Abstract
Exposure to metallic environmental toxicants has been demonstrated to induce a variety of oxidative stress responses in mammalian cells. The transcription factor Nrf2 is activated in response to oxidative stress and coordinates the expression of antioxidant gene products. In this study, we describe the development of an Nrf2-specific reporter gene assay that can be used to study the oxidative stress response in multiple cell types. Using five different cell lines, the Nrf2-activating potency of twenty metals was assessed across a range of concentrations. While ten of the metals tested (cadmium, cobalt, copper, gold, iron, lead, mercury, silver, sodium arsenite and zinc) stimulated Nrf2-dependent transcriptional activity in at least three of the engineered cell lines, only three (cadmium, copper and sodium arsenite) were active in all five cell lines. A comparison of metal-induced Nrf2 transcriptional activation revealed significant differences in the absolute magnitude of activation as well as the relative potencies between the cell lines tested. However, there was no direct correlation between activity and potency. Taken together, these results show that the capacity to stimulate Nrf2 activity and relative potencies of these test compounds are highly dependent on the cell type tested. Since oxidative stress is thought to be involved in the mode of action of many toxicological studies, this observation may inform the design of paradigms for toxicity testing for toxicant prioritization and characterization.
Keywords: Nrf2, metals, oxidative stress, reporter gene, cell lines, toxicity testing.
INTRODUCTION
Metazoan cells expend a significant amount of energy to maintain a reducing intracellular environment through the constitutive expression of antioxidant enzymes, metabolic maintenance of reducing equivalents (NAPDH, NADH), protein and peptide thiols, and highly reducing mitochondrial redox potentials. Environmental stressors such as chemical toxicants can create oxidizing imbalances in the cellular redox state resulting in a loss of reducing potential, a condition termed “oxidative stress”. Heavy metals are pervasive environmental toxicants that have been shown to exert oxidative stress on living systems through the production of reactive oxygen species (ROS), which overwhelm the cell’s capacity to maintain a reduced state [1,2]. Metal-induced ROS cause damage to cellular proteins, nucleic acids and lipids, leading to a variety of cellular dysfunctions including cell death. Mammalian cells have developed multiple homeostatic systems to counteract the effects of oxidative stress by scavenging free radicals and repairing oxidant damage to biomolecules. The activation of this antioxidant stress response pathway is a reliable indicator of oxidative perturbation.
In large part, the mammalian oxidative stress response is coordinated by a transcription factor, nuclear factor erythroid 2-related factor 2 (Nrf2) [3-5]. Under normal cellular conditions, Nrf2 is held inactive in the cytoplasm by a protein known as Keap1 [3]. Keap1 negatively regulates Nrf2 activity through interactions with Roc1 and Cul3 which recruit E2 ubiquitin ligase, driving the proteasomal degradation of Nrf2 [6]. This tight regulation of Nrf2 transcriptional activity permits only a minimal, basal level of Nrf2-directed gene expression. Oxidative stress leads to the activation of numerous transducers such as mitogen-activated protein kinases (MAPK, ERK, p38), protein kinase C (PKC), and phosphatidylinositol 3 kinase (PI3K) which phosphorylate both Keap1 and Nrf2 [7, 8]. Reactive electrophiles also directly attack the sulfhydryl-rich Keap1 protein [9], leading to conformational changes in Keap1 structure. Both series of events disrupt the Keap1-Nrf2 complex and stimulate the translocation of stabilized, transcriptionally active Nrf2 to the nucleus where Nrf2 partners with small Maf proteins [10]. These resulting Nrf2/Maf heterodimers bind directly to anti-oxidant response elements (AREs) located within promoters of Nrf2 target genes, displacing the transcriptional repressor Bach1 [11, 12]. This de-repression/activation dramatically increases the transcriptional expression of Nrf2-inducible genes such as those encoding heme oxygenase-1 (HMOX1), NAD(P)H quinone oxidoreductase (NQO1), and glutathione-S transferase A2 (GSTA2). Products of these genes serve to neutralize reactive oxygen species and electrophiles, biosynthesize glutathione, direct xenobiotic efflux and recycle oxidized proteins [13]. In concert these proteins scavenge ROS and conjugate electrophiles to ultimately limit oxidative damage and to detoxify the cell.
Given that several metals are known to generate ROS, and that mammalian cells activate Nrf2-mediated transcription in response to ROS, it is not surprising that metal exposure activates the Nrf2 pathway. Nrf2 activation has been demonstrated in response to a variety of metals [14-16]. Metal exposure has been shown to exert a number of effects on the Nrf2 pathway including reduction of sulfhydryl groups in Keap-1, MAPK activation and resultant Nrf2 phosphorylation, and inhibition of proteasomal pathways which stabilizes Nrf2. The cumulative impact of these events is stabilization and activation of Nrf2 and transcriptional upregulation of anti-oxidant genes. While metals have been previously shown to induce Nrf2-mediated oxidative stress response, these studies have employed a variety of different exposure conditions and different human cell lines such as skin fibroblasts [17], hepatoma cells [18], monocytes [19] and retinal pigment epithelial cells [20]. Both the cell type used and the experimental endpoints can lead to variability in oxidative stress response.
A number of methods have been developed to measure Nrf2 transcriptional activity as a means to monitor cellular response to oxidative stressors including real time PCR [21, 22]. While real-time PCR quantitatively measures endogenous target gene expression, reporter genes offer a higher-throughput, cost-effective assay platform that permits a rapid and more defined analysis of the oxidative stress response at the transcriptional level. Moreover, promoter reporter gene assays provide an assessment of the transcriptional activity associated with the expression of a gene, while quantitative PCR only measures steady-state levels of the transcript, which is influenced by transcriptional activity as well as mRNA stability. Here, we describe the development of a novel Nrf2 reporter gene assay engineered in a portable universal vector that facilitates the rapid generation of various stable reporter cell lines from either established or primary cells. With this Nrf2-specific reporter gene, we established stable reporter cell lines using immortalized/tumor cells derived from human kidney, liver, lung, breast and brain. These stable reporter cell lines were then challenged with a panel of twenty transition-, alkali-, and heavy metals to compare their Nrf2 activating potential in the engineered cell types. Contrary to our expectation of similar Nrf2 activation among cells derived from various human tissues, we found significant differences in Nrf2 activation in response to metal exposure among the cell types tested. Distinct subsets of the test metals elicited Nrf2 responses among the cell lines used; also, the relative potencies of each metal differed with each cell type. We conclude that cells differ dramatically in either their ability to mount an effective oxidative stress response to metal toxicity. These findings suggest that the properties of Nrf2 activation are cell-type specific and may not be predictive of response patterns in other cell types.
MATERIALS AND METHODS
Cell Lines and Growth Conditions
The HEK293T cell line was purchased from GenHunter (Nashville, TN). The HepG2, MCF7, A549 and A172 cell lines were purchased from American Type Culture Collection (Manassas, VA). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Atlanta Biologicals, Lawrenceville, GA) and penicillin-streptomycin (HyClone; Waltham, MA) at 37°C in a humidified 5% CO2 atmosphere.
