Abstract
The ability to determine expression levels of multiple genetic and/or proteomic signatures simultaneously from the same sample volume is of tremendous importance in a wide range of biomedical applications including disease diagnostics, drug discovery, and fundamental studies of complex biological processes. Unfortunately, many of the tremendously enabling genomic and proteomic technologies available to the academician are not applicable in the clinical setting. Combining traditional chemical and biological insight with cutting-edge concepts from materials science, physics, and engineering, our group brings an interdisciplinary approach to developing new tools to tackle longstanding, refractory problems in the biological sciences—technologies that will enable fundamental biological discovery, and in some applications, expedite the transfer of biomolecular insight to the hands of the physician where it can directly impact patient care.
Why multiparameter analysis?
Personalized medicine: The concept that a patient's disease can be most effectively treated utilizing individualized diagnostic information.
Multiparameter analysis: Simultaneous determination of the expression levels from multiple genomic, transcriptomic, or proteomic signatures of disease.
This is an exciting time in biomedical science. With the sequencing of the human genome, researchers are armed with the fundamental blueprint of human life. Advances in enabling genomic and proteomic technologies have continued to add more clarity to the complex picture of human biology, and the perpetually growing body of available data coupled with advances in computational approaches for data integration and correlation have effectively ushered biomedical science into a golden age of information.
Global genomic and proteomic analyses have dramatically advanced our knowledge of the molecular bases of many human diseases. Our understanding of cancer biology, in particular, has greatly benefited from this “information boom.” We now have an extensive list of causative genetic and epigenetic factors and purported proteomic disease biomarkers, but equally if not more important is our improved understanding of biomolecular interconnectivity. Cascades of biomolecular interactions (or lack thereof) at the level of transcription and translation, in concert with post-transcriptional and post-translational events propagate genetic perturbations that eventually result in the diseased phenotype. Referred to in different contexts as pathways, networks, or systems, scientists are increasingly studying entire groups of interacting biomolecules to glean greater insight into the mechanism of disease onset and progression. The significance of this approach cannot be overstated since it highlights the defects or mistakes within the biological system that represent “weak links” or targets to which therapeutic agents can be effectively directed. Stated another way, multiparameter analysis not only answers the primary question, “Is this cancer?”, but can also provide insight towards answering the essential question, “How can most effectively treat this disease?”
Disease pathway: A collection of biomolecules functioning together and propagating signals that either promote or inhibit disease. The expression levels of these biomolecules can be correlated or anti-correlated and they often act directly on one another, i.e. protein phosphorylation by kinases.
WANTED: New multiparameter analysis technologies
Our detailed understanding of the pathways, networks, and/or systems that drive disease has been assembled using high throughput genomic and proteomic technologies in concert with immortalized cell lines and animal models. However, there can be significant differences between even the best models and “real” human disease on account of heterogeneity—both population heterogeneity and more subtle heterogeneities within patients themselves. To circumvent these complications diagnoses need to be performed at the level of each individual. Accordingly, individualized diagnosis may well be the most critical development needed to implement personalized approaches to medicine.
Unfortunately, many of the same measurement approaches are not applicable in the clinic where sample sizes and high technology-based instrumentation and expertise are more limited. As a result, despite our increased knowledge of the molecular bases of human disease, the translation to clinical medicine has sadly lagged significantly behind. This is a measurement limitation, owing to the fact that current analytical techniques lack the compatibility, sensitivity, and/or quantitative abilities to simultaneously monitor expression levels of many genes and/or proteins from readily accessible body fluids or small, solid tumor samples. By developing new analytical tools to perform personalized multiparameter analysis on clinically accessible samples, patient survival and quality of life will be greatly enhanced by 1) reducing the need for invasive biopsy procedures, and 2) providing an informative diagnosis that will guide appropriate targeted molecular therapies to individuals.
A further consideration in the design of new multiparameter analysis technologies is the information that one would ideally wish to obtain from a single patient sample. Traditional genomic, transcriptomic, and proteomic technologies are inherently limited to independently analyzing DNA, RNA, or proteins. However it is becoming increasingly clear that information flow between levels and the correlations or discordance in expression contains an incredible amount of disease-specific information. Attempts to understand information flow are currently hindered by the requirements for multiple analysis methodologies and sample volumes. This in turn leads to difficulties in data correlation and interpretation in light of differences in sample preparation, tissue heterogeneity, and cross-platform data correlation. Therefore, a “wish list” for new technology development might include the ability to monitor, on a single analytical platform, expression of DNA, RNA, miRNA, proteins, and post-translationally-modified proteins.
MicroRNAs (miRNAs): Small (19-24 nucleotides), single-stranded, non-protein coding RNA molecules that are powerful regulators of gene expression. Widely implicated in many human cancers, miRNAs are emerging targets for informative, multiparameter disease diagnostics.
New multiparameter analysis technologies in development by the Bailey group
Motivated by practical challenges associated with performing multiparameter biological analysis, our group is developing a suite of powerful analysis tools that will be applicable in clinical settings and beyond. Our goal is to facilitate personalized diagnosis and individualized treatment by providing a more detailed picture of the biomolecular signatures of disease from a single patient. On account of their simplicity, scalability, and molecular generality, these tools also have broad applicability to many aspects of pharmaceutical research and fundamental biological studies.
Silicon photonic microring resonators
Silicon photonics: A technology that uses mass producible, integrated circuit manufacturing tools to fabricate optical circuits that steer light around a silicon chip. Originally developed for applications in telecommunications, silicon photonics are now being explored for highly multiplexed biosensing.
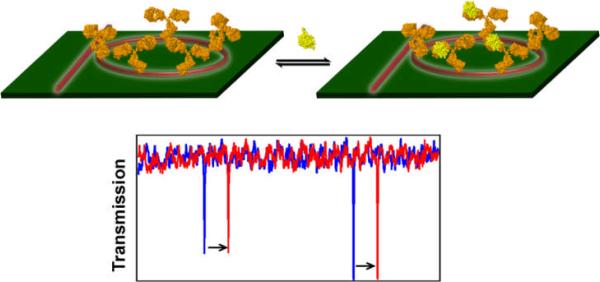
We are developing an incredibly enabling biological analysis technology based upon high density arrays of silicon photonic microring resonators. The basis of operation for this technology is quite simple, as illustrated in Figure 2. Optical modes supported within the microcavities are very sensitive to the local refractive index near the microring surface. When biomolecules of DNA, RNA, or proteins bind to an appropriate capture probe immobilized onto the microring surface, the refractive index changes and the frequencies of optical resonances shift in a well-defined manner. It is important to note that this is a label-free detection technology, meaning that no fluorescent, enzymatic, or radioactive probes are needed. From a practical perspective label-free methods are often advantageous in terms of reduced assay cost and complexity and also avoid deleterious effects that the signaling moiety itself might have on the biomolecular interaction being studied.
Figure 2.
Schematic illustration of a microring resonator biosensor. The microring on left is functionalized with a capture agent (an antibody, as shown here) and has optical resonance spectrum shown in blue. Binding of the target biomolecule changes the local refractive index surrounding the microring at right that in turn causes a shift in the optical resonances (red trace). By monitoring the frequency shift of resonant peaks accompanying sample introduction, the concentration of biomolecular species can be quantitatively determined.
Many thousands of microring resonators can be fabricated with tremendous precision on the wafer scale using commercially validated semiconductor processing techniques. Furthermore, each microring sensor can be uniquely encoded with a different capture agent (cDNA, antibody, etc.) using conventional microarraying tools and operation of these devices in a frequency region used by existing optical telecommunications technologies allows us to construct a robust and easy-to-use sensing methodology with on-chip integrated optical components, microfluidic fluid actuation, and biofouling resistant coating strategies. The ability to leverage these existing technologies for an entirely different application leads towards an extremely sensitive, highly multiplex-able detection platform that should be applicable to many clinical and pharmaceutical discovery applications.
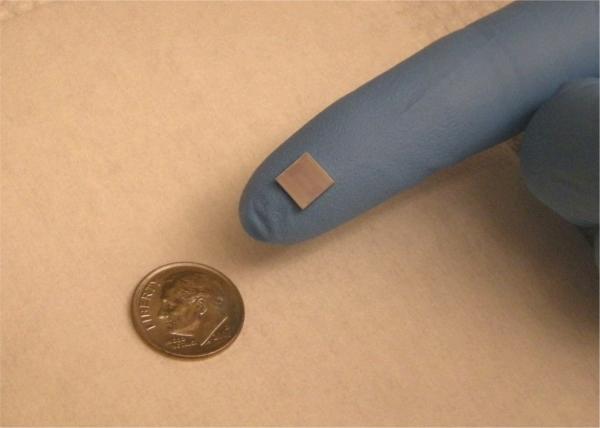
Still in the early stages of development, we have demonstrated the applicability of the technology to the detection of DNA, miRNAs, and protein biomarkers using chips similar to that shown in Figure 3. This version of the sensor chip has 64-uniquely addressable sensors that can be independently interrogated. The presence of many sensors on the same chip allows different modes of operation—many unique assays can be run in parallel or a smaller number of assays can be performed multiple times in parallel. The ability to perform many identical measurements in the same small (< 100 μL) sample volume increases measurement precision and reduces the number of false positive responses. Also, additional microrings in the assay chamber that are not functionalized to be selective for a particular target analyte can greatly help alleviate matrix effects by serving as control sensors to correct for biofouling, small temperature variations, and other sources of non-specific bulk response.
Figure 3.
Photograph of a silicon photonics chip that contains 64 microring optical resonators that can be uniquely functionalized with multiple capture agents against different biomolecular targets. Each microring sensor can be individually addressed to determine the concentration of different biomolecules from within a single sample. Optimization of device geometry should enable sensor densities in excess of 10,000/cm2.
As an initial demonstration of the technology for proteomic applications, we have performed label-free quantitation of the cancer biomarker carcinoembryonic antigen (CEA) with a limit of detection comparable to and precision exceeding that of commercial ELISA assays. We have also shown that assay dynamic ranges can exceed 4 orders of magnitude and that measurements can be made in media as complex as undiluted blood serum with only a small reduction in absolute sensitivity. We have applied this technology to the sensitive quantitation of cytokines and have also shown that label-free, multiplexed assays utilizing six different cancer biomarker-specific capture agents arrayed in triplicate can be performed with absolutely no loss in sensitivity—each reaching clinically-relevant levels. Another important proteomic application of the technology lies in the multiplexed screening of protein-protein interactions. The ability to simultaneously determine the kinetic association and dissociation constants and equilibrium constants of multiple protein-protein interactions has applications throughout drug target determination, antibody screening, and fundamental biology.
We have also shown that microring resonators can be effectively utilized to detect nucleic acids with high sensitivity. In particular, we are developing a hybridization-based multiplexed assay for profiling the expression levels of multiple miRNAs without additional steps of ligation, amplification, or labeling. Preliminary experiments show that the platform is significantly more sensitive than traditional Northern blotting methods of detection, and the sample size requirements are smaller than many commercial miRNA analysis kits. Additionally, we have begun to utilize our microring resonator platform to investigate protein-nucleic acid and protein-carbohydrate binding interactions.
Based on the platform's sensitivity and scalability, as well as the ability to detect both proteins and nucleic acids, we feel label-free microring resonator sensors are a promising tool for multiparameter analysis in clinical, pharmaceutical, and fundamental biological applications. Much work remains to be performed, but this technology development effort is, in part, aimed at translating biological insight into improvements in clinical medicine by empowering physicians with the same biomolecular information available to academics.
Multicomponent strategies for assessing tissue heterogeneity
One challenge underlying many aspects of tissue-based disease diagnosis is sample heterogeneity. It is well established that cell-to-cell heterogeneity is present within even the simplest biological systems and is a critical determinant in processes such as embryonic development and disease onset and progression. Therefore, the critical biomolecular signatures of disease can only be elucidated from homogeneous samples. One of the most pronounced examples of dynamic tissue heterogeneity lies in the metastasis of cancerous tumors. Metastasis, as in normal cellular adhesion, is controlled by interactions with many hundreds of different biomolecules including proteins, carbohydrates, and lipids. Substrates that present only a single biomolecule can be very valuable in understanding disease-altered adhesion; however, we feel that a more complete picture will be obtained by more complex models that investigate multicomponent biomolecular contributions.
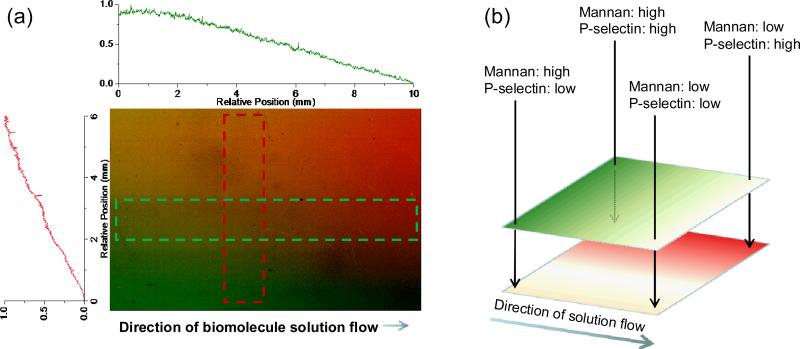
To help understand the molecular determinants that dictate normal and disease-induced perturbations in cell adhesion, we are developing methods of controlling the spatial presentation and density of multiple biomolecules on model substrates. Our method, based upon photochemical attachment, is molecularly universal (works equally well on proteins and carbohydrates) and can create surfaces with overlapping patterns and gradients of multiple biomolecules, as shown in Figure 4. Importantly, gradient structures present continually varying compositions of multiple different receptors. We envision several applications of the resulting substrates including: 1) A heterogeneous sample might stratify according to small differences in phenotype due to different cell-surface interactions at different spatial locations, and 2) The effects of competing surface interactions on a clonal population of cells could be observed in a single experiment by culturing the population on a multicomponent gradient surface.
Figure 4.
Fluorescence scan of a 10 × 5 mm, two-component photopatterned gradient surface and corresponding line traces to illustrate the gradient geometry. From left to right, the concentration of mannan (green, visualized by secondary staining with a lectin) decreases in a controllable manner. Similarly, from top to bottom, the concentration of P-selectin (red, visualized by secondary staining with an antibody) is systematically controlled. The simultaneous control over density of both immobilized biomolecules is significant because a large number of concentration combinations are represented on one substrate, and can thus be interrogated in a single experiment, from a single sample.
Surface gradients: Substrates patterned so that the presented density of bound biomolecules is controllably and continuously varied across the surface.
Outlook for the future
The future of biomedicine lies in the ability to measure and integrate the expression levels of multiple biomolecular signatures to better understand normal human biology as well as disease onset and progression. The powerful genomic and proteomic technologies that have provided the motivation for these studies are ill equipped, at present, to take the next step—particularly in clinical settings. Most likely, new methodologies will need to be developed to meet these emerging measurement challenges. The Bailey group is developing two such multiparameter analysis technologies that we hope will develop into valuable tools to provide physicians with personalized disease fingerprints. Appropriate multiparameter analysis technologies will also contribute to our understanding of how pharmaceuticals can be developed and prescribed most effectively. In the future, we envision that personalized diagnostics will transition cancer classification from organ of origin to a characteristic set of specific biomolecular perturbations that are known to respond to a particular treatment regimen for that individual. The development of multiparameter assays will likely become increasingly coupled to pharmaceutical development, leading to more effective clinical trials conducted on the most appropriate subset of patients and ultimately improving patient survival and quality of life.
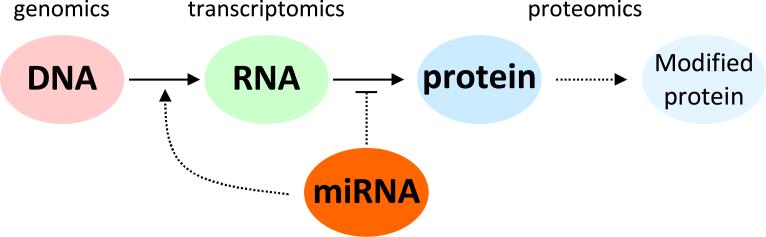
Figure 1.
Biological information flows from the level of DNA to RNA and on to proteins. Post-transcriptional effects from miRNAs and post-translational modifications of proteins add additional layers of biological complexity. Information about disease onset and progression can be extracted from conventional measurements of DNA, RNA, or proteins, but valuable insight can also be extracted from analyzing the correlations or discordance as information flows between levels. New analytical methods that can simultaneously perform measurements across multiple levels of biomolecular complexity will be extraordinarily valuable in understanding the propagation of perturbations that result in diseased phenotypes.
Acknowledgements
We acknowledge financial support for our efforts in developing new multiparameter biomolecular analysis methods from the following agencies: the NIH Director's New Innovator Award Program, part of the NIH Roadmap for Medical Research, through grant number 1-DP2-OD002190-01; the Roy J. Carver Charitable Trust; a New Faculty Grant from the Camille and Henry Dreyfus Foundation; and a 3M Non-Tenured Faculty Grant.
Biography
Prof Ryan C. Bailey is an Assistant Professor in the Department of Chemistry and the Institute for Genomic Biology and an affiliate of the Micro and Nanotechnology Laboratory at the University of Illinois at Urbana-Champaign. Prof Bailey received his B.S. from Eastern Illinois University and his Ph.D. in Chemistry from Northwestern University where he worked with Prof Joseph Hupp to create a new chemical and biological sensing methodology based upon optical diffraction gratings. He then worked jointly as a postdoctoral fellow with Prof James Heath at the California Institute of Technology and with Dr Leroy Hood at the Institute for Systems Biology and developed the DNA-Encoded Antibody Library (DEAL) strategy for multiplexed immunoassays, the core technology behind the Integrated Blood-Barcode Chip (IBBC). Research in Prof Bailey's lab at Illinois is broadly focused on developing new tools to enable multiparameter analysis of complex biological samples with applications in individualized disease diagnostics and theragnostics, drug discovery, and fundamental biology.
Selected publications
- Washburn AL, Gunn LC, Bailey RC. Label-Free Quantitation of a Cancer Biomarker in Complex Media using Silicon Photonic Microring Resonators. doi: 10.1021/ac902006p. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh CR, Fraterman TA, Walker DA, Bailey RC. Direct Biophotolithographic Method for Generating Substrates with Multiple Overlapping Biomolecular Patterns and Gradients. Langmuir. 2009 doi: 10.1021/la9019537. advanced on-line publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qavi AJ, Washburn AL, Byeon J-Y, Bailey RC. Label-Free Technologies for Quantitative Multiparameter Biological Analysis. Anal. & Bioanal. Chem. 2009;394:121–135. doi: 10.1007/s00216-009-2637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey RC, Washburn AL, Qavi AJ, Iqbal M, Gleeson M, Tybor F, Gunn LC. A Robust Silicon Photonic Platform for Multiparameter Biological Analysis. Proc. SPIE. 2009;7220:72200N. [Google Scholar]
- Bailey RC, Kwong G, Radu CG, Witte ON, Heath JR. DNA-Encoded Antibody Libraries: A Unified Platform for Multiplexed Cell Sorting and Detection of Genes and Proteins. J. Am. Chem. Soc. 2007;129:1959–1967. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey RC, Parpia M, Hupp JT. Sensing via Optical Interference. Materials Today. 2005;8:46–52. [Google Scholar]