Abstract
The P2Y14 receptor is a G protein-coupled receptor activated by uridine-5′-diphosphoglucose and other nucleotide sugars that modulates immune function. Covalent conjugation of P2Y14 receptor agonists to PAMAM (polyamidoamine) dendrimers enhanced pharmacological activity. Uridine-5′-diphosphoglucuronic acid (UDPGA) and its ethylenediamine adduct were suitable functionalized congeners for coupling to several generations (G2.5–6) of dendrimers (both terminal carboxy and amino). Prosthetic groups, including biotin for avidin complexation, a chelating group for metal complexation (and eventual magnetic resonance imaging), and a fluorescent moiety, also were attached with the eventual goals of molecular detection and characterization of the P2Y14 receptor. The activities of conjugates were assayed in HEK293 cells stably expressing the human P2Y14 receptor. A G3 PAMAM conjugate containing 20 bound nucleotide moieties (UDPGA) was 100-fold more potent (EC50 2.4 nM) than the native agonist uridine-5′-diphosphoglucose. A molecular model of this conjugate docked in the human P2Y14 receptor showed that the nucleotide-substituted branches could extend far beyond the dimensions of the receptor and be available for multivalent docking to receptor aggregates. Larger dendrimer carriers and greater loading favored higher potency. A similar conjugate of G6 with 147 out of 256 amino groups substituted with UDPGA displayed an EC50 value of 0.8 nM. Thus, biological activity was either retained or dramatically enhanced in the multivalent dendrimer conjugates in comparison with monomeric P2Y14 receptor agonists, depending on size, degree of substitution, terminal functionality, and attached prosthetic groups.
Introduction
G protein-coupled receptors (GPCRs)1 are cell membrane-spanning receptors that respond to extracellular signaling molecules to control cell function via specific signaling pathways. Direct or indirect modulation of GPCRs serves as the basis of many disease treatments (1). The P2 receptors for purine and pyrimidine nucleotides have diverse biological roles (2–4). P2 receptors are divided into two structurally unrelated subfamilies–the P2X receptors, which are ligand-gated ion channels (5), and the P2Y receptors (2), which are GPCRs. P2Y receptors are divided into two subgroups. The first subgroup consisting of the P2Y1, P2Y2, P2Y4, P2Y6, and P2Y11 receptors activate Gq and promote phospholipase C (PLC)-dependent inositol lipid signaling. The second consists of P2Y12, P2Y13, and P2Y14 receptors, which preferentially activate Gi and inhibit adenylyl cyclase (2). P2Y receptors are activated by adenine and/or uracil nucleotides, while P2X receptors are principally activated by adenine nucleotides. P2Y receptors are widely distributed, for example, in the immune system (and widely distributed on hematopoietic cells), cardiovascular system, central and peripheral nervous system, endocrine system, and the renal and pulmonary systems (3–10).
The P2Y14 receptor is activated by uridine-5′-diphosphoglucose (UDPG) 1 (Chart 1) and other UDP-sugars (11, 12). The P2Y14 receptor affects immune function, but the physiological function of the receptor is not clearly established (13, 14). Extracellular 1 also acts as a weak full agonist at the P2Y2 receptor (EC50 10 μM) (15). We have studied the SAR of nucleotide derivatives at this receptor, which is among the most structurally restrictive of the P2Y family (15–18). Activity of synthetic analogues of 1 at the P2Y14 receptor has been followed through the stimulation of phosphoinositide hydrolysis by coexpression in COS-7 cells of a PLC-activating chimeric G protein that responds to Gi-coupled receptors (18).
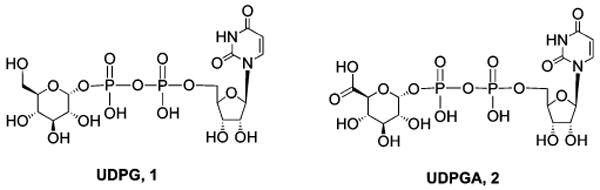
Chart 1. Structures of a Native Agonist of the P2Y14 Receptor 1 and a Carboxylic Acid Derivative 2 Suitable for Covalent Coupling to Carriers while Preserving Biological Activity.
Dendrimers are tree-like polymeric macromolecular nanostructures,which can serve as nanocarriers for drug delivery (19–21). Dendrimer chemistry offers versatility in the control of the component functional groups and in feasibility for conjugation of multiple functional units at both the periphery and interior. Dendrimers recently have attracted considerable attention in biomedical research in the context of drug delivery (targeted/ controlled release, encapsulation, or covalent/electrostatic attachment) (22–24), protein–carbohydrate interactions (multivalent effect) (25–30), medical diagnostics (signal amplification) (31, 32), and tissue engineering (33, 34).
Polyamidoamine (PAMAM) dendrimers have well-defined chemical structures in which a core is surrounded by successively added (and bifurcating) layers of methyl acrylate and ethylenediamine, which form each dendrimeric shell (or “generation”) of the polymer. Different generations of PAMAM dendrimers have either primary amine (integral generations) or carboxylic acid (half-integral generations) groups on their surface, and the number of surface groups for conjugation to multiple functional units depends on the generation of the dendrimer. PAMAM dendrimers are relatively biocompatible, which allows many applications in biomedicine (35–38).
We recently reported the first example of application of dendrimers to multivalent GPCR-promoted signal transduction in a study of adenosine receptor-targeted dendrimers (39–41). Various adenosine receptor agonists were attached covalently to PAMAM dendrimers, and the resulting nucleoside conjugates displayed distinct biological properties, including potent inhibition of platelet aggregation.
Here, we report the first example of application of PAMAM dendrimers to covalently conjugate a P2Y receptor ligand resulting in the enhancement of its pharmacological activity. The P2Y14 receptor agonist uridine-5′-diphosphoglucuronic acid (UDPGA) 2 and its ethylenediamine adduct 3a were utilized as appropriately functionalized congeners to couple with several generations of PAMAM dendrimers with the goal of modulating pharmacological interaction with this receptor. We also attached prosthetic groups, such as biotin, AlexaFluor488, and the metal chelating group diethylenetriaminepentaacetic acid (DTPA) on the dendrimer-nucleotide conjugates for targeting the P2Y14 receptor in vivo. Our long-term goal is to initiate the genesis of receptor probes that lead to improved biomedical diagnostics and possibly to novel conjugates for drug treatment.
