Abstract
High-throughput sequencing has recently begun to revolutionize the study of structural variants in the genomes of humans and other species. More recently, this technology and others have been applied to the study of human retrotransposon insertion polymorphisms (RIPs), yielding an unprecedented catalog of common and rare variants due to insertional mutagenesis. At the same time, the 1000 Genomes Project has released an enormous amount of whole-genome sequence data. In this article, we present evidence for 1016 L1 insertions across all studies to date that are not represented in the reference human genome assembly, many of which appear to be specific to populations or groups of populations, particularly Africans. Additionally, a cross-comparison of several studies shows that, on average, 27% of surveyed nonreference insertions is present in only one study, indicating the low frequency of many RIPs.
Retrotransposable elements have been described as the drivers of genome evolution (Kazazian 2004; Cordaux and Batzer 2009) due to their impressive ability to insert, delete, and rearrange genomic material through their copy-and-paste mechanism of propagation. Along with this ability comes the potential for deleterious mutation in the human genome, as observed in a small, but ever-increasing, number of cases, presently about 70 (Belancio et al. 2008). The majority of these insertion events are neutral mutations either passed from generation to generation when they occur in the parental germ cells or occupying some fraction of genomes in somatic tissue.
In this article, we focus on the only autonomous active retrotransposon in humans, the LINE-1 (L1) element. The vast majority of L1 sequences, occupying ∼17% of the human genome reference sequence (Lander et al. 2001), are inactive genomic fossils useful for studies of human evolution (Vincent et al. 2003). In humans, there is one active family of L1 elements (Skowronski et al. 1988) referred to here as L1Hs. It has been estimated from study of the reference genome that the average diploid genome contains 80–100 active L1s, of which only a small number are highly active or “hot” (Sassaman et al. 1997; Brouha et al. 2003). Recently, Beck et al. (2010) analyzed six additional genomes and found 37 more “hot” L1s not present in the reference genome. Thus, the number of highly active L1s in the human population may be quite large. Because active elements are still contributing copies of themselves to locations throughout the genome, individual humans differ considerably with respect to presence or absence of various L1 insertions (Xing et al. 2009; Beck et al. 2010; Ewing and Kazazian 2010; Huang et al. 2010; Iskow et al. 2010). These variants are known as retrotransposon insertion polymorphisms (RIPs). Some RIPs are present in the human reference genome assembly, but the vast majority is not likely to be shared with this reference. These are termed nonreference RIPs.
Whole-genome sequences have also proved useful in locating RIPs in both human (Bennett et al. 2004; Konkel et al. 2007; Xing et al. 2009; Hormozdiari et al. 2010) and mouse (Akagi et al. 2008; Quinlan et al. 2010) genomes. The challenges of dealing with next-generation sequence data necessitate the development of new algorithms to detect RIPs, and there are now a few examples of applications that are suited for this task (Hormozdiari et al. 2010; Quinlan et al. 2010). A number of human genomes have been sequenced using these next-generation technologies, yielding insights into human variation and the genetic basis of human disease (Lupski et al. 2010). The 1000 Genomes Project (The 1000 Genomes Project Consortium 2010) aims to capture human genomic variation on an unprecedented scale through whole-genome sequencing. Much of the data from the 1000 Genomes Project consist of individual genomes sequenced to 2×–4× coverage depth, with six individuals currently sequenced much more deeply. Here, we present an analysis of L1 insertions present in these genomes that are not represented in the reference genome assembly.