Reagents
Hydroquinone (HQ), Sodium Chloride (Na), Gold (I) Chloride (Au), Sodium Arsenite (As), Silver Chloride, (Ag), Barium (II) Chloride (Ba), Calcium Chloride (Ca), Cadmium Chloride (Cd), Cobalt Chloride (Co), Chromium (VI) Oxide (Cr), Cesium Chloride (Cs), Copper (II) Chloride (Cu), Iron (II) Chloride (Fe), Mercury (II) Chloride (Hg), Manganese Chloride (Mn), Nickel Chloride (Ni), Osmium (III) Chloride (Os), Lead (II) Chloride (Pb), Rubidium Chloride (Rb), Thallium (I) Nitrate (Tl), Zinc (II) Chloride (Zn), and o-phenylenediamine (OPD) were purchased from Sigma-Aldrich (St. Louis, Mo). Aqueous stock solutions (16mM) were aliqouted and stored in air-tight vials at -80°C until use. Thawed stock aliquots were discarded after each use.
Reporter Gene Construction
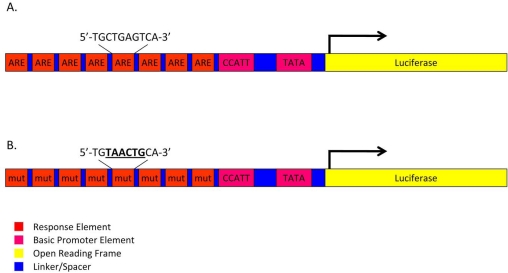
To measure Nrf2 transcriptional activity, we engineered a novel Nrf2-responsive reporter gene by engineering a chimeric enhancer containing seven tandem repeats of the human Nrf2 consensus DNA-binding site, termed the antioxidant response element (ARE: 5’-TGCTGAGTCA-3’; [23] or a mutated ARE (AREmut: 5’-TGTAACTGCACA-3’, mutated bases underlined) separated by 12-bp spacers was custom synthesized (IDT; Coralville, IA) (Fig. 1). The ARE and AREmut enhancers were cloned upstream of a synthetic 97-bp basic promoter element with a CCAAT site (-90 relative to the transcriptional start site) and a TATA binding site (-35 relative to the transcriptional start site) which was subsequently optimized through an iterative process using Tfsitescan (http://www.ifti.org/cgi-bin/ifti/Tfsitescan.pl) to eliminate predicted binding sites for nearly all other known mammalian DNA-binding proteins. The resulting ARE and AREmut promoters were subsequently cloned into the multiple cloning site of a lentiviral transfer vector (pTZV, Thermo Fisher Scientific Inc.,) modified to contain the firefly luciferase open reading frame (derived from pGL3, Promega, Madison, WI) and a secondary tetracycline-inducible turboRED fluorescent reporter. ARE-luciferase and AREmut-luciferase transfer vector constructs were verified by fluorescent DNA capillary sequencing.
Fig. (1).
Schematics of ARE-luciferase and AREmut-luciferase reporter genes. ARE-luciferase reporter gene (A) contains seven multimerized consensus AREs upstream of a basic promoter containing CCAAT and TATA boxes that drive the expression of firefly luciferase. The AREmut-luciferase reporter is identical to the ARE-luciferase reporter except that six core nucleotides within each ARE have been mutated (mutated bases underlined).
shRNA Construction
shRNA hairpin oligonucleotides shown below were designed by selecting an 18-bp site from the human NFE2L2 complete mRNA (NCBI Reference Sequence: NM_006164.3) optimized for siRNA targeting of the NFE2L2 mRNA (S001) or a random 18-bp sequence (scramble) with no predicted homology to human genomic or transcript sequences.:
S001: 5’-TCAAATCCATGTCCTGCT-3’
Scramble: 5’-ACTCTCGCCCAAGCGAGA-3’
Single stranded synthetic 97-bp oligonucleotides (Invitrogen Corp., Valcencia, CA) incorporating the sense/antisense sequences in a stem-loop motif were PCR amplified using the primers forward: 5’-AGTCACTCGAGTGCTGTTGACAGTGAG-3’ and reverse: 5’-AAGTCAGGATCCTCCGAGGCAGTAGG-3.’ The resulting shRNA PCR products were subcloned into a modified lentiviral transfer vector, GIPZ (Thermo Fisher Scientific Inc., Birmingham, AL) between the XhoI and BamHI sites and the constructs were verified by fluorescent DNA capillary sequencing. This cloning strategy nests the shRNA fragments between 5’ and 3’ miRNA30 adaptors within the 3’ untranslated region of a green fluorescent protein reporter gene under the control of a CMV promoter.
Lentiviral Vector Production and Titering
HEK293T cells were co-transfected in 10 cm dishes with purified ARE-luciferase, AREmut-luciferase, pGIPZ-scamble or pGIPZ-S001 transfer vector plasmids and lentiviral packing mix (Open Biosystems; Huntsville, AL) according to manufacturer’s instructions. Sixteen hours post-transfection, cell culture medium was replaced with 12 ml fresh DMEM and cells were incubated for an additional 48 h at 37°C. Medium was then harvested and detached cells were pelleted by centrifugation for 10 minutes at 5,000g. The resulting supernatants from the individual transfections were concentrated once by low-speed centrifugation through an Amicon Ultra 100kD centrifuge filter unit (Millipore; Billerica, MA), and the retentates were aliquoted and stored at -80°C. To determine viral titers, 50,000 HEK293T cells stably expressing the TetOff (rtTA3; Clontech, Mountain View, CA) transactivator were transduced with 50 µl of lentiviral stock dilutions ranging from 1:10 to 1:781,250. Viral titers for ARE-luciferase and AREmut-luciferase (expressed as transducing units per ml viral stock) were determined 96 hours post-transduction by counting red fluorescent colonies by fluorescent microscopy (red colonies form due to rTTA3-mediated activation of the secondary tetracycline-inducible turboRED fluorescent reporter) and multiplying the colony count by the dilution and volume factors. Scramble and S001 shRNA vectors were titered in the same manner using green fluorescent protein (encoded by the vector) to quantify colonies.
Generation of Stable Reporter Gene Assay Cell Lines
HEK293T, HepG2, MCF7, A549 and A172 cell lines were transduced with ARE-luciferase or AREmut-luciferase lentiviral vector at a multiplicity of infection (MOI) of 10. Cells were allowed to grow in culture for seven days post-transduction to amplify the transduced cell lines. Stable HepG2 ARE-luciferase cells were subsequently transduced with either scramble or S001 shRNA vectors at an MOI of 50 in order to ensure maximal Nrf2 knockdown, and were cultured an additional seven days. Cells were then plated in 384-well white culture plates (Nunc, Pittsburgh, PA) at the following densities: HEK293T (104 cells/well), HepG2 (104 cells/well), MCF7 (8x103 cells/well), A549 (8x103 cells/well) and A172 (5x103 cells/well) in DMEM supplemented with 1% FBS and cultured overnight at 37°C. Cells were then exposed for 16 hours with test compounds over a 14-point concentration range spanning 10nM to 1mM for 16 hours. After treatment, culture media were aspirated and cells were lysed with a non-denaturing lysis buffer (25mM Tris phosphate, 2mM trans-1,2-diaminocyclohexane-N,N’N’,N’-tetraacetic acid monohydrate, 10% glycerol, 0.5% Triton-X 100, 2mM Dithiothreitol, pH 7.8) for 15 minutes at ambient temperature. Luciferase activity was detected by adding 25ul Luciferase Detection Buffer (20mM Tricine, 1.07mM (MgCO3)4·Mg(OH)2·5H20, 2.67mM MgSO4, 100μM EDTA, 33.3mM Dithiothreitol, 270μM Coenzyme A, 470μM D-luciferin, 530μM ATP, pH 7.8). Plates were read on a BMG FluoStar (Durham, NC) plate reader with 1 second integration time.