Experimental Procedures
Chemical Synthesis
Materials and Methods
All reactions were carried out under a nitrogen atmosphere. G2.5 PAMAM (10 wt % solution in methanol), G3 PAMAM (20 wt % solution in methanol), G5.5 PAMAM (5 wt % solution in methanol), and G6 PAMAM (5 wt % solution in methanol), dendrimer with an ethylenediamine core, EDC, UDPGA, Gd(OAc)3, and ethylenediamine were purchased from Aldrich. Sulfo-NHS-LC-biotin was purchased from Pierce (Rockford, IL), and Alex-aFluor488-carboxylic acid 2,3,5,6-tetrafluorophenyl ester (AlexaFluor488–5-TFP) was purchased from Invitrogen Corp. (Carlsbad, CA) and 2-(4-isothiocyanatobenzyl)-diethylenetriaminepentaacetic acid (p-SCN-Bn-DTPA) purchased from Macrocyclic (Dallas, TX). Dialysis membranes (Spectra/Pore Membrane, MWCO 3500, flat width 18 mm) were purchased from Spectrum Laboratories, Inc. (Rancho Dominguez, CA).
Nuclear magnetic resonance (NMR) spectra were recorded on a Bruker DRX-600 spectrometer using D2O as a solvent. The chemical shifts are expressed as relative ppm from HOD (4.80).
The electrospray ionization mass spectrometry (ESI MS) and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) MS experiments were performed on a Waters LCT Premier mass spectrometer at the Mass Spectrometry Facility, NIDDK, NIH.
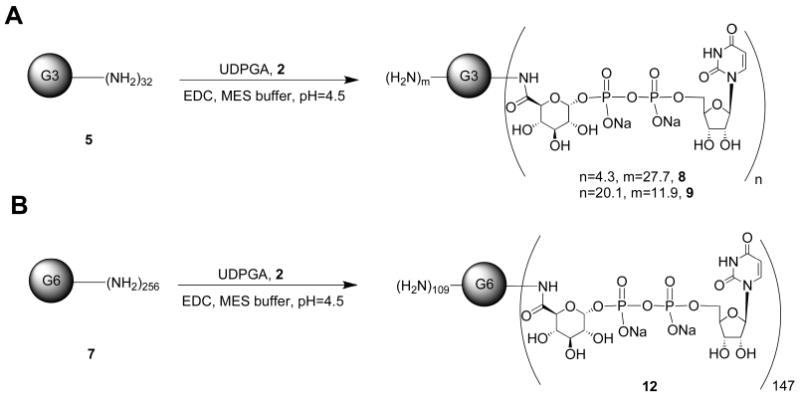
Synthesis of G3 or G6 PAMAM-UDPGA Conjugates 8, 9, and 12
A commercial solution of G3 PAMAM 5 (100 μL, 2 μmol) or G6 PAMAM 7 (167 μL, 0.11 μmol) was added to a round-bottom flask, and the methanol was evaporated using a rotary evaporator. The dendrimer was treated with EDC-HCl (2.1 mg, 5.5 equiv for compound 8; 13.8 mg, 36 equiv for compound 9; and 5.4 mg, 256 equiv for compound 12) and then dissolved with minimum volume of 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES), pH 5 (1 mL). The reaction mixture was stirred and UDPGA 2 (7.7 mg, 6 equiv for compound 8; 82.7 mg, 64 equiv for compound 9; and 27.2 mg, 384 equiv for compound 12) was added. The pH of the reaction was adjusted to the range 4.5–5.0 using 0.1 M HCl. The reaction mixture was stirred at room temperature under nitrogen atmosphere for 2 days and then diluted with water (1 mL). After that, the product was purified by extensive dialysis in water. The mixture was then lyophilized to give the pure product of the conjugates 8, 9, and 12.
G3 PAMAM-UDPGA Conjugate 8 (Low Loading)
Compound 8 (13.5 mg, 71%) was obtained as a white solid following the general procedure. The product was analyzed by NMR and MALDI-TOFF MS, which indicated approximately 4.3 UDPGA moieties attached per dendrimer. 1H NMR (D2O) δ 7.84 (d, J = 8.8 Hz, 4.3 H), 5.91 (d, J = 5.7 Hz, 8.6 H), 5.53 (m, 4.3 H), 4.28 (m, 8.6 H), 4.17 (m, 8.6 H), 4.08 (m, 8.6 H), 3.85 (m, 4.3 H), 3.63 (m, 8.6 H), 3.44 (m, 180 H), 3.3 (m, 120 H), 3.14 (m, 60 H), 2.72 (m, 120 H); m/z (M +ESI MS) found 9848.0; calc 9519.0.
G3 PAMAM-UDPGA Conjugate 9 (High Loading)
Compound 9 (26 mg, 68%) was obtained as a white solid following the general procedure. The product was analyzed by NMR and MALDI-TOFF MS, which indicated approximately 20.1 UD-PGA moieties attached per dendrimer.1H NMR (D2O) δ 7.88 (d, J = 7.4 Hz, 20.1 H), 5.81 (d, J = 5.4 Hz, 40.2 H), 5.51 (dd, J = 3.3, 6.1 Hz, 20.1 H), 4.3 (m, 40.2 H), 4.2 (m, 40.2 H), 4.1 (m, 40.2 H), 3.65 (m, 20.1 H), 3.52 (m, 40.2 H), 3.41 (m, 180 H), 3.3 (m, 120 H), 3.14 (m, 60 H), 2.72 (m, 120 H); m/z (M +ESI MS) found 18721.4; calc 19113.7.
G6 PAMAM-UDPGA Conjugate 12
Compound 12 (9.86 mg, 61%) was obtained as a white solid following the general procedure. The product was analyzed by NMR and MALDI-TOFF MS, which indicated approximately 147 UDPGA moieties attached per dendrimer. 1H NMR (D2O) δ 7.87 (d, J = 7.5 Hz, 147 H), 5.89 (d, J = 5.4 Hz, 294 H), 5.51 (m, 147 H), 4.29 (m, 294 H), 4.17 (m, 294 H), 4.09 (m, 294 H), 3.81 (m, 147 H), 3.7 (m, 294 H), 3.51 (m, 512 H), 3.41 (m, 512 H), 3.28 (m, 1020 H) 3.12 (m, 504 H), 3.07 (m, 504 H), 2.78 (m, 1020 H). The molecular weight was unable to be determined using ESI or MALDI-TOF MS, possibly due to stacking of the PAMAM dendrimer or because of the high molecular weight.
