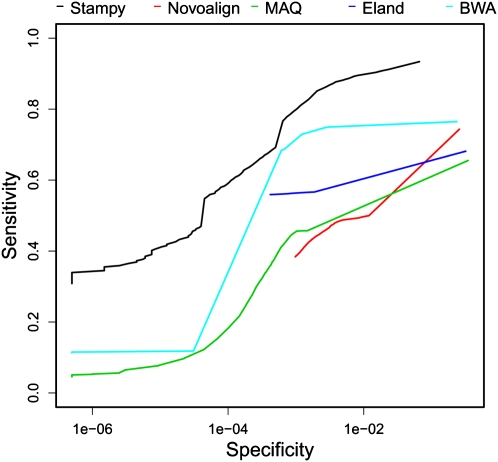
In all three criteria Stampy shows superior performance. Recall rates are high even for larger indels (e.g., 80%–95% of 72-bp paired-end reads are mapped correctly; Fig. 2), and the ROC curve shows that a good balance between sensitivity and specificity is achievable (Fig. 3). Similar conclusions hold for shorter and single-end reads (Supplemental Figs. S8, S9, S3). A good proportion of reads overlapping the ends of very large insertions or deletions was mapped to either breakpoint (Supplemental Fig. S1; Supplemental Table S3), suggesting that Stampy may be helpful in identifying structural variants, although the sequence context of such variants are likely to be more complex than in our simulation. Mapping quality calibration is a challenge for this data set, because the large number of possible combinations of mapping loci and indels cannot all be considered by the probabilistic model. Consequently, the reported mapping quality is somewhat overconfident, particularly for single-end reads, something to keep in mind in downstream indel calling pipelines. Nevertheless, Stampy mapping qualities are more consistent than are those of the other mappers we tested (Supplemental Fig. S6), and in particular for paired-end reads, mapping qualities are well calibrated. Combined with a good sensitivity, this will allow indels to be inferred with confidence if sufficient paired-end coverage is available.
Figure 2.
Recall rates for simulated 72-bp paired-end reads, one of which overlaps a single insertion (A) or deletion (B) of various lengths (horizontal axes). For results for shorter and single-end reads, see Supplemental Figures S8 and S9. A read was required to overlap at least one correct base, but the indel was not required to be correctly called; for indel call rates, see Supplemental Figures S10 and S11.
Figure 3.
Receiver operator characteristics for 72-bp paired-end reads, each of which overlaps a single insertion or deletion of 1–30 bp. For results for shorter and single-end reads, see Supplemental Figure S3.
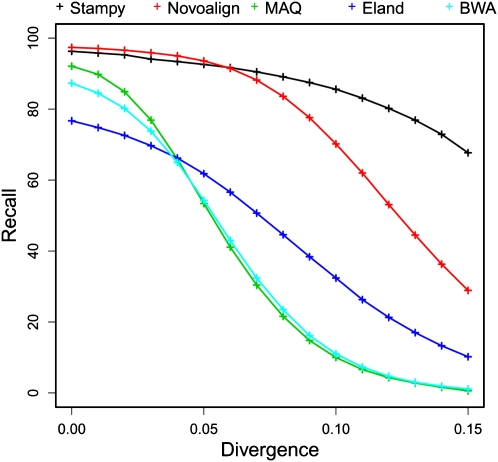
With increasing read lengths, it becomes possible, in principle, to map short reads to a divergent reference. In relevant cases this may be an alternative to de novo assembly, which is challenging particularly for large genomes with a high repetitive sequence content. The ability to map divergent reads is also important to reduce mapping biases for reads containing SNP variation and in order to deal with highly diverse haplotypes such as in the mammalian major histocompatibility complex. Both Novoalign and Stampy show good performance for longer paired-end reads; for instance at 5% divergence, both programs were able to map 93% of the 72-bp paired-end reads to their correct locations (Fig. 4). For these reads, mapping quality calibration is good, but it deteriorates for shorter and single reads, and long paired-end reads are recommended for this application (Supplemental Figs. S7, S12).
Figure 4.
Recall rates for 72-bp paired-end reads at a range of divergences to the human reference (horizontal axes; average number of substitutions per site). For results for shorter and single-end reads, see Supplemental Figure S12.
Finally, we assessed the performance of read mappers on real data. To address the lack of a ground truth, we mapped the paired-end sequence as single reads and calculated the concordance as the fraction of reads that was mapped to consistent locations (Li and Durbin 2009). Although this procedure only assesses the single-end mapping algorithms, it does so with reads containing the true spectrum of polymorphisms, substitutions, and read errors. In addition, current paired-end mapping algorithms are built on top of a single-end mapping stage, so the results are expected to be indicative of paired-end mapping performance. To represent mildly polymorphic whole-genome data, we used two human individuals sequenced to 4× coverage in the 1000 Genomes Project (The 1000 Genomes Project Consortium 2010). We also included a data set consisting of short reads from human mRNA transcript data, which we mapped against the nuclear genome; this is relevant for de novo transcript discovery and may also be regarded as testing the ability of mappers to deal with large variation, with introns and poly-A tails taking the roles of large deletions and large insertions, respectively. Finally, to test mapping to a divergent reference, we mapped 59 lanes of short reads (∼28× coverage) from a sample of Mus spretus (Algerian mouse) to the Mus musculus (C57BL/6J) reference (NCBI build 37); these subspecies are about 2% divergent (Zhang et al. 2005). Because of available CPU resources, we focused on a comparison between Stampy, MAQ, and BWA.