Statistical Analysis, Curve Fitting and Clustering Analysis
All experimental results are aggregates of 3-4 independent experiments. Unless otherwise indicated, all data are normalized to in-plate vehicle controls and are presented as mean fold change over vehicle + SE. A two-tailed paired Student’s t-test was used to evaluate differences between vehicle and treated groups; values of p < 0.05 were considered statistically significant. Concentration-responses were fitted to four-parameter, non-parametric curves using a least squares (ordinary) fit method using GraphPad Prism 5 (San Diego, CA). A Talalay’s CD (CD) value, defined as the concentration required to elicit a doubling of the baseline response [24], was calculated from the fitted curves using GraphPad’s log (agonist) vs. response—Find EC anything function where F = 2/maximal Nrf2 activity x 100% (Table 2). All data points from concentrations higher than the ECmax were masked to minimize the effects of overt cytotoxicity on curve fitting. In cases where the ECmax was equal to the highest concentration tested (1mM), the response value at 1mM was assumed to be the maximum response level for the purposes of curve fitting. Unsupervised hierarchical clustering was performed on the chemical (row) by cell type (column) matrix of either CD or maximal Nrf2 activity values using MultiExperiment Veiwer v4.3 [25] with Manhattan distance as the distance metric and average linkage in clusters.
Table 2.
Maximum Nrf2 Induction Activity of Metals in Reporter Cell Lines
| Compound | HEK293T | HepG2 | MCF7 | A172 | A549 |
|---|---|---|---|---|---|
| HQ | 6.9# | 10.9 | 2.0 | 10.0 | NR* |
| Au | 3.9 | 7.1 | 3.2 | 25.9 | NR |
| As | 14.0 | 32.9 | 6.0 | 61.2 | 6.0 |
| Ag | 3.9 | 6.9 | 68.8 | 9.1 | NR |
| Cd | 62.5 | 4.0 | 23.1 | 13.8 | 3.3 |
| Co | 3.0 | 4.5 | 5.3 | NR | NR |
| Cu | 4.2 | 8.3 | 55.7 | 15.5 | 12.2 |
| Fe | 2.2 | 2.4 | NR | 5.1 | NR |
| Hg | 8.1 | 5.1 | 4.1 | 6.8 | NR |
| Pb | 3.7 | 3.8 | 1.9 | 1.9 | NR |
| Zn | 3.4 | 2.6 | 2.1 | 2.6 | NR |
| OPD | 5.3 | 3.4 | 2.5 | 6.4 | NR |
fold change over vehicle treated control;
No response.
RESULTS
ARE-Luciferase Reporter Gene Accurately Reflects Nrf2-Mediated Cellular Oxidative Stress Response
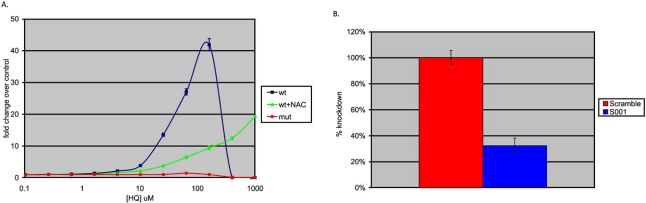
To determine whether the ARE-luciferase reporter was functional, human HepG2 cells were transduced with the ARE reporter vector and were subsequently passaged for seven days to allow for transgene stabilization. The resulting HepG2 ARE-luciferase reporter cells were treated with hydroquinone (HQ), a compound known to induce oxidative damage in HepG2 cells, at concentrations ranging from 10nM-1mM for 16 hours and then assayed for luciferase activity. The luciferase activity increased in a concentration-dependent manner (Fig. 2), indicating that the ARE-luciferase reporter is functionally inducible [26]. In parallel, HepG2 reporter cells were pre-treated with 5mM N-acetylcysteine (NAC), a widely used sulfhydryl antioxidant 2 hours prior to HQ treatment [27]. As shown in Fig. (2), NAC pre-treatment clearly reduces the potency of HQ (EC50=48.1µM vs EC50=208µM with pre-treatment), but also reduces HQ-induced Nrf2 maximal activity as shown by a 4.4-fold reduction in the magnitude of activation at 160µM, which was the maximal effective concentration (ECmax) for HQ in the absence of NAC. These results demonstrate that the ARE reporter responds to oxidative stress and this response is ameliorated by antioxidant (NAC) treatment.
Fig. (2).
ARE response to oxidative stress requires functional AREs and Nrf2. (A) ARE-luciferase response to 16-hour hydroquinone (HQ) in HepG2 cells (blue), attenuation of the response by pre-treatment with a antioxidant n-acetyl cysteine (NAC) (green) and lack of response of AREmut-luciferase reporter to HQ treatment (red). (B) Reduction in Nrf2 levels via Nrf2-specific shRNA inhibition impairs response to HQ (100 M, 16 hours) in HepG2 ARE-luciferase stable reporter cells (blue) whereas the response is unaffected by scrambled shRNA control (red).
To further confirm the Nrf2 specificity of the ARE reporter response described above, a parallel HepG2 line was generated harboring a luciferase reporter identical to ARE-luciferase in every respect except that six of the consensus nucleotides of the AREs were mutated so as to alter the binding affinity of the transcription factor (Fig. 1B). This AREmut-luciferase cell line was produced by lentiviral transduction using an identical MOI to permit direct comparison of the response of ‘wild-type’ ARE to the mutated version. As shown in Fig. (2), the mutated ARE reporter did not respond to HQ-treatment, indicating that functional AREs are sufficient and necessary for the ‘wild-type’ reporter construct responsiveness to oxidative stressors such as hydroquinone.
In order to determine whether the HQ-induced increase in reporter activity was driven by Nrf2, we next stably transduced HepG2-ARE/luciferase cells with a constitutively active gene expressing an anti-Nrf2 shRNA (S001) or one expressing a negative control shRNA (scrambled). As shown in Fig. (2), HepG2 reporter cells expressing the Nrf2-targeting shRNA had a markedly lower luciferase activity when challenged with 100µM HQ as compared with cells expressing a negative control shRNA, indicating that the ARE-luciferase reporter response to HQ treatment results from activated Nrf2. Taken together, these results show that the synthetic ARE–luciferase reporter responds to oxidative treatment by Nrf2-specific transcriptional activity signaling through a functional antioxidant response element, and this response can be blunted by antioxidant pre-treatment.