Synthesis of G2.5 or G5.5 PAMAM–UDPGA Conjugates 10 and 11
A commercial solution of G2.5 PAMAM 4 (13.76 μL, 0.2 μmol) or G5.5 PAMAM 6 (115 μL, 0.1 μmol) was added to a round-bottom flask, and the methanol was evaporated. EDC-HCl (1.4 mg, 36 equiv, for compound 8; and 1.9 mg, 100 equiv for compound 9) was added to the residue, and then the mixture was dissolved in a minimum volume of 0.1 M MES, pH 5 (0.5 mL), and stirred under a nitrogen atmosphere. UDPGA-ethylenediamine (3a) was added to the reaction mixture (6.5 mg, 50 equiv, for compound 10; and 13.1 mg, 200 equiv, for compound 11). The pH of the reaction mixture was maintained within the range 4.5–5.0 using 0.1 M HCl. After 48 h, the small molecule impurities were removed by extensive dialysis in water. After dialysis, the mixture was lyophilized to give conjugate 10 or 11.
G2.5 PAMAM-UDPGA Conjugate 10
Compound 10 (2.4 mg, 66%) was obtained as a white solid following the general procedure. The product was analyzed by NMR and MALDI-TOF MS, which indicated approximately 17.3 UDPGA moieties attached per dendrimer. 1H NMR (D2O) δ 7.82 (d, J = 7.7 Hz, 17.34 H), 5.84 (d, J = 5.5 Hz, 34.68 H), 5.51 (dd, J = 3, 5.1 Hz, 17.34 H), 4.29 (m, 52.0 H), 4.15 (m, 52.0 H), 3.7 (m, 17.34 H), 3.51 (m, 98.7 H), 3.41 (m, 120 H), 3.31 (m, 90.7 H), 3.14 (m, 60 H), 2.68 (m, 34.7 H), 2.51 (m, 56 H); m/z (M +ESI MS) found 18151.4, calc 18477.9.
G5.5 PAMAM-UDPGA Conjugate 11
Compound 11 (4.5 mg, 63%) was obtained as a white solid following the general procedure. The product was analyzed by NMR and MALDI-TOF MS, which indicated approximately 29.9 UDPGA moieties attached per G5.5 dendrimer. 1H NMR (D2O) δ 7.85 (d, J = 8.3 Hz, 29.94 H), 5.88 (d, J = 6.5 Hz, 59.9 H), 5.57 (dd, J = 3.1, 5.7 Hz, 29.9 H), 4.26 (m, 89.8 H), 4.14 (m, 89.8 H), 3.72 (m, 29.94 H), 3.44 (m, 571 H), 3.23 (m, 1075.88 H), 2.84 (m, 504 H), 2.67 (m, 508 H), 2.48 (m, 564 H); m/z (M +ESI MS) found 72626, calc 71651.
Synthesis of G3 PAMAM-UDPGA-Biotin Conjugate 13
A mixture of G3 PAMAM complex 9 (20 mg, 1.04 μmol) and Sulfo-NHS-LC-Biotin 17 (361 μg, 0.65 μmol) in a flask was dissolved in bicarbonate buffer (1 mL, pH 8.5, 0.002 M Na2CO3, 0.048 M NaHCO3, 0.15 M NaCl). After 48 h of stirring at room temperature, the reaction mixture was diluted with 1 mL water, and the product was purified by dialysis against water. The solution was then lyophilized, which provided G3 PAMAMbotin 13 (14.4 mg, 67%) as a white solid. The product was analyzed by NMR and MALDI-TOF MS, which indicated that approximately 4.87 biotin moieties attached per dendrimer. 1H NMR (D2O) δ 7.87 (d, J = 7.4 Hz, 20.1 H), 5.82 (d, J = 5.4 Hz, 40.2 H), 5.51 (m 20.1 H),4.55 (m, 9.74 H) 4.28 (m, 40.2 H), 4.18 (m, 40.2 H), 4.09 (m, 40.2 H), 3.67 (m, 20.1 H), 3.53 (m, 40.2 H), 3.42 (m, 180 H), 3.28 (m, 135 H), 3.11 (m, 60 H), 2.91(m, 9.74 H) 2.71 (m, 120 H), 2.12 (m, 19.5 H), 1.52 (m, 39.0 H), 1.27 (m, 19.5 H); m/z (M +ESI MS) found 21114.0, calc 20767.0.
Synthesis of G3 PAMAM-UDPGA-AlexaFluor488 Conjugate 14
G3 PAMAM complex 9 (20 mg, 1.04 μmol) was reacted with AlexaFluor488-5-TFP 18 (575 μg, 0.65 μmol) in bicarbonate buffer (1 mL, pH 8.5, 0.002 M Na2CO3, 0.048 M NaHCO3, 0.15 M NaCl). The reaction mixture was stirred for 2 days at room temperature and then diluted with 1 mL of water. The mixture was purified by dialysis with water. Lyophilization of the solution gave the G3 PAMAM–AlexaFluor488 complex 14 (13.4 mg, 62%) as a red solid. The product was analyzed by NMR and MALDI-TOFF MS, which indicated approximately 2.3 AlexaFluor488 moieties attached per dendrimer. 1H NMR (D2O) δ 8.28 (m, 2.3 H), 8.01 (d, J = 7.4 Hz, 20.1 H), 7.85 (m, 4.6 H), 6.1 (m, 2.3 H), 6.9 (m, 2.3 H), 5.9 (d, J = 5.4 Hz, 40.2 H), 5.75 (m, 2.3 H), 5.53 (dd, J = 3.1, 5.9 Hz, 20.1 H), 5.3 (m, 2.3 H), 4.32 (m, 40.2 H), 4.21 (m, 40.2 H), 4.18 (m, 40.2 H), 3.94 (m, 40.2 H), 3.68 (m, 20.1 H), 3.43 (m, 180 H), 3.28 (m, 120 H), 3.15 (m, 60 H), 2.63 (m, 120 H); m/z (M +ESI MS) found 22336.0, calc 20766.3.
Synthesis of G3 PAMAM-UDPGA-DTPA Conjugate 15 and G3 PAMAM-UDPGA-DTPA-Gd Conjugate 16
G3 PAM-AM complex 9 (20 mg, 1.04 μmol) was stirred with p-SCN-Bn-DTPA 19 (351 μg, 0.65 μmol) in bicarbonate buffer (1 mL, pH 8.5, 0.002 M Na2CO3, 0.048 M NaHCO3, 0.15 M NaCl). The reaction mixture was stirred for 48 h, at room temperature, and then the small molecule impurities were removed by extensive dialysis in water followed by lyophilization to yield the G3 PAMAM-DTPA complex 15 (15.2 mg, 68%) as a white solid. The product was analyzed by NMR and MALDI-TOF MS, which indicated approximately 4.5 DTPA moieties attached per dendrimer. 1H NMR (D2O) δ 7.89 (d, J = 7.4 Hz, 20.1 H), 7.21 (m, 9 H), 7.31 (m, 9 H), 5.85 (d, J = 5.4 Hz, 40.2 H), 5.53 (dd, J = 3.3, 6.1 Hz, 20.1 H), 4.31 (m, 40.2 H), 4.24 (m, 40.2 H), 4.15 (m, 80.4 H), 3.71 (m, 20.1 H), 3.64 (m, 49.5 H), 3.44 (m, 180 H), 3.33 (m, 120 H), 3.16 (m, 69 H), 2.63 (m, 120 H), 2.66 (m, 27 H); m/z (M +ESI MS) found 22744.0, calc 21546.1.