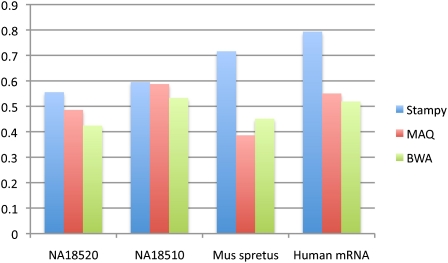
Consistent with the simulation results, in all four cases Stampy finds the highest fraction of concordant reads. The difference is most pronounced for the mRNA data (Stampy 79%, vs. MAQ 55% and BWA 52%) and for the divergent M. spretus data set (Stampy 72%, MAQ 39%, BWA 45%) (Fig. 5). The level of concordance underestimates the fraction of accurately mapped paired-end reads in a practical setting, because both reads are counted as discordant if either is mapped incorrectly; in addition, in paired-end mapping, a fraction of reads can be rescued by using the mate as an anchor.
Figure 5.
Pairwise concordance of independently mapped reads. The data (two human samples from the 1000 Genomes Project (The 1000 Genomes Project Consortium 2010); a divergent mouse subspecies; and human mRNA from an MCF-7 cell line; see text) were mapped to the human or mouse reference genomes (both NCBI build 37) by considering each read of a pair independently. Concordance was calculated as the proportion of reads that mapped to within 500 bp (for genomic DNA) or 10,000 bp (for the mRNA data set) of its mate.
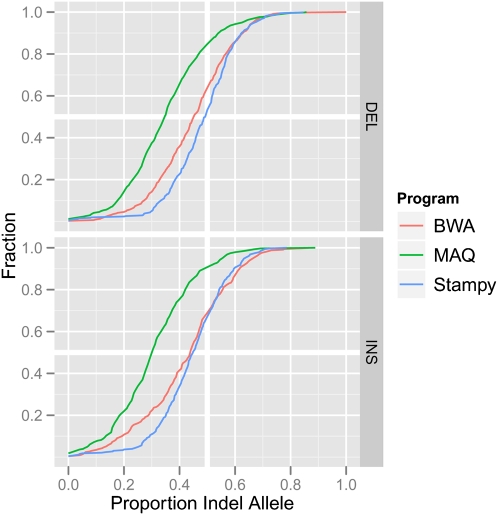
Finally, we looked at whether these algorithms show any bias toward the reference allele in the presence of an indel. This may be expected if algorithms have a lower sensitivity for reads overlapping indels. To do this, we identified sites in a human sample (NA12878) where a heterozygous indel was called with high confidence (see Supplemental material), recorded the number of reads supporting the reference or the alternative allele, and plotted the distribution of the allele ratio. All mappers show a bias toward the reference allele. The effect is most noticeable in MAQ, while it is weaker in BWA and weaker still in Stampy (Fig. 6). A reduced mapping bias indicates that a higher proportion of reads containing indels are mapped correctly. This should prevent false negatives, and will help reduce errors in genotype calls.
Figure 6.
Reference bias at heterozygous indel sites. The plot shows the cumulative distribution of the proportion of reads supporting the non-reference allele in an individual (NA12878) sequenced to high coverage in the 1000 Genomes Project (The 1000 Genomes Project Consortium 2010), and mapped using MAQ, BWA, and Stampy, across high-confidence heterozygous indel sites (see Supplemental material). A left shift of the curve indicates a bias toward the reference allele.
Discussion
It is now straightforward to produce large quantities of DNA and RNA sequence data. Making effective use of much of this data requires the sensitive, accurate, and unbiased mapping of sequence reads to a reference genome. By mapping a larger proportion of reads to their correct location, particularly when reads contain sequence variants, and reporting well-calibrated mapping quality scores, Stampy will help to increase the efficacy of downstream analyses in most standard workflows.
Methods
Broadly, the read mapper algorithm comprises the fast hash table data structure and lookup algorithm, an SIMD-vectorized linear-time Smith-Waterman aligner, an algorithm for generating paired-end candidates, and a Bayesian error model. Full details are provided in section 1 of Supplemental material. Section 2 of that document details the simulation experiment, and section 3 discusses the methods used for real-data comparisons.
Acknowledgments
We thank David Adams for use of the M. spretus data, Ioannis Ragoussis for use of the MCF7 mRNA-seq data, and Cornelis A. Albers and Zam Iqbal for helpful discussions. This work was supported by the Wellcome Trust, grant no. 075491/Z/04.
Footnotes
[Supplemental material is available for this article. Stampy is available at http://www.well.ox.ac.uk/project-stampy.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.111120.110.
References
- The 1000 Genomes Project Consortium 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AJ 2007. ELAND: Efficient large-scale alignment of nucleotide databases. Illumina, San Diego [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J 2009. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Metzker ML 2010. Sequencing technologies: The next generation. Nat Rev Genet 11: 31–46 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Stewart DA, Stromberg MP, Marth GT 2008. Pyrobayes: An improved base caller for SNP discovery in pyrosequences. Nat Methods 5: 179–181 [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wheeler DA, Yakub I, Wei S, Sood R, Rowe W, Liu PP, Gibbs RA, Buetow KH 2005. SNPdetector: A software tool for sensitive and accurate SNP detection. PLoS Comput Biol 1: e53 doi: 10.1371/journal.pcbi.0010053 [DOI] [PMC free article] [PubMed] [Google Scholar]