Results
Mining whole-genome paired-end sequence data for L1 RIPs
Using the approach outlined in Figure 3 (see below), we identified 953 putative nonreference L1 insertion sites from a collection of Illumina short-insert paired-end sequence data encompassing 310 individuals. The vast majority of these individuals were sequenced to 2×–4× coverage, a depth generally insufficient to call L1 insertion variants. Therefore, most of the insertions detected are likely to be somewhat more common in terms of allele frequency, as reads are pooled across all 310 individuals prior to detection of L1 insertions. By considering reads from all individuals at once, we can detect insertions with greater confidence than if we handled low-coverage resequencing data sets one-by-one. Insertions detected by this strategy are more likely to appear in a number of genomes for a sufficient number of read pairs to indicate their presence. In order to estimate the underdetection of L1 insertions due to low sequence coverage, we considered elements present in the reference genome and compared this to our previous study (Ewing and Kazazian 2010). In total, 717 distinct elements are detected with short-insert paired-end reads spanning both the 3′ and 5′ junctions, compared with 772 elements detected by L1-targeted resequencing. Any one individual analyzed from whole-genome resequencing data yielded an average of 556 insertions (95% confidence interval [CI], 393 to 719), significantly lower than in our previous study where we found an average of 628 insertions per individual (95% CI, 462 to 793) shared with the reference genome (P = 0.005, Welch two-sample t-test). Based on this, we estimate that this analysis of whole genomes underestimates the number of insertions by 7.1% in total and by 11.5% on a per-individual basis. The calculation of this underestimate is based on the number of L1 insertions detected in our previous study versus the total number of L1 insertions detected in this study, either in total for reference insertions or per individual for all insertions.
Figure 3.
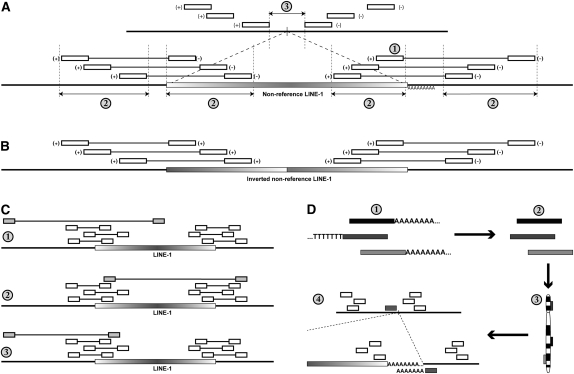
Bioinformatic procedures for identifying nonreference L1 insertions from whole-genome resequencing data. (Open boxes) Mapped reads indicating the presence of a nonreference L1; (gradient boxes) nonreference L1 insertions; (thicker horizontal lines) genomic sequence. (A) Identification of a nonreference L1 insertion from short-insert paired-end sequence reads. Short-insert paired-end reads where one end matches the reference genome and the other matches an L1 reference are clustered based on mapping location to the human genome reference assembly (top). The criteria for detection as discussed in Methods are labeled with numbers: (1) The 3′ end of the L1 insertion must be represented. (2) Reads must form tight clusters based on the locations of reads mapping to both the reference genome and the reference L1. (3) The minimum distance between the locations of genomic reads must be <100 bp, this interval contains the L1 insertion site (vertical bar). The orientation of the reads is annotated next to the open boxes representing the mapped read positions. (B) L1 insertions may be inverted on the 5′ end (Ostertag and Kazazian 2001), resulting in reads aligning to the reference L1 in the same orientation at the 5′ and 3′ ends of the L1 element. (C) Examples of outlier reads that are filtered as described in Methods. (1) The shaded paired read is an outlier because the locations of the reads corresponding to the L1 and the reference genome do not satisfy criteria 2 in panel A. (2) The shaded paired read is an outlier in terms of the reference L1 location. (3) The location of the shaded paired read is an outlier in terms of the reference genome relative to other reads in the cluster. (D) Identifying reads corresponding to the 3′ junction between the L1 poly-A tail and the reference genome sequence. Reads with 5′ or 3′ poly-T or poly-A stretches of at least six bases (1) are trimmed (2) and aligned to the reference genome assembly (3). Trimmed reads aligning to locations within the predicted L1 insertion (A, 3) site are identified (4).