The chelating prosthetic group on conjugate 15 was used to complex Gd(III) by addition of Gd(OAc)3. G3 PAMAM-UDPGA-DTPA 15 (10 mg, 0.46 μmol), Gd (OAc)2 (94 μg, 0.28 μmol) were dissolved in 0.3 M citrate buffer (1 mL, pH 4.5). The mixture was stirred at room temperature for 48 h in dark. The mixture was dialyzed sequentially with 0.1 M NH4OAc (pH 7.0) to remove the uncomplexed Gd(III) and then extensively against water. Lyophilization of the solution gave the product 16 (7.5 mg, 74%). Since the compound 16 contains Gd(III), we could not analyze the NMR spectra. We assume that all the DTPA moieties would complex with Gd(III), because we reacted 15 with excess Gd(OAC)3 in this theoretically quantitative reaction. So, we assumed that compound G3 PAMAM-UDPGA-DTPA-Gd(III) 16 contained approximately 4.5 Gd(III) moieties attached per dendrimer based on the stoichiometry of the precursor, which was comparable to mass spectral data. m/z (M +ESI MS) found 24175.0, calc 22311.2.
Biological Methods
Materials
IBMX was purchased from SigmaAldrich (St. Louis, MO). Compounds 1 and 2 were manufactured by Fluka and were purchased from SigmaAldrich (St. Louis, MO). [3H]Adenine was purchased from American Radiolabeled Chemicals (St. Louis, MO). All cell culture media and sera were from Gibco (Invitrogen, Carlsbad, CA).
Cell Culture
Human embryonic kidney-293 cells stably expressing the human P2Y14-R (P2Y14–HEK293 cells) were generated as previously described by Fricks et al. (46). P2Y14–HEK293 cells were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% genticin (Gibco), and 1% antibiotic–antimocotic (Gibco) at 37 °C in a 5% CO2 environment.
Cyclic AMP Accumulation
P2Y14–HEK293 cells were grown in 24-well plates and incubated with 1 μCi [3H]adenine/well in serum-free DMEM for at least 2 h prior to assay. Assays were initiated by addition of HEPES-buffered, serum-free DMEM containing 200 μM 3-isobutyl-1-methyl-xanthine (IBMX) and 30 μM forskolin, with or without drugs, and incubation continued for 15 min at 37 °C. Incubations were terminated by aspiration of the medium and addition of 450 μL ice-cold 5% trichloroacetic acid. [3H]Cyclic AMP was isolated by sequential Dowex and alumina chromatography and quantified by liquid scintillation counting as previously described by Harden et al. (55).
Data Analysis
The concentrations of the dendrimer–ligand complexes were measured by the concentration of the dendrimer, not the attached ligand. EC50 values were determined using Prism software (GraphPad, San Diego, CA) and are presented as mean ± SE. All experiments were repeated at least three times.
Results
Chemical Synthesis
We recently investigated the SAR (structure–activity relationship) of analogues of UDPG 1 at the P2Y14 receptor (15, 16), but few of the ligands displayed comparable pharmacological properties to those of the native ligand, and nearly all modifications of the uracil and ribose moieties abolished activity. We also reported that functionalized congeners of UDPGA 2 in which an amide-linked chain was extended from the terminal carboxylic acid group via N-acetylethylenediamine (e.g., 3b) or N-(t-butyloxycarbonyl)ethylenediamine moieties did not significantly differ from 1 in their pharmacological activity (15). This suggests that it is possible to incorporate structural modifications of the carboxylic acid chain in 2 that do not substantively affect interaction with the P2Y14 receptor.
In this study, we first coupled 2 with the peripheral amino groups of a third-generation (G3) PAMAM dendrimer to yield 8 and 9 and to a sixth-generation (G6) PAMAM dendrimer to yield 12. The coupling conditions used were the water-soluble carbodiimide reagent EDC in aqueous medium at pH 4.5 in 0.1 M MES buffer and under a nitrogen atmosphere (Scheme 1). The degree of substitution of the G3 dendrimer was varied to determine how the fraction of drug loading on the PAMAM surface would affect the pharmacological activity. Compound 8 contained an average of 4.3 bound nucleotide moieties (2) per dendrimer, and compound 9 an average of 20.1 bound nucleotide moieties per dendrimer, out of a theoretical 32. The degree of substitution was calculated using mass spectroscopy and integration of the 600 MHz 1H NMR spectra. For compound 12, we added excess nucleotide monomer to derivatize the maximum number of nucleotide moieties on the surface of a G6 dendrimer, in which the average number of drug moieties attached was found to be an average of 147 out of a theoretical 256. Following the coupling reaction, we removed all residual compound 2 that remained unreacted by extensive dialysis in water. Thus, we successively varied the number of covalently attached ligands, determined to be an average of 4.3, 20.1, and 147 per dendrimer in compounds 8, 9, and 12, respectively.
Scheme 1. Covalent Coupling of the Carboxylic Acid Functionalized Congener UDPGA 2 to PAMAM Dendrimers of Integral Generations 3 and 6, by Carbodiimide Condensation with the Surface Amino Groups of the Dendrimer.
A G3 PAMAM-biotin conjugated UDPGA complex 13 also was prepared. Since biotin and its amide derivatives are known to bind strongly with the tetrameric protein avidin, compound 13 was designed as a multifunctional chemical probe of the P2Y14 receptor. For the preparation of compound 13, PAMAM-UDPGA 9 was reacted with a water-soluble N-hydroxysuccinimide (NHS) ester of a chain-elongated biotin 17 in bicarbonate buffer at pH 8.5 (Scheme 2) and the product 13 was subjected to dialysis in water. From the 1H NMR and mass spectra, we confirmed that the average number of biotin units attached in compound 13 was 4.9 per dendrimer.
Scheme 2. Multiple Functionalization of a Covalent Conjugate 9 of UDPGA 2 on a PAMAM G3 Dendrimer, For the Purpose of Fluorescent Detection (14), Avidin Binding (13), and Chelation with Heavy Metal Gadolinium (16).