Heavy Metals Activate Different Nrf2 Responses within a Cell Type
To enable the comparison of metal-induced Nrf2 transcriptional activities in different cell types derived from different tissues, stable ARE reporter cells lines were established from five commonly-used human cell models: HEK293T (kidney), A172 (brain), A549 (lung), HepG2 (liver) and MCF7 (breast). Each stable reporter cell line was generated using an identical MOI. These cells were then challenged with a battery of 20 metal salts (Fig. 3), hydroquinone (positive control) or a reference compound, o-phenylenediamine (OPD). Cells were treated for 16 hours, and luciferase activity was measured. Concentration-response curves were then generated; the resulting CD values and maximal fold increases in Nrf2 activities are shown in Tables 1 and 2, respectively.
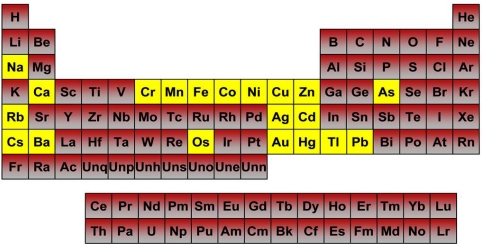
Fig. (3).
Metals studied for oxidative stress induction potential and their grouping in the periodic table. The twenty alkali, transition and heavy metals tested in cell lines are highlighted in yellow. Doses ranged from 10nM to 1mM and the cells were exposed for 16 hours. All test compounds were cation salts except for sodium arsenite (NaAsO2).
Table 1.
Nrf2 Activation Potency of Metals in Reporter Cell Lines CD (uM)
| Compound | HEK293T | HepG2 | MCF7 | A172 | A549 |
|---|---|---|---|---|---|
| HQ | 7.12 | 20.8 | # | 33.2 | NR* |
| Au | 76.1 | 169 | 740 | 146 | NR |
| As | 1.88 | 16.5 | 9.05 | 15.9 | 123 |
| Ag | 11.8 | 2.52 | 4.73 | 5.54 | NR |
| Cd | .907 | .954 | 11 | 6.03 | 54.7 |
| Co | 484 | 185 | 532 | NR | NR |
| Cu | 281 | 455 | 295 | 136 | 392 |
| Fe | # | # | NR | 239 | NR |
| Hg | 2.82 | 19.5 | 6.35 | 6.18 | NR |
| Pb | # | 426 | # | # | NR |
| Zn | 84.8 | 249 | 256 | 100 | NR |
| OPD | 16.8 | 110 | 42.7 | 46.6 | NR |
No response,
Calculated CD exceeds empirically tested concentration range.
Half of the tested metals failed to induce any appreciable Nrf2 activity compared to vehicle-treated controls in any of the five stable reporter cell models. All of the Group I and Group II metals tested in this study (sodium, calcium, rubidium, cesium and barium) failed to produce either a significant Nrf2 response or cytotoxicity in any of the cell lines at the concentrations used. Similarly, chromium (VI), manganese (II), osmium (III), nickel (II) and thallium (I) did not stimulate Nrf2 activity in any cell model tested; however, with the exception of osmium, all of these induced overt cytotoxicity at the highest concentrations (data not shown). The specific responses of individual cell lines to metal expsoure are described in detail below:
HEK293T
In HEK293T cells, derived from human embryonic kidney, there was a marked increase in Nrf2 activity in response to ten of the twenty test compounds (Tables 1 and 2). Most of the ten Nrf2-inducing metals stimulated luciferase activities ranging from 2- to 8-fold over vehicle-treated controls. The two notable exceptions were sodium arsenite (14.0-fold) and cadmium (62.5-fold). There was a wide disparity among the potencies of positive compounds. Heavy metals cadmium (CD=0.907μM) and mercury (CD=2.82μM) were very potent, as was silver (CD=11.8μM) and sodium arsenite (CD=1.88μM). Other metals in this group elicited milder Nrf2 responses at the higher concentrations tested: zinc (CD=84.8μM), cobalt (CD=484μM) and copper (CD=281μM).
HepG2
HepG2 cells, derived from a human hepatocellular carcinoma, also detected appreciable Nrf2 activity induction for the same subset of ten test compounds as HEK293T cells. Also like HEK293T cells, the level of Nrf2 activities ranged from 2- to 8-fold over vehicle-treated controls. A striking exception in this cell line was sodium arsenite, which induced a 32.9-fold increase in activity. Interestingly, the response to cadmium was muted in this cell line in comparison to HEK293T cells. The relative potencies of the positive test compounds showed an even wider disparity than seen in HEK293T cells. Like HEK293T cells, cadmium was the most potent inducer of Nrf2 activity (CD=0.954μM) and metals such lead (CD=426μM), cobalt (CD=185μM) and copper (CD=455μM) elicited Nrf2 stimulation at only the higher tested concentrations.
MCF7
MCF7 human breast tumor cells responded to nine of the twenty test compounds; all nine of these metals tested positive in the HEK293T and HepG2 reporter cell lines. In contrast to HEK293T and HepG2 cells, MCF7 cells failed to respond to iron with a significant Nrf2 activation. Another interesting finding in the MCF7 assay was that silver (CD=4.73μM), not cadmium (CD=11.0μM), was the most potent Nrf2 inducing test compound; in fact, gold had the lowest CD value in MCF7 cells than the other cell models tested. With respect to maximal Nrf2 activities, by far the most dramatic fold increases over controls were observed for silver (68.8), copper (55.7) and cadmium (23.1); these values were the highest observed for any cell model tested. Most other positive metals had maximal activities in the 2- to 6-fold range.
A172
Nine of the twenty test compounds stimulated significant Nrf2 activity in stable reporter cells derived from the A172 human glioblastoma cell line; these nine were also a subset of the ten positives from the HEK293T and HepG2 assays. Interestingly, cobalt did not stimulate Nrf2 activity in these cells, as it had in HEK293T, HepG2 and MCF7 cells. Like the MCF7 cells, silver was the most potent inducer of Nrf2 transcriptional activity (CD=6.03μM) in A172 cells. The relative potencies of other positive test agents ranked in the middle compared to the other reporter cell models. Among the five cell lines tested, the maximal fold activations of gold (25.9) and sodium arsenite (61.2) were highest in A172 cells. Cadmium (13.8) and copper (15.5) being significantly more active than the other metals.
A549
The A549 reporter cells only detected significant Nrf2 activities in three of the twenty test agents: sodium arsenite, cadmium and copper. That three test agents had effects provides evidence that the Nrf2 signaling pathway is functional in these cells; however, the CD values for all three positives were the highest, or ranked with the highest, observed in any cell model, suggesting that these cells are relatively resistant to oxidative insult induced by metals. Also noteworthy is the observation that the magnitudes of the responses to exposure to sodium arsenite and cadmium were lower in A549 cells than any of the other models in this study, and A549 cells were the only tested model that failed to respond to hydroquinone control. Interestingly, copper induced the highest Nrf2 activity level observed in these cells.
Heavy Metals Activate Different Nrf2 Responses Across Cell Types
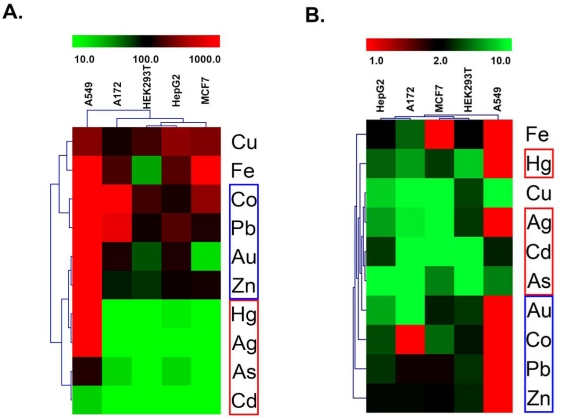
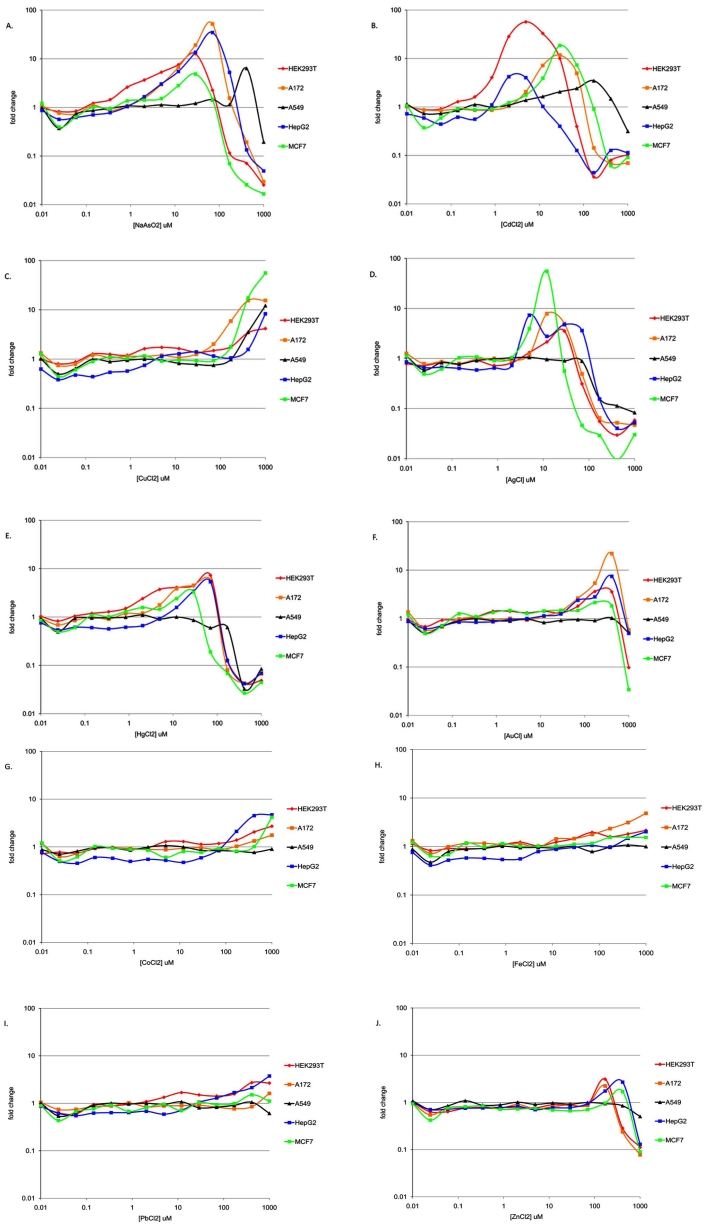
In the course of examining Nrf2 response profiles within a particular cell type, some interesting patterns of response across cell types emerged. Hierarchical clustering was used to better organize these response patterns (Fig. 4). Response patterns were clustered using two different metrics of the dose response curves for the ten positive test compounds, viz., CD values (Fig. 4A) and maximal activity (Fig. 4B). In both response maps, it is clear that A549 cells were the most atypical among the reporter lines tested. The two activity maps also reveal a common pattern of the positive compounds into three distinct response classes. Response class I compounds (Fig. 4, red boxes) include sodium arsenite, cadmium and copper. These metals grouped together because they were active in all five cell models tested. The individual response curves for each of these compounds are shown in Fig. (5A-C). Each of these metals was most active in a different cell model (As-A172, Cd-HEK293T, Cu-MCF7). Response class II compounds (Fig. 4, blue boxes) elicited the most significant Nrf2 activities and did so at concentrations below 100µM, but only in a subset of the cell models tested. Silver, mercury and gold were in response class II. The individual response curves for each of these compounds are shown in Fig. (5D-F). Each of these metals was most active in a different cell model (Ag-MCF7, Hg-HEK293T, Au-A172). The third response class (Fig. 4, violet boxes) includes cobalt, iron, lead and zinc. These compounds generally stimulated milder Nrf2 activities than those in response class II, however at the highest concentrations tested, between 100 and 1000µM (Fig. 5G-J). Again, each of these compounds had the highest activity in a different cell model- cobalt in MCF7 cells, iron in A172 cells, lead in HepG2 cells and zinc in HEK293T cells. Although gold was included in response class II, it did not neatly fall within either response class II or III because it strongly stimulated Nrf2 activity (like response class II metals), but only at the highest tested concentrations like response class III metals.
Fig. (4).
Hierarchical clustering of Nrf2 response patterns. Cell responses for the ten active metals were clustered using CD (A) and maximal activity (B) derived from the concentration-response curves. Response class I compounds (red boxes) elicited Nrf2 activities in all tested cell models. Response class II compounds (blue boxes) elicited the most significant Nrf2 activities at the lowest concentrations in a subset of cell models. Response class III (violet boxes) generally stimulated milder Nrf2 activities than those in response classes I and II and only at the highest concentrations tested.
Fig. (5).
Concentration-response curves for Response Class I, II and III metals in the five reporter cell lines. Luciferase activities (expressed as fold change over vehicle control) in response to each member of (A-C) Response Class I, (D-F) II and (G-J) III were compiled for all five cell lines.
DISCUSSION
Oxidative stress, one of the most common mechanisms of cellular injury, is measured using a broad array of techniques ranging from measurement of ROS, to monitoring reduced glutathione levels, and assaying for products of DNA, lipid, or protein oxidation [28]. Often the methods are technically laborious thus calling for a rapid and economical assay that can be used to assess oxidative stress injury with improved throughput. Such a method is vital to realizing the vision of the future of toxicology enunciated in the recent NRC Report [29], wherein assays in human-derived cells are used as proxies for animal tests. The findings that we describe in this study represent the development and characterization of a rapid and economical cell-based assay for the measurement of oxidative stress that is amenable to ultra high-throughput screening.
The transcription factor Nrf2 is one of the key coordinators of an effective cellular response to oxidative stress. This study describes the development of a novel cell-based transcriptional reporter assay to monitor Nrf2-dependent transcriptional activation. The ARE-luciferase reporter makes use of multimerized consensus AREs placed upstream of a synthetic promoter engineered to be devoid of putative sites for DNA-binding proteins other than Nrf2. This design offers the distinct advantage of being highly specific relative to existing reporter genes used to monitor Nrf2 activity. Reporter genes driven by promoter elements derived from genomic sources (i.e., target genes) which have been used previously do utilize relevant response element(s). However, these response elements are found in the context of binding sites for other transcription factors which facilitates complex combinatorial interactions between transcription factors. For instance, the human HMOX1 promoter is commonly used in reporter genes to measure Nrf2 activity because it has perhaps the best-characterized AREs of any Nrf2 target gene studied to date [30]. However, a recent study showed that HMOX1 can be stimulated by cadmium independently of Nrf2 through an element termed the cadmium response element (CdRE), which is nested between two AREs [31]. HMOX1 is also known to be activated by other ROS-responsive transcription factors such as AP-1 and NFkB [32-35], complicating the mechanistic interpretation of gene activation by toxicants when using these gene-based reporters.
The specificity of transcriptional activation of genomic promoter reporter genes can be further investigated through the use of response element mutants or other methods to ensure that the observed responses observed result from activation of the transcription factor under study; however these approaches are laborious and are complicated by the combinatorial interactions of the elements being studied. This problem of specificity can be addressed more elegantly through the use of multimerized response element reporters, which are designed to eliminate non-specific binding sites and therefore largely isolate the activity of a single transcription factor.
A recent development in assay design employed in this study is the use of lentiviral vectors to efficiently deliver reporter genes to a range of cell lines enabling the rapid generation of stable reporter cell lines [36]. Lentiviral vectors effectively delivered the ARE luciferase transgenes to the cell types used in this study evidenced by the robust luciferase signal observed in all five stable reporter cell lines. Although the basal reporter gene activity levels for these cell lines were within a three-fold range of one another (data not shown), normalization to vehicle-treated controls makes meaningful comparisons of metal-induced Nrf2 activity across different cell models possible.
Five of the metals belonging to Group I and Group II of the periodic table (sodium, calcium, rubidium, cesium and barium), tested negative in all of the cell lines used. This was not unexpected, as no evidence for Group I or Group II Nrf2-inducing cations have been reported to date. Another group of Nrf2-inactive metals included four that were clearly cytotoxic: thallium (I), chromium (VI), manganese and nickel. The lack of Nrf2 activation preceding cytotoxicity (data not shown) suggests that these compounds induce cytotoxicity through a mechanism other than oxidative stress. Alternatively, it may be that the oxidative stress response to these metals does not involve Nrf2 transcriptional activity. This latter observation is seemingly at odds with the findings of recent studies. He et al. [37] demonstrated that hexavalent chromium exposure in mouse Hepa1c1c7 cells stimulated Nrf2 nuclear translocation and activation of target genes Hmox1 and Nqo1 via their respective AREs. Another study showed significant Nrf2 nuclear localization in the liver cells of rats exposed to manganese [38]. Lastly, a 2006 study by Lewis et al. [39] showed that nickel exposure led to increased Nrf2 nuclear accumulation in human THP1 monocytes. The studies by Casalino et al. and Lewis et al. [38, 39] not only used different cell models, but did not measure Nrf2 transcriptional activity, but rather nuclear translocation/accumulation.
Ten of the twenty metals tested triggered Nrf2 activity in at least three or more cell types. Among the ten Nrf2-inducing metals, cadmium, copper and sodium arsenite were positive in all of the cell lines. Five among the ten were active in all of the reporter cell lines except A549 cells. A549 cells, derived from a human alveolar basal epithelial adenocarcinoma, are widely used to dissect signaling mechanisms related to inflammation in human lung, but have been shown to have remarkably high levels of intracellular glutathione and to be more resistant to glutathione depletion than normal human lung fibroblasts [40]. This “buffering” effect of the high levels of antioxidant glutathione likely accounts for the relatively low number of Nrf2-stimulating metals observed in A549 cells, especially when one considers that for all three A549-positive test compounds, the observation that the CD values for the three metals active in this cell line were among the highest when compared to the other cell lines in this study. Also, A549 cells harbor a mutation in the gene encoding Keap1 [41]. The resulting mutant Keap1 protein is ineffective at negatively regulating Nrf2 activity resulting in a reduced capacity to induce Nrf2 activity in these cells. Differences in maximal Nrf2 activities and relative potencies of the positive metals showed pronounced differences among the other four cell lines tested. Although the reasons for these differences are not immediately clear, it is noteworthy that such differences in potency and efficacy across multiple cell models have been noted for other endpoints such as cell viability and proliferation [42, 43].
The clustering analysis approach used to mine for patterns of Nrf2 response across the five cell models may be somewhat constricted by the number of cell types and positive test compounds, but despite these limitations, clear patterns of response were observed. A549 cells were clearly the most atypical cell model used in this study with respect to the number of Nrf2-inducing metals detected and relative potencies and activities of the Nrf2 responses. The clustering analysis also segregated the ten Nrf2-stimulating metals into three groups. Sodium arsenite, cadmium and copper were effective at stimulating Nrf2 activity in all five cell models tested, including A549 cells. Silver, mercury and gold were potent and highly efficacious inducers of Nrf2 activity in all of the tested cell models except A549 cells. Cobalt, iron, lead and zinc were less potent than class II compounds and elicited cell type specific variations in efficacy. One interesting observation stemming from the clustering analysis is that despite the significant cell type specific differences in both potency and efficacy, a similar biological response pattern emerged irrespective of which metric was used for clustering. Therefore, while potency and efficacy may not be strictly predictive of one another across cell models for a single compound, these two parameters seem to be generally proportional across multiple models, such that a decrease in potency generally corresponds to decreased efficacy and vice versa.
Studies on nearly all of the metals that tested positive have been reported in the literature in at least one of the cell lines used here albeit in limited studies examining one or two metals in a single cell model. Our study is the first to comprehensively compare the impact of heavy metal exposure on Nrf2 activity using multiple test agents across multiple cell models. An analogous study conducted in 1993 by Fischbach et al. examined the induction of a human heat shock 70 promoter by 31 metals using a human growth hormone reporter assay in mouse NIH3T3 cells [44]. Of the fifteen metals that overlapped between the present study and the Fishbach study, nine were positive and six were negative in the ARE-luciferase assay. Copper, cadmium and sodium arsenite were the most efficacious inducers of heat shock response in the Fischbach study; interestingly, these metals were the only three that stimulated Nrf2 activity in all five cell lines tested in this study. However, the Fischbach study reported heat shock activities for several metals that tested negative in the Nrf2 activity assay such as nickel, manganese, thallium (I), and barium indicating that heat shock response and oxidative stress response may not overlap for either mechanistic or cell line-related differences.
Given the level of phylogenetic conservation of the Nrf2 signaling pathway, from nematodes to humans [45], it could be expected that Nrf2 activation in response to toxicants would be uniformly manifested. However, the central finding in the present study is that Nrf2 activity profiles vary greatly between cell models. Differences in responses between cell models could be attributed to factors such as tissue of origin, cellular subtype, inter-individual variability in the source, and culture conditions. Differences in response of cells from different organs to heavy metals have been noted in vivo and these differences are attributable to pharmacokinetic factors such as absorption and distribution as well as to pharmacodynamic actions at a cellular level such as oxidative stress induction [2]. Although many of the metals play vital roles in biology as enzyme cofactors and protein stabilizers, being charged entities metal ions do not passively cross the hydrophobic biological membranes, leading to differences in both their pharmacokinetic and pharmacodynamic properties [46]. However, cells have developed a variety of ways for the uptake and storage of metals such as transporters and binding proteins, etc. Mechanisms have also been developed to counteract excessive levels of the required metals such as induction of protective proteins, transporter downregulation and efflux pumps. These pathways are also used by nonessential toxic metals (e.g. cadmium, using zinc binding proteins). Thus the differences in responses between cells derived from different tissues can be attributed to differences in the activities or abundance of critical proteins [46]. Further activity differences could arise from the idiosyncrasies that come about from immortalization, such as pathway dysregulation and genetic instability.
Cell-based assays such as the one described here provide a rapid, inexpensive and high-throughput approach to prioritize and characterize potential toxicants. One of the key findings of this study is that significant differences exist between various in vitro models, even for endpoints and pathways thought to be ubiquitously expressed and highly conserved. It is routine in toxicology to study oxidative stress induction by metals and other environmental toxicants using a variety of cell types. This study reinforces the conclusion that no single in vitro model can provide a complete picture of biological function or its perturbation by toxicants. A more comprehensive approach would involve the use of multiple parallel models whereby confidence in assessing toxic potential and chemical mode of action is increased through overlapping observations.
ACKNOWLEDGEMENTS
The authors are grateful to Drs. Chris Corton, James Samet, James Allen and Julian Preston of the National Health and Environmental Effects Research Laboratory of the US EPA for critical review of the manuscript and helpful comments and to Keith Tarpley of SRA International, Inc. for assistance with graphics.
FUNDING
U.S. Environmental Protection Agency.
DISCLAIMER
This document has been reviewed by the National Health and Environmental Effects Research Laboratory of US EPA and approved for publication. Approval does not signify that the contents reflect the views of the Agency, nor does mention of trade names or commercial products constitute the endorsement of recommendation for use.
REFERENCES
- 1.Ercal N, Gurer-Orhan H, Aykin-Burns N. Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage. Curr Top Med Chem. 2001;1(6):529–39. doi: 10.2174/1568026013394831. [DOI] [PubMed] [Google Scholar]
- 2.Stohs SJ, Bagchi D. Oxidative mechanisms in the toxicity of metal ions. Free Radic Biol Med. 1995;18(2):321–36. doi: 10.1016/0891-5849(94)00159-h. [DOI] [PubMed] [Google Scholar]
- 3.Kang KW, Lee SJ, Kim SG. Molecular mechanism of nrf2 activation by oxidative stress. Antioxid Redox Signal. 2005;7(11-12):1664–73. doi: 10.1089/ars.2005.7.1664. [DOI] [PubMed] [Google Scholar]
- 4.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 5.Osburn WO, Kensler TW. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutat Res. 2008;659(1-2):31–9. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J Biol Chem. 2005;280(36):31768–75. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- 7.Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem. 2002;277(45):42769–74. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 8.Kang KW, Ryu JH, Kim SG. The essential role of phosphatidylinositol 3-kinase and of p38 mitogen-activated protein kinase activation in the antioxidant response element-mediated rGSTA2 induction by decreased glutathione in H4IIE hepatoma cells. Mol Pharmacol. 2000;58(5):1017–25. doi: 10.1124/mol.58.5.1017. [DOI] [PubMed] [Google Scholar]
- 9.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99(18):11908–13. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kataoka K, Handa H, Nishizawa M. Induction of cellular antioxidative stress genes through heterodimeric transcription factor Nrf2/small Maf by antirheumatic gold(I) compounds. J Biol Chem. 2001;276(36):34074–81. doi: 10.1074/jbc.M105383200. [DOI] [PubMed] [Google Scholar]
- 11.Dhakshinamoorthy S, Jain AK, Bloom DA, Jaiswal AK. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J Biol Chem. 2005;280(17):16891–900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- 12.Reichard JF, Motz GT, Puga A. Heme oxygenase-1 induction by NRF2 requires inactivation of the transcriptional repressor BACH1. Nucleic Acids Res. 2007;35(21):7074–86. doi: 10.1093/nar/gkm638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klaassen CD, Reisman SA. Nrf2 the rescue: effects of the antioxidative/electrophilic response on the liver. Toxicol Appl Pharmacol. 2010;244(1):57–65. doi: 10.1016/j.taap.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Massrieh W, Derjuga A, Blank V. Induction of endogenous Nrf2/small maf heterodimers by arsenic-mediated stress in placental choriocarcinoma cells. Antioxid Redox Signal. 2006;8(1-2):53–9. doi: 10.1089/ars.2006.8.53. [DOI] [PubMed] [Google Scholar]
- 15.Korashy HM, El-Kadi AO. Transcriptional regulation of the NAD(P)H:quinone oxidoreductase 1 and glutathione S-transferase ya genes by mercury, lead, and copper. Drug Metab Dispos. 2006;34(1):152–65. doi: 10.1124/dmd.105.005397. [DOI] [PubMed] [Google Scholar]
- 16.Alam J, Wicks C, Stewart D, et al. Mechanism of heme oxygenase-1 gene activation by cadmium in MCF-7 mammary epithelial cells. Role of p38 kinase and Nrf2 transcription factor. J Biol Chem. 2000;275(36):27694–702. doi: 10.1074/jbc.M004729200. [DOI] [PubMed] [Google Scholar]
- 17.Cortese-Krott MM, Munchow M, Pirev E, et al. Silver ions induce oxidative stress and intracellular zinc release in human skin fibroblasts. Free Radic Biol Med. 2009;47(11):1570–7. doi: 10.1016/j.freeradbiomed.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 18.Hou WH, Rossi L, Shan Y, Zheng JY, Lambrecht RW, Bonkovsky HL. Iron increases HMOX1 and decreases hepatitis C viral expression in HCV-expressing cells. World J Gastroenterol. 2009;15(36):4499–510. doi: 10.3748/wjg.15.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis JB, Messer RL, Pitts L, Hsu SD, Hansen JM, Wataha JC. Ni(II) ions dysregulate cytokine secretion from human monocytes. J Biomed Mater Res B Appl Biomater. 2009;88(2):358–65. doi: 10.1002/jbm.b.31063. [DOI] [PubMed] [Google Scholar]
- 20.Rezaei KA, Chen Y, Cai J, Sternberg P. Modulation of Nrf2-dependent antioxidant functions in the RPE by Zip2, a zinc transporter protein. Invest Ophthalmol Vis Sci. 2008;49(4):1665–70. doi: 10.1167/iovs.07-0959. [DOI] [PubMed] [Google Scholar]
- 21.Ade N, Leon F, Pallardy M, et al. HMOX1 and NQO1 genes are upregulated in response to contact sensitizers in dendritic cells and THP-1 cell line: role of the Keap1/Nrf2 pathway. Toxicol Sci. 2009;107(2):451–60. doi: 10.1093/toxsci/kfn243. [DOI] [PubMed] [Google Scholar]
- 22.Dhakshinamoorthy S, Jaiswal AK. Small maf (MafG and MafK) proteins negatively regulate antioxidant response element-mediated expression and antioxidant induction of the NAD(P)H:Quinone oxidoreductase1 gene. J Biol Chem. 2000;275(51):40134–41. doi: 10.1074/jbc.M003531200. [DOI] [PubMed] [Google Scholar]
- 23.Tsuji Y. JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress. Oncogene. 2005;24(51):7567–78. doi: 10.1038/sj.onc.1208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fahey JW, Stephenson KK, Dinkova-Kostova AT, Egner PA, Kensler TW, Talalay P. Chlorophyll, chlorophyllin and related tetrapyrroles are significant inducers of mammalian phase 2 cytoprotective genes. Carcinogenesis. 2005;26(7):1247–55. doi: 10.1093/carcin/bgi068. [DOI] [PubMed] [Google Scholar]
- 25.Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 26.Luo L, Jiang L, Geng C, Cao J, Zhong L. Hydroquinone-induced genotoxicity and oxidative DNA damage in HepG2 cells. Chem Biol Interact. 2008;173(1):1–8. doi: 10.1016/j.cbi.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Lauterburg BH, Corcoran GB, Mitchell JR. Mechanism of action of N-acetylcysteine in the protection against the hepatotoxicity of acetaminophen in rats in vivo. J Clin Invest. 1983;71(4):980–91. doi: 10.1172/JCI110853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halliwell B, Whiteman M. Measuring reactive species and oxidative damage in vivo and in cell culture: how should you do it and what do the results mean? Br J Pharmacol. 2004;142(2):231–55. doi: 10.1038/sj.bjp.0705776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Research Council. Toxicity Testing in the 21st Century: A vision and a strategy. Washington DC: National Academies Press; 2007. [DOI] [PubMed] [Google Scholar]
- 30.Prestera T, Talalay P, Alam J, Ahn YI, Lee PJ, Choi AM. Parallel induction of heme oxygenase-1 and chemoprotective phase 2 enzymes by electrophiles and antioxidants: regulation by upstream antioxidant-responsive elements (ARE) Mol Med. 1995;1(7):827–37. [PMC free article] [PubMed] [Google Scholar]
- 31.Sikorski EM, Uo T, Morrison RS, Agarwal A. Pescadillo interacts with the cadmium response element of the human heme oxygenase-1 promoter in renal epithelial cells. J Biol Chem. 2006;281(34):24423–30. doi: 10.1074/jbc.M602287200. [DOI] [PubMed] [Google Scholar]
- 32.Hsieh HL, Wu CY, Wang HH, Yang CM. ROS-dependent c-Fos/AP-1 Induction Up-regulates Heme Oxygenase-1 Expression by Bradykinin in Brain Astrocytes. Antioxid Redox Signal. 2010;13(12):1829–44. doi: 10.1089/ars.2009.2957. [DOI] [PubMed] [Google Scholar]
- 33.Ferrandiz ML, Devesa I. Inducers of heme oxygenase-1. Curr Pharm Des. 2008;14(5):473–86. doi: 10.2174/138161208783597399. [DOI] [PubMed] [Google Scholar]
- 34.Lavrovsky Y, Schwartzman ML, Levere RD, Kappas A, Abraham NG. Identification of binding sites for transcription factors NF-kappa B and AP-2 in the promoter region of the human heme oxygenase 1 gene. Proc Natl Acad Sci USA. 1994;91(13):5987–91. doi: 10.1073/pnas.91.13.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alam J, Den Z. Distal AP-1 binding sites mediate basal level enhancement and TPA induction of the mouse heme oxygenase-1 gene. J Biol Chem. 1992;267(30):21894–900. [PubMed] [Google Scholar]
- 36.Mitta B, Rimann M, Ehrengruber MU, et al. Advanced modular self-inactivating lentiviral expression vectors for multigene interventions in mammalian cells and in vivo transduction. Nucleic Acids Res. 2002;30(21):e113. doi: 10.1093/nar/gnf112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He X, Lin GX, Chen MG, Zhang JX, Ma Q. Protection against chromium (VI)-induced oxidative stress and apoptosis by Nrf2. Recruiting Nrf2 into the nucleus and disrupting the nuclear Nrf2/Keap1 association. Toxicol Sci. 2007;98(1):298–309. doi: 10.1093/toxsci/kfm081. [DOI] [PubMed] [Google Scholar]
- 38.Casalino E, Calzaretti G, Landriscina M, Sblano C, Fabiano A, Landriscina C. The Nrf2 transcription factor contributes to the induction of alpha-class GST isoenzymes in liver of acute cadmium or manganese intoxicated rats: comparison with the toxic effect on NAD(P)H:quinone reductase. Toxicology. 2007;237(1-3):24–34. doi: 10.1016/j.tox.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 39.Lewis JB, Messer RL, McCloud VV, Lockwood PE, Hsu SD, Wataha JC. Ni(II) activates the Nrf2 signaling pathway in human monocytic cells. Biomaterials. 2006;27(31):5348–56. doi: 10.1016/j.biomaterials.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 40.Russo A, DeGraff W, Friedman N, Mitchell JB. Selective modulation of glutathione levels in human normal versus tumor cells and subsequent differential response to chemotherapy drugs. Cancer Res. 1986;46(6):2845–8. [PubMed] [Google Scholar]
- 41.Singh A, Misra V, Thimmulappa RK, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3(10):e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia M, Huang R, Witt KL, et al. Compound cytotoxicity profiling using quantitative high-throughput screening. Environ Health Perspect. 2008;116(3):284–91. doi: 10.1289/ehp.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costa LG, Fattori V, Giordano G, Vitalone A. An in vitro approach to assess the toxicity of certain food contaminants: methylmercury and polychlorinated biphenyls. Toxicology. 2007;237(1-3):65–76. doi: 10.1016/j.tox.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Fischbach M, Sabbioni E, Bromley P. Induction of the human growth hormone gene placed under human hsp70 promoter control in mouse cells: a quantitative indicator of metal toxicity. Cell Biol Toxicol. 1993;9(2):177–88. doi: 10.1007/BF00757579. [DOI] [PubMed] [Google Scholar]
- 45.An JH, Blackwell TK, SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17(15):1882–93. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lyons TJ, Eide DJ. In: Transport and storage of metal ions in biology. Gray HB, Stiefel EI, Valentine SJ, Bertini I, editors. University Science Books; 2003. pp. 57–78. [Google Scholar]