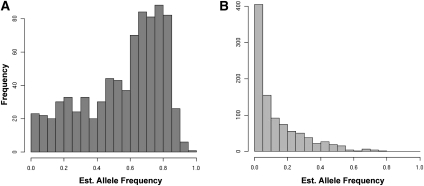
Currently, our bioinformatic pipeline does not yield diploid genotype information for the detected insertions, so the allele frequencies of the insertions cannot be directly measured. However, we have estimated the allele frequencies through the application of an expectation-maximization algorithm, assuming the diploid L1 insertion alleles are in Hardy-Weinberg equilibrium (HWE). Previous studies of polymorphic L1 insertion alleles found little deviation from HWE (see Supplemental Methods) (Myers et al. 2002; Badge et al. 2003; Seleme et al. 2006; Witherspoon et al. 2006). This allele frequency distribution generally agrees with previous studies of nonreference L1 RIPs, in that the majority of alleles are present at relatively low frequency (Fig. 1A). Insertions present in the reference genome are more likely to be present at higher allele frequencies, since all fixed insertions will be present (Fig. 1B). The combined estimated allele frequency histogram for both reference and nonreference insertions is shown in Supplemental Figure S2d. Because of the aforementioned under-ascertainment of L1 insertions due to low-coverage genome sequences, the true allele frequencies of the L1 insertions are likely slightly higher than those estimated here. Previously, we identified 367 nonreference L1 insertions in a diverse sample of 15 unrelated individual genomes (Ewing and Kazazian 2010). These insertions were validated by site-specific PCR, and we noted that there were a potentially significant number of other insertions in the data set for which PCR validation either had not been attempted or had yielded a false-negative result. Indeed, cross-referencing these data yielded 183 overlaps that had not been validated by site-specific PCR. To ascertain whether these overlaps were meaningful, we chose 84 overlapping sites at random for PCR validation (for primers, see Supplemental Table 1). For the 72 primer pairs that yielded PCR amplification products, 58 reactions amplified a product of the expected size corresponding to the predicted insertion, bringing the number of PCR-validated insertions from our previous study to 429 (four other sites were validated for other reasons, since the publication of Ewing and Kazazian [2010]).
Figure 1.
Histograms of filled-site allele frequencies estimated from L1 insertion site presence/absence counts for reference and nonreference L1 insertion sites (Supplemental Fig. S2a,b). (A) Estimated allele frequencies for detected L1 insertions shared with the reference genome. (B) Estimated allele frequencies for detected L1 insertions not present in the reference genome assembly.
Using these data, we are also able to determine some of the characteristics of the L1 insertion itself. Of 953 putative insertions, 227, or 24%, are ∼6 kb or greater in length, representing full-length products of L1 retrotransposition (Supplemental Fig. S1). L1 insertions can also be inverted at their 5′ ends due to a mechanism termed twin-priming (Ostertag and Kazazian 2001). We find 206 (∼21%) detected insertions appear to have an inversion, similar to the 25% of elements inverted in the reference genome. Additionally, we can determine the 3′ junction between the L1 insertion's genomic poly-A tail and the reference genome by remapping potentially polyadenylated reads back to the reference genome after trimming off the putative homopolymer (see Methods). We find evidence for 485 of the 953 insertion sites using the criteria that at least one previously unmappable read re-maps to the predicted insertion 3′ L1-genome junction (generally a ∼40-bp window). We also require that the re-mapped read be derived from a genome whose reads also contributed to the read cluster representing the possible insertion site.
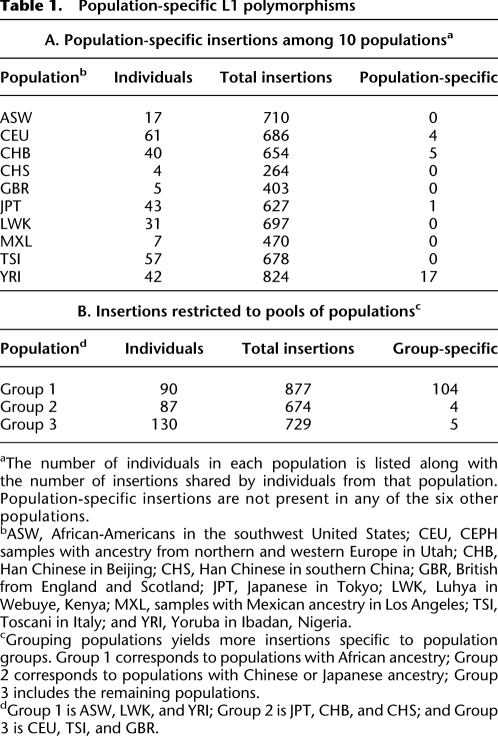
Because these nonreference insertions are polymorphic in human populations, we sought to determine whether any were restricted to certain populations. The genomes studied here are from individuals representing 13 populations, three of which are represented by only one member as these data were derived from publications of personal genomes (Ahn et al. 2009; Schuster et al. 2010). Of the 10 populations represented by more than a single individual, there are four for which an L1 insertion exists that is found only in one of those populations (Table 1A). Notably, the YRI population has more population-specific insertions per individual than any of the others. We also looked for insertions restricted to pools of populations (Table 1B). In this case, the population subset consisting of the ASW, LWK, and YRI populations, representing populations with African ancestry, has by far the highest number of group-specific L1 insertions. In total, 104 insertions were found among African populations that were not present in members of other populations. To assess the significance of this result, we looked at all 120 possible combinations of three populations from the 10 populations studied and compared this to 100 instances where populations were randomly assigned to individual genomes (Supplemental Fig. S3). Based on this random distribution, we might expect to see at most two insertions specific to any possible grouping of three populations from this data set. Taken together, these results suggest a significant level of retrotransposition-induced interpopulation variation, although the majority of insertions are shared among populations.
Table 1.
Population-specific L1 polymorphisms
aThe number of individuals in each population is listed along with the number of insertions shared by individuals from that population. Population-specific insertions are not present in any of the six other populations.
bASW, African-Americans in the southwest United States; CEU, CEPH samples with ancestry from northern and western Europe in Utah; CHB, Han Chinese in Beijing; CHS, Han Chinese in southern China; GBR, British from England and Scotland; JPT, Japanese in Tokyo; LWK, Luhya in Webuye, Kenya; MXL, samples with Mexican ancestry in Los Angeles; TSI, Toscani in Italy; and YRI, Yoruba in Ibadan, Nigeria.
cGrouping populations yields more insertions specific to population groups. Group 1 corresponds to populations with African ancestry; Group 2 corresponds to populations with Chinese or Japanese ancestry; Group 3 includes the remaining populations.
dGroup 1 is ASW, LWK, and YRI; Group 2 is JPT, CHB, and CHS; and Group 3 is CEU, TSI, and GBR.
Comparison of L1 RIP data yields over 1000 nonreference insertions
To further characterize these results in the context of all known L1 insertions, we cross-referenced data from five studies: two L1-targeted sequencing studies (Ewing and Kazazian 2010; Iskow et al. 2010), a study where full-length L1 insertions were derived and thoroughly characterized from mapping fosmid end sequences generated from the genomes of diverse individuals back to the reference genome (Beck et al. 2010), data from dbRIP that encompasses a number of previous studies (Wang et al. 2006), and finally the insertion sites derived from massively parallel paired-end sequencing of human genomes presented here. Another study using high-density tiling arrays of the human genome in conjunction with vectorette PCR (Huang et al. 2010) also catalogs many novel nonreference insertions, but this study is not included here because it focuses mostly on the X chromosome, and the addition of many extra insertion sites on one chromosome would bias the subsequent analysis of genome-wide L1 RIP distribution.
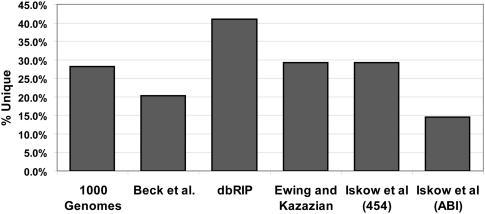
Cross-referencing the data from all of the aforementioned studies yields 1680 possible insertion sites, 1016 of which have some corroborating evidence for their existence. This evidence comes in the form of PCR validation, the presence of poly-A stretches resulting in unmappable reads in the specific breakpoint region, or the presence of a given insertion in more than one data set. Of the 1016 insertions, 486 are present in more than one data set, and a detailed table of these overlaps is available in the Supplemental material (Supplemental Table S2). The histogram of filled site frequencies shown in Figure 1A indicates that nonreference insertion sites have a tendency toward low allele frequencies across all populations. This is consistent with the number of validated insertions present in only one data set: On average, 27% of the validated L1 insertions found in each study are unique to that study (Fig. 2).
Figure 2.
L1 insertions present in only one data set. The columns represent the fraction of insertions present in only the indicated data set out of all validated insertions in the data set (unique). Insertions can be validated by site-specific PCR, sequencing spanning the element, or presence in another independent data set, depending on the study. Iskow et al. (2010) presented data generated using two different sequencing methods as noted in the column labels. We analyzed the paired-end Illumina data from the 1000 Genomes Project (The 1000 Genomes Project Consortium 2010), Beck et al. (2010) employed a fosmid-end resequencing strategy, dbRIP cross-references data sets generated using a wide variety of techniques (Wang et al. 2006), and Ewing and Kazazian (2010) used Illumina sequencing.
Discussion
In this article, we derive information about L1 insertion sites and about the L1 itself from paired-end next-generation sequencing data, including low-coverage data sets when considered in conjunction with other data sets. In doing so, we have identified more nonreference L1 insertions than in any single study to date, an unsurprising result given the number of individual genomes analyzed. Many insertions appear to be specific either to a specific population or to a group of populations with common geographic origin (Table 1), indicating that these insertions arose recently in human evolutionary history. This concurs with the observation that the estimated allele frequencies are skewed toward rare variants for nonreference L1 insertions (Fig. 1A), along with the observation that for all studies of nonreference L1 RIPs, a significant fraction of those RIPs are unique to that study (Fig. 2). In contrast, reference L1 insertion sites are more common at higher frequency (Fig. 1B) as expected. We were surprised by the number of L1 insertions present only in populations with African ancestry (Table 1B) coupled with the slightly higher, although not significantly so, minimum estimate for L1 retrotransposition rate in these populations (see Supplemental materials). We suspect this is due to the age of these populations relative to the more recent bottlenecks, which occurred during human migrations into Eurasia. African populations have higher levels of genomic diversity than populations that have experienced founder effects (Tishkoff and Williams 2002; Jakobsson et al. 2008; Lohmueller et al. 2008), and the levels of African-specific L1 diversity seem to be no exception.
When L1 RIPs present in the reference sequence are included, we have a collection totaling 1400–2100 L1 insertions across a combined data set of roughly 400 individuals. As we identify an increasing number of polymorphic L1 insertion loci, other efforts are underway to identify polymorphisms due to Alu retrotransposition that are likely to outnumber L1 RIPs given their copy number in the reference genome (Lander et al. 2001) and previous estimates of their activity (Xing et al. 2009). A recent study reporting a highly efficient approach for identifying mobile element insertions by sequence capture identified 487 nonreference AluYb8/Yb9 loci from four genomes, 223 of which overlap with insertions present in dbRIP (Witherspoon et al. 2010). In combination, all forms of retrotransposon polymorphism constitute a significant force inducing genomic variation between human individuals. Except in the limited number of cases where retrotransposon insertions were associated with genetic disease, the phenotypic consequences of this variation are unknown, largely due to our previous inability to assess its extent on a genome-wide scale. With increasing numbers of RIP markers, such as those cross-referenced and cataloged in this study, it will soon be reasonable to genotype common RIP variants on a large scale using array-based technologies in order to identify genotype-phenotype associations, especially in complex human genetic diseases. We believe that RIPs should be considered alongside SNPs and CNVs in studies of human genomic variation.
Methods
Data sources
Sequence data for 307 individual genomes was obtained from the 1000 Genomes Project FTP site (ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/). Three additional genomes: ABT, KB1 (Schuster et al. 2010), and SJK (Ahn et al. 2009) were obtained from the NCBI SRA (accession nos. SRA010356 for ABT and KB1 and SRA008175 for SJK). Locations of L1 elements were obtained from the supplemental material of articles described in the introduction (Beck et al. 2010; Iskow et al. 2010) and from dbRIP (Wang et al. 2006).
Genomic DNA sources and PCR RIP genotyping
Genomic DNA was obtained from sources previously described (Ewing and Kazazian 2010). Detection of L1 RIP filled and/or empty sites was carried out in the manner described in that article. Briefly, two PCR primers were ordered for each predicted RIP flanking the expected insertion site. A primer in the L1 3′ UTR was used along with a primer 3′ of the predicted L1 3′ UTR to detect the L1 insertion, and primers flanking the 5′ and 3′ ends were used to detect the empty insertion site, if present. Primers for new insertion sites are included in the Supplemental material (Supplemental Table 1).
Bioinformatics
The MySQL DBMS was used throughout this study for storage, retrieval, and analysis of data. All computation was carried out on a 102-node SGE cluster operated by the Penn Genomics Frontiers Institute (PGFI). Manipulation of sequence data was performed using bowtie with the options --best -m 1 -n 2 (Langmead et al. 2009), and Perl and Unix shell scripts.
Detection of nonreference RIPs from short-insert paired-end sequence reads
Starting from short-insert paired-end Illumina sequence data, each read was aligned against a reference L1 element (L1RP), allowing two mismatches. For each read whose mate did not also match the reference L1, that read was aligned against the reference genome (hg18), again allowing two mismatches. Only uniquely mappable reads were retained. Each resulting L1-genome pair was added to a table, along with the identity of the genome from which the read was derived, as reads from all individuals are added to the same table for subsequent analysis. Next, reads were grouped based on their genomic location: The location of each read was averaged with its nearest neighbor within a 1-kb window iteratively to build clusters. For each read, iteration continues until there are no more neighbors within a 1-kb window to be added. The center of the 1-kb window is defined by the average location of all reads in the cluster and is updated following each iteration. These clusters of L1-paired reads were then refined to determine whether they are likely to represent a nonreference L1 insertion event using the following criteria (Fig. 3A): (1) Both the 5′ and 3′ ends of the element must be represented based on the locations of reads within the L1 reference sequence. Additionally, the 3′ most read mapped to the L1 reference must occur within 100 bp of the end of the element (>5900 bp) to ensure the presence of the 3′ end of the element, as 3′ truncations are rare (Fig. 3A, 1). At least two read-pairs must span each junction, and each insertion must be represented by a minimum of eight read-pairs. This coverage requirement was determined empirically based on our ability to detect nonreference insertions known to exist from other studies. (2) The mapped locations of all reads representing a given insertion must fall into 300-bp windows both on the genomic flanks and on the L1 reference sequence (Fig. 3A, 2). This may be adjusted depending on the insert size of the paired-end library: In general, if two read-pairs map to locations >300 bp apart, they cannot refer to the same L1 insertion. (3) The 5′ most and 3′ most positions of the reads mapping to the reference genome must be <100 bp from each other, but non-overlapping (Fig. 3A, 3). The L1 element itself must be at least 150 bp in length as inferred from the positions of reads mapped onto the reference L1 element, although in principle the algorithm should be able to detect smaller insertions. (4) The mapped read-pairs must not correspond to L1s present in the reference genome. For example, if the nonreference L1 depicted in Figure 3A corresponded to an L1 annotated in the reference genome assembly, this read-pair cluster would be discarded. Combined with criteria 3, this eliminates any small (∼100 bp) L1 insertions that may generate the pattern of alignments consistent with a nonreference insertion (Fig. 3A).
Although the error rate due to mispairing of paired-end reads is relatively low, when many millions of paired reads are considered this yields a significant number of “noise” reads mixed in with the “signal” reads that result from nonreference L1 insertions. Additionally, features of the reference genome sequence, especially existing L1 insertions in the reference sequence, can yield spurious clustering of L1-genome paired reads that do not correspond to true nonreference insertion sites. In order to filter out these reads, we consider the possible orientations of mapped reads, and employ a filtering method to remove “outlier” mappings.
Here, we use the term orientation pair in reference to a set of paired-end reads, each with one end aligning to the reference genome and the other end aligning to the L1 sequence where all reads in the set have the same reference genome and L1 orientations. However, the orientation of the genomic read does not have to match that of the L1 read. There are four possible orientation pairs for mapped L1-genome paired reads: +/+, −/−, +/−, and −/+, where the first + or – in each pair represents the orientation of aligned reads to the reference genome and the second represents the orientation of the aligned reads to the reference L1 element. As an example, Figure 3A shows two sets of paired reads with +/− orientation pairs, one spanning the 5′ L1-genome junction and another spanning the 3′ junction. Since each putative insertion is represented by read-pairs that span both the 5′ and 3′ L1-genome junctions, there are two orientation pairs associated with each insertion. Read sets from each of the four possible orientation pairs might be present in a cluster prior to refinement, so each possible pairing is considered. For a putative L1 insertion, not all pairs of paired-end orientations are possible: The genomic ends must be in opposite orientations due to the directionality of the sequencing method. The orientations of the L1 ends are not subject to this restriction as ends with the same read orientation can occur due to inversions (Fig. 3B) that occur as a result of twin-priming (Ostertag and Kazazian, 2001). For each valid orientation pairing, the criteria listed above are considered. In practice, we have not encountered a case where more than one valid orientation pairing passes all four criteria, but it is possible if two insertions exist in the same 1-kb window with opposite orientations. Prior to being subjected to the criteria listed above, each possible orientation pair is filtered for “outlier reads” (Fig. 3C) that occur by chance due to the large number of reads considered. This is accomplished by considering the interquartile ranges (IQRs) of the genomic or L1 locations to which reads are mapped. Anything outside the interval (first quartile – 1.5*IQR, third quartile + 1.5*IQR) is eliminated. If >25% of the paired reads for any given orientation pair are eliminated in this way, the pair is discarded.
Detection of reference insertions from short-insert paired-end sequence reads
Similar to the detection of nonreference L1 insertions from paired-end sequence data, we required that both the 3′ and 5′ L1-genome junctions be spanned by paired reads. The genomic and L1 mapping locations for each L1-genome paired read were considered to identify reads corresponding to L1 insertions present in the human genome reference assembly, keeping track of the genome from which each paired read was sequenced.
Remapping polyadenylated reads
The described method for detecting RIPs from short-insert paired-end sequence reads narrows the insertion site to within a window of 1–100 bp (43 bp on average). This is a good start, but for many reads, we can narrow down the exact breakpoint between the L1 insertion and the genomic DNA sequence even further by considering unmapped reads containing possible polyadenosine stretches that could correspond to the poly-A tails associated with nonreference L1 3′ ends (Fig. 3D). This is done by obtaining the unmapped reads for genomes represented in the 1000 Genomes Project, selecting reads that contain either 5′ An or 3′ Tn, where n is at least 6. These A or T nucleotides are trimmed off, the remaining sequence is re-aligned to the reference genome, and the results are stored in a table. Insertions predicted from paired-end reads are then cross-referenced against this table to find instances where these putative nonreference poly-A sequences could indicate the 3′ junction between the L1's inserted poly-A tail and the flanking genomic sequence. This is similar to the method used to detect nonreference poly-A stretches in our L1-targeted resequencing technique (Ewing and Kazazian 2010).
Acknowledgments
We thank Christine R. Beck for discussions regarding the initial conception of the analyses presented here, John V. Moran for revisions, and Sridhar Hannenhalli for revisions and suggestions regarding the analysis and significance of group-specific insertions. We thank three anonymous reviewers for their comments and suggestions.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.114777.110.
References
- The 1000 Genomes Project Consortium 2010. A map of human genome variation from population scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SM, Kim TH, Lee S, Kim D, Ghang H, Kim DS, Kim BC, Kim SY, Kim WY, Kim C 2009. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res 19: 1622–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi K, Li J, Stephens RM, Volfovsky N, Symer DE 2008. Extensive variation between inbred mouse strains due to endogenous L1 retrotransposition. Genome Res 18: 869–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badge RM, Alisch RS, Moran JV 2003. ATLAS: A system to selectively identify human-specific L1 insertions. Am J Hum Genet 72: 823–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV 2010. LINE-1 retrotransposition activity in human genomes. Cell 141: 1159–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P 2008. Mammalian non-LTR retrotransposons: For better or worse, in sickness and in health. Genome Res 18: 343–358 [DOI] [PubMed] [Google Scholar]
- Bennett EA, Coleman LE, Tsui C, Pittard WS, Devine SE 2004. Natural genetic variation caused by transposable elements in humans. Genetics 168: 933–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH Jr 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci 100: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA 2009. The impact of retrotransposons on human genome evolution. Nat Rev Genet 10: 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD, Kazazian HH Jr 2010. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res 20: 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, Hajirasouliha I, Dao P, Hach F, Yorukoglu D, Alkan C, Eichler EE, Sahinalp SC 2010. Next-generation VariationHunter: Combinatorial algorithms for transposon insertion discovery. Bioinformatics 26: 350–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CR, Schneider AM, Lu Y, Niranjan T, Shen P, Robinson MA, Steranka JP, Valle D, Civin CI, Wang T, et al. 2010. Mobile interspersed repeats are major structural variants in the human genome. Cell 141: 1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE 2010. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141: 1253–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R 2008. Genotype, haplotype and copy-number variation in worldwide human populations. Nature 451: 998–1003 [DOI] [PubMed] [Google Scholar]
- Kazazian HH Jr 2004. Mobile elements: Drivers of genome evolution. Science 303: 1626–1632 [DOI] [PubMed] [Google Scholar]
- Konkel MK, Wang J, Liang P, Batzer MA 2007. Identification and characterization of novel polymorphic LINE-1 insertions through comparison of two human genome sequence assemblies. Gene 390: 28–38 [DOI] [PubMed] [Google Scholar]
- Lander ES, Heaford A, Sheridan A, Linton LM, Birren B, Subramanian A, Coulson A, Nusbaum C, Zody MC, Dunham A, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Indap AR, Schmidt S, Boyko AR, Hernandez RD, Hubisz MJ, Sninsky JJ, White TJ, Sunyaev SR, Nielsen R, et al. 2008. Proportionally more deleterious genetic variation in European than in African populations. Nature 451: 994–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, et al. 2010. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med 362: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JS, Vincent BJ, Udall H, Watkins WS, Morrish TA, Kilroy GE, Swergold GD, Henke J, Henke L, Moran JV, et al. 2002. A comprehensive analysis of recently integrated human Ta L1 elements. Am J Hum Genet 71: 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH Jr 2001. Twin priming: A proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res 11: 2059–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Clark RA, Sokolova S, Leibowitz ML, Zhang Y, Hurles ME, Mell JC, Hall IM 2010. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res 20: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH Jr 1997. Many human L1 elements are capable of retrotransposition. Nat Genet 16: 37–43 [DOI] [PubMed] [Google Scholar]
- Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, Harris RS, Petersen DC, Zhao F, Qi J, et al. 2010. Complete Khoisan and Bantu genomes from southern Africa. Nature 463: 943–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seleme MC, Vetter MR, Cordaux R, Bastone L, Batzer MA, Kazazian HH Jr 2006. Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity. Proc Natl Acad Sci 103: 8036–8041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski J, Fanning TG, Singer MF 1988. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol 8: 1385–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM 2002. Genetic analysis of African populations: Human evolution and complex disease. Nat Rev Genet 3: 611–621 [DOI] [PubMed] [Google Scholar]
- Vincent BJ, Myers JS, Ho HJ, Kilroy GE, Walker JA, Watkins WS, Jorde LB, Batzer MA 2003. Following the LINEs: An analysis of primate genomic variation at human-specific LINE-1 insertion sites. Mol Biol Evol 20: 1333–1348 [DOI] [PubMed] [Google Scholar]
- Wang J, Song L, Grover D, Azrak S, Batzer MA, Liang P 2006. dbRIP: A highly integrated database of retrotransposon insertion polymorphisms in humans. Hum Mutat 27: 323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherspoon DJ, Marchani EE, Watkins WS, Ostler CT, Wooding SP, Anders BA, Fowlkes JD, Boissinot S, Furano AV, Ray DA, et al. 2006. Human population genetic structure and diversity inferred from polymorphic L1(LINE-1) and Alu insertions. Hum Hered 62: 30–46 [DOI] [PubMed] [Google Scholar]
- Witherspoon DJ, Xing J, Zhang Y, Watkins WS, Batzer MA, Jorde LB 2010. Mobile element scanning (ME-Scan) by targeted high-throughput sequencing. BMC Genomics 11: 410 doi: 10.1186/1471-2164-11-410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, Zhou Q, Kirkness EF, Levy S, Batzer MA, et al. 2009. Mobile elements create structural variation: analysis of a complete human genome. Genome Res 19: 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]