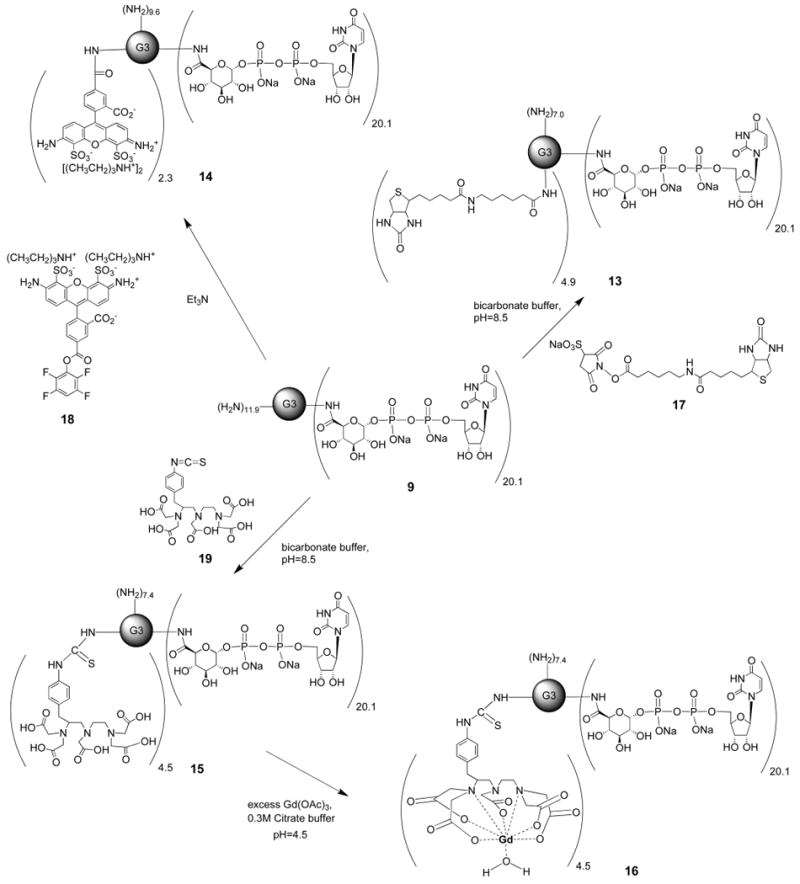
With the eventual goal of direct microscopic visualization of the P2Y14 receptor in biological systems, we prepared a fluorescent dendrimer–UDPGA derivative 14. Compound 14 was synthesized by reaction of 9 and an equimolar amount of 5-carboxy tetrafluorophenyl ester of AlexaFluor488 18 in the presence of triethylamine in a minimum volume of water. AlexaFluor488 was chosen, because although its fluorescent properties are similar to fluorescein, it exhibits higher photostability, pH insensitivity, and good water solubility (42). We subjected compound 14 to dialysis in water to remove low MW impurities. The conjugated product was analyzed by 1H NMR and mass spectroscopy, which showed that the average number of AlexaFluor488 units attached per dendrimer molecule was 2.3.
PAMAM dendrimers recently were used for magnetic resonance imaging (MRI) (43). MRI is one of the fastest growing diagnostic methods in medical technology because of its effectiveness in visualizing soft tissues with good resolution. Ionic gadolinium is a widely used reagent for MRI, and a chelated Gd(III) complex recently was covalently attached to PAMAM dendrimers for molecular imaging (44). As such, we derivatized the PAMAM–UDPGA conjugate 9 with the same MRI-active reagent for chelation of Gd(III). Compound 9 was reacted with an electrophilic reactive derivative (aryl isothiocyanate) of diethylenetriaminepentaacetic acid (DTPA) 19 in bicarbonate buffer at pH = 8.5 for two days to obtain compound 15, which was purified by dialysis in water. The molecular weight of 15 was >10 000 D, and 1H NMR and mass spectroscopic analyses revealed that the average number of DTPA moieties attached in compound 15 was 4.5 per dendrimer. We subsequently reacted compound 15 with excess Gd(OAc)3 in the presence of 0.3 M citrate buffer (pH = 4.5) to obtain compound 16, which was subjected to dialysis using 0.1 M ammonium acetate to remove low MW impurities. It was not possible to obtain a 1H NMR spectrum for compound 16, so we have assumed that the number of Gd(III) ions in compound 16 was 4.5 based on quantitative complexation of the attached DTPA with excess ionic Gd. We used Gd(OAc)3 as a biological control to ensure that Gd(III) does not exhibit activity in the system examined and to anticipate eventual replacement of nonradioactive Gd(OAc)3 with radioactive Gd(OAc)3 for in vivo imaging.
The unreacted dendrimer surface in compounds 8, 9, and 12–16 contains amino groups, which are known to be associated with cell toxicity (45). Therefore, we also produced conjugates of carboxylic acid-containing PAMAM dendrimers by coupling an amine-functionalized congener of UDPGA 3a to G2.5 (4) and G5.5 (6) PAMAM dendrimers to form compounds 10 and 11. Compound 3a was further purified by a subsequent HPLC step for subsequent biological testing. Compound 3a was reacted with PAMAM dendrimer G2.5 4 and G5.5 6 by EDC coupling in 0.1 M MES buffer under a nitrogen atmosphere and at pH 4.5 to 5.0 to form products 10 and 11, respectively. Each polymeric product was subjected to dialysis in water to remove low MW impurities. Compounds 10 and 11 were analyzed by 1H NMR and mass spectroscopy, which showed that the average number of monomers 3a in compounds 10 and 11 was 17.3 and 29.9 per dendrimer, respectively.
Biological Activity
The activities of dendrimer conjugates were tested in human embryonic kidney (HEK293) cells stably expressing the human P2Y14 receptor. Concentration effect curves were generated as previously described (46) comparing the relative capacities of UDPG 1 and the denrimer conjugates to inhibit forskolin (30 μM)-stimulated cyclic AMP accumulation. Neither compound 1 (46) nor any of the dendrimers tested (data not shown) exhibited activity in wild-type HEK293 cells.
The control dendrimers 4 and 5 were inactive in this test system. In contrast, compound 9, derived from the G3 dendrimer and containing free residual amino groups, was a very potent agonist that exhibited an EC50 of 2.4 nM at the P2Y14 receptor (Figure 1). This potency was approximately 100-fold greater than that of the native agonist UDPG 1, which had an EC50 of 261 nM. UDPGA 2 was approximately equipotent to 1 in this assay. The corresponding dendrimer 8 (Figure 1), which had a lower degree of loading of the nucleotide on a G3 dendrimer, was considerably less potent than 9, but similar in potency to 1. Therefore, a distinct multivalent effect occurs in these compounds.
Figure 1.
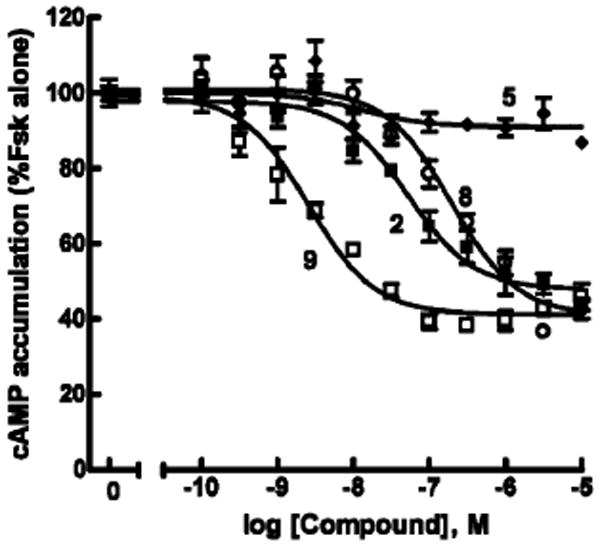
Comparison of G3 dendrimer conjugates as agonists at the human P2Y14 receptor. The capacities of UDPGA 2 (■) and the G3 dendrimer conjugates 5 (◆), 8 (○), and 9 (□) to inhibit forskolin (30 μM) stimulated cyclic AMP accumulation were quantified in P2Y14–HEK293 cells as described in Methods. Assays were carried out in triplicate, and all experiments were repeated at least three times. Data were pooled and normalized using forskolin-stimulated cyclic AMP accumulation as 100% and the phosphodiesterase inhibitor IBMX alone as 0%.
Compounds 10 and 11, both of which were derived from half-integral dendrimers and contain free residual carboxylate groups, were very potent at the P2Y14 receptor with EC50 values of 3.2 and 3.1 nM, respectively (Figure 2). The corresponding amine derivative 3a of UDPGA, which also served as the synthetic precursor of conjugates 10 and 11, was considerably less potent.
Figure 2.
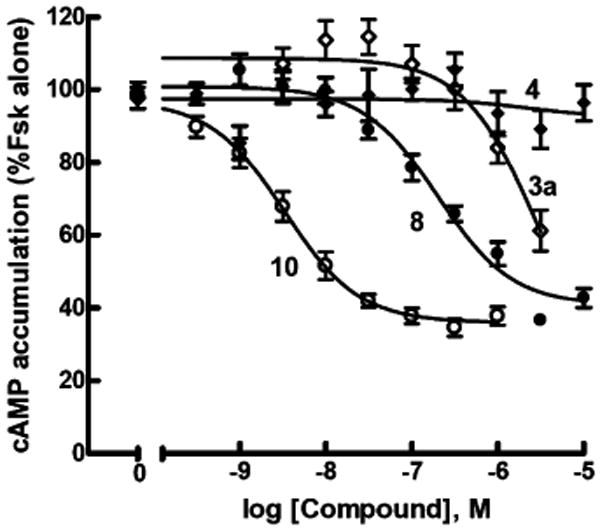
Comparison of G2.5 dendrimer conjugates as agonists at the human P2Y14 receptor. The capacities of G2.5 dendrimer conjugates 3a (◊), 4 (◆), 8 (●), and 10 (○) to inhibit forskolin (30 μM) stimulated cyclic AMP accumulation were quantified in P2Y14–HEK293 cells as described in Methods. Assays were carried out in triplicate, and all experiments were repeated at least three times. Data were pooled and normalized using forskolin-stimulated cyclic AMP accumulation as 100% and IBMX alone as 0%.
The higher molecular weight G6 dendrimer conjugate 12 was similar in relative composition to the smaller conjugate 9, which had a high degree of loading of nucleotide. Compound 12 (EC50 = 0.8 nM) was significantly more potent than 9 (Figure 3). Both 9 and 12 contain significant fractions (but less than 50%) of residual amino groups.
Figure 3.
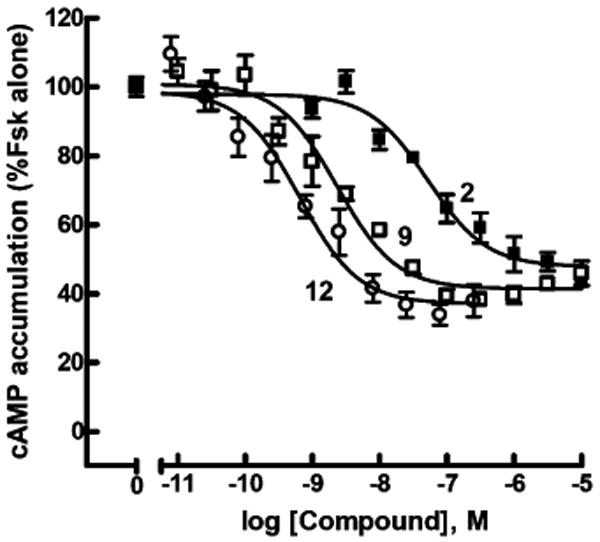
Comparison of larger dendrimer conjugates as agonists at the human P2Y14 receptor. The capacities of UDPGA 2 (■), G3 dendrimer conjugate 9 (□), and G6 dendrimer conjugate 12 (○) to inhibit forskolin (30 μM)-stimulated cyclic AMP accumulation were quantified in P2Y14–HEK293 cells as described in Methods. Assays were carried out in triplicate, and all experiments were repeated at least three times. Data were pooled and normalized using forskolin-stimulated cyclic AMP accumulation as 100% and IBMX alone as 0%.
The G3 dendrimer conjugates 13 and 15 were equivalent to the precursor dendrimer conjugate 12, except that they both contain covalently attached prosthetic groups in addition to the nucleotide. Each of these dendrimers was essentially as potent as 9. The G3 dendrimer conjugate 14, which had approximately two covalently attached AlexaFluor488 moieties, was 17-fold less potent than the corresponding 9 but 7-fold more potent than compound 1. However, when the DTPA conjugate 15 was complexed with gadolinium, the potency was greatly reduced, to the level of 1, the native agonist.
Molecular Modeling
A molecular model of compound 3b docked to the human P2Y14 receptor recently was constructed on the basis of the crystal structure of the human A2A adenosine receptor (15, 47). This model was utilized to build a complex of the P2Y14 receptor bound to a PAMAM G3 dendrimer using the same approach as reported for the docking of a polyvalent dendrimer–nucleoside conjugate to the human A2A adenosine receptor (48). In particular, our published model of the conjugate of PAMAM (G3) with an A2A receptor agonist (2-[p-(2-carboxyethyl)phenyl-ethylamino]-5′-N-ethylcarboxamidoadenosine, CGS21680) was utilized to build a model of dendrimer–nucleotide conjugate 9, which was partially substituted with nucleotide moieties. Initially, all fragments of the carboxylic acid CGS21680 were removed from the model of PAMAM-CGS21680, and hydrogen atoms were added to the remaining PAMAM structure. Twenty moieties of UDPGA 2 then were coupled to randomly selected amino groups of the PAMAM chains. The remaining 12 unacylated chains of the dendrimer were substituted with protonated amino groups.
The model of 9 was minimized in the OPLS2005 force field. The Polak–Ribier conjugate gradient (PRCG) minimization method with a maximum of 5000 iterations and with a convergence threshold of 0.05 kJ · mol−1 · Å−1 was applied. One randomly selected UDPGA moiety attached to the dendrimer then was removed from the PAMAM model, and the UDPGA docked to the P2Y14 receptor was attached to the dendrimer. Thus, an initial complex of 9 docked to the P2Y14 receptor was obtained. The orientation of a few UDPGA moieties was manually adjusted to avoid overlays with the amino acid residues of the P2Y14 receptor. The model was subjected to geometry optimization using the same protocol as mentioned above. After minimization, the binding mode of the UDPGA moiety located inside the P2Y14 receptor was found to be similar to the binding mode of 1 itself (15). The ethylenediamine chain of PAMAM connected to the receptor-bound UDPGA moiety was located between extracellular loops of P2Y14 receptor. In particular, the nucleotide moiety was surrounded by Ile173 (EL2), and Leu175 (EL2), Arg274 (EL3), and Lys277 (7.35). In addition, Lys277 formed an H-bond with the amide oxygen atom of the chain of PAMAM. The molecular model of compound 9 docked in the human P2Y14 receptor (Figure 4) showed that the nucleotide-substituted branches of the dendrimer extended far beyond the dimensions of the receptor protein and thus would be available for multivalent docking to receptor dimers and higher-order aggregates (49).
Figure 4.
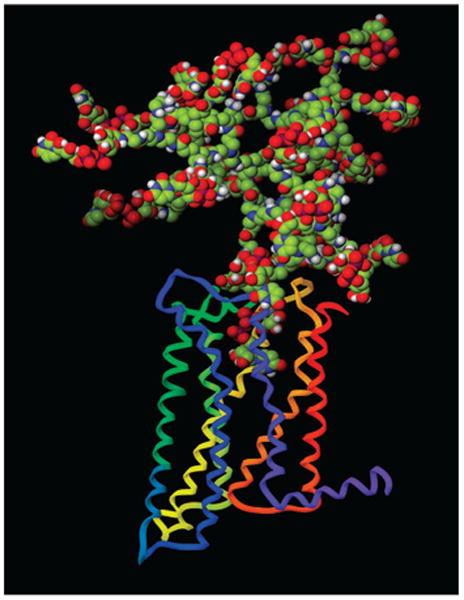
Molecular model of compound 9 docked in the human P2Y14 receptor, obtained after docking to the receptor homology model built based on the crystal structure of the A2A adenosine receptor (47). The receptor helices are colored by residue position: N-terminus in red, TM 1 in orange, TM 2 in ochre, TM 3 in yellow, TM 4 in green, TM 5 in cyan, TM 6 in blue, and TM 7 and C-terminus in purple.
Discussion
A functionalized ethylenediamine-containing chain was incorporated in agonists of the human P2Y14 receptor, and this chain provided a conjugation strategy for coupling nucleoside agonists to PAMAM dendrimer carriers. Uridine-5′-diphosphoglucuronic acid and its ethylenediamine adduct were suitable functionalized congeners (50) for coupling to several generations (G2.5–6) of dendrimers (both terminal carboxy and amino) to modulate potency of the intact conjugates. The observed biological activity of the dendrimer conjugates was clearly P2Y14 receptor-dependent; the control dendrimers 4 and 5 lacking nucleotides were inactive. A G3 PAMAM conjugate containing 20 bound nucleotide moieties was 100-fold more potent than the native agonist 1. Larger dendrimer carriers and those with greater loading favored higher potency as P2Y14 receptor agonists. All of the dendrimer–nucleotide conjugates were full agonists at the receptor.
The molecular mechanism whereby these dendrimers exhibit increased activity relative to the native cognate agonist UDPG is unclear. One possible explanation is that these multivalent compounds may bridge multiple P2Y14 receptor binding sites simultaneously. Molecular modeling has indicated this possibility for an analogous G3 PAMAM dendrimer conjugate of an A2A adenosine receptor agonist (48). The dramatic enhancement in potency, especially in the more heavily substituted dendrimer derivatives, is highly suggestive of a multivalent effect. The ability to span multiple receptor binding sites would be further promoted by the possible aggregation of dendrimer derivatives (56). Nanomolar and subnanomolar potencies have been achieved, which are unprecedented for any monomeric nucleotides acting at the P2Y14 receptor. Dendrimer conjugates 10 and 11, which are both more than half substituted with nucleotide moieties, are more than 800-fold more potent than a corresponding monomer, i.e., the ethylenediamine analogue 3a. Lower loading of the same dendrimer with nucleotides resulted in a conjugate (compound 9) that exhibited relatively lower potency. Variability of the potency depending on degree of loading will be the subject of future systematic studies. Such studies also might benefit from quantification of the relative activities of these compounds across a wide range of receptor expression levels in the same cell system.
It is possible that conjugation to dendrimers might protect the nucleotides against enzymatic degradation, but this is not the basis for the enhancement of potency in cAMP-inhibition with the multivalent dendrimers. Our earlier studies (51) indicated that little metabolism of extracellular 1 occurs under the conditions of these assays. Moreover, the concentration effect curve of 1 for decreasing cyclic AMP levels is similar whether quantified at times of <5 min or times up to 30 min.
Dendrimers containing several different prosthetic groups also were synthesized. Dendrimer conjugate 14, which is tagged with AlexaFluor488, potentially will be useful for receptor visualization. Although the potency of 14 was 17-fold lower than that of the corresponding dendrimer lacking the fluorescent group (i.e., 9), the EC50 of approximately 40 nM was comparable to previously introduced fluorescent ligand probes for other GPCRs (52). A similar approach in a previous study, using a fluorescent AlexaFluor488-tagged G3 PAMAM dendrimer, was used to detect the expression of the A3 adenosine receptor in cultured cells (41). Gd(III) is a paramagnetic ion that is used for magnetic resonance imaging (44). Dendrimer conjugate 15, which is tagged with the chelating group DTPA, is substantially less potent as its Gd(III) complex, thus limiting the potential utility of this derivative. The in vivo application of dendrimer conjugates is normally limited to i.p. or i.v. injection, but novel approaches for gaining greater bioavailability of dendrimers by other routes of administration have been discussed (53, 54).
In conclusion, we have identified a site on a nucleotide agonist of the P2Y14 receptor for chemical tethering to a macromolecular carrier without losing the ability to activate the receptor. Moreover, we have shown that the potency of the native ligand is greatly enhanced in these multivalent ligands. These biologically active drug conjugates do not require cleavage or cellular internalization for their action at the GPCR; in fact, these processes would reduce activity. Covalent conjugation of P2Y14 receptor agonists to PAMAM dendrimers qualitatively altered their pharmacological activity. Thus, potency was either retained or dramatically enhanced in the multivalent dendrimer conjugates in comparison to the monomeric P2Y14 receptor agonists, depending on size, degree of substitution, terminal functionality, and attached prosthetic groups. The ability to modulate the potency of a given GPCR ligand by the mode and stoichiometry of attachment to dendrimer carriers promises to have general applicability to this therapeutically important class of receptor proteins.
Supplementary Material
Scheme 3. Synthesis of an Amine-Functionalized Congener UDPGA 3a and Its Covalent Coupling to PAMAM Dendrimers of Half-Generations 2.5 and 5.5, by Carbodiimide Condensation with the Surface Carboxylic Groups of the Dendrimer.
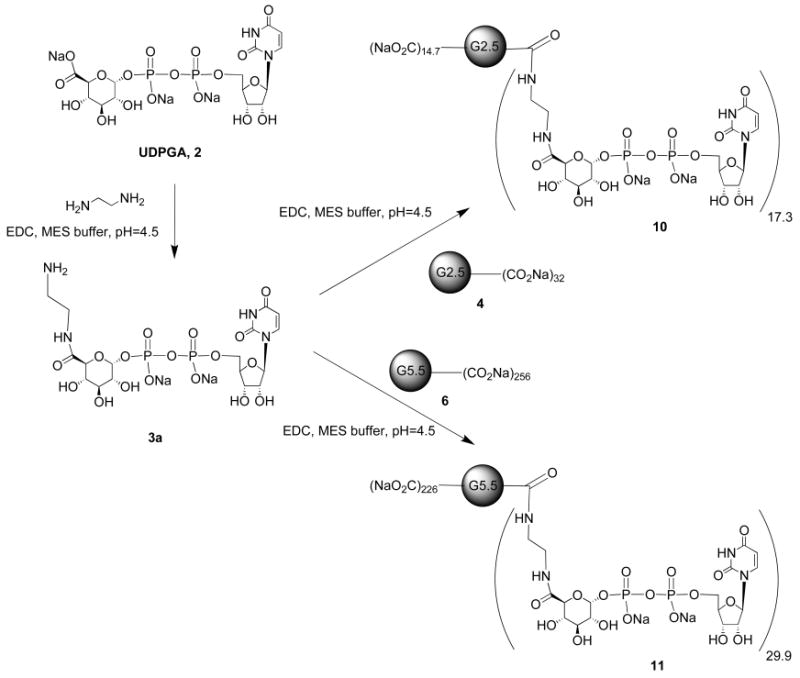
Table 1. In Vitro Pharmacological Data for Various Nucleotides and Their Complexes with PAMAM Dendrimers.
| compound | name/composition | molecular weight (kD) | EC50 at hP2Y14 receptor, nM |
|---|---|---|---|
| 1 | UDPG | 0.61 | 261 ± 53 |
| 2 | UDPGA | 0.65 | 370 ± 70 |
| 3a | UDPGA-ethylenediamine | 0.82 | 2590 ± 1590 |
| 3b | UDPGA-(N-Ac-ethylenediamine) | 0.87 | 496 ± 67 |
| 4 | G2.5 dendrimer | 6.27 | NEa |
| 5 | G3 dendrimer | 6.91 | NE |
| 8 | G3-(UDPGA)4.3 | 9.52 | 159 ± 14 |
| 9 | G3-(UDPGA)20.1 | 19.11 | 2.4 ± 0.1 |
| 10 | G2.5-(UDPGA-ethylenediamine)17.3 | 18.48 | 3.2 ± 1.4 |
| 11 | G5.5-(UDPGA-ethylenediamine)29.9 | 71.65 | 3.1 ±0.8 |
| 12 | G6-(UDPGA)147 | 147.31 | 0.8 ± 0.4 |
| 13 | G3-(UDPGA)20.1-(Biotin)4.9 | 20.77 | 3.4 ± 0.8 |
| 14 | G3-(UDPGA)20.1-(AlexaFluor488)2.3 | 20.77 | 39.8 ± 7.4 |
| 15 | G3-(UDPGA)20.1-(DTPA)4.5 | 21.55 | 4.1 ± 1.2 |
| 16 | G3-(UDPGA)20.1-(DTPA-Gd(III))4.5 | 22.31 | 421 ± 104 |
NE, no effect.
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, NIDDK, and by NIH grant GM38213 to T.K.H. We thank Dr. Herman Yeh and Dr. Athena M. Keene-Klutz of NIDDK for helpful advice on the NMR experiments and chemical synthesis. We thank Dr. John Lloyd (NIDDK) for mass spectral measurements.
Footnotes
Abbreviations: DTPA, diethylenetriaminepentaacetic acid; EDC, N-ethyl-N′-dimethylaminopropylcarbodiimide; GPCR, G protein-coupled receptor; HEK, human embryonic kidney; IBMX, 3-isobutyl-1-methylxanthine; MES, 2-(N-morpholino)ethanesulfonic acid; MRI, magnetic resonance imaging; NHS, N′-hydroxysuccinimide; PAMAM, polyamidoamine; SAR, structure activity relationship; p-SCN-Bn-DTPA, 2-(4-isothiocyanatobenzyl)-diethylenetriaminepentaacetic acid; UDPG, uridine-5′-diphosphoglucose; UDPGA, uridine-5′-diphosphoglucuronic acid.
Supporting Information Available: 1H NMR and MALDI spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
Literature cited
- 1.Bridges TM, Lindsley CW. G-protein-coupled receptors: from classical modes of modulation to allosteric mechanisms. ACS Chem Biol. 2008;3:530–541. doi: 10.1021/cb800116f. [DOI] [PubMed] [Google Scholar]
- 2.Abbracchio MP, Burnstock G, Boeynaems JM, Barnard EA, Boyer JL, Kennedy C, Fumagalli M, King BF, Gachet C, Jacobson KA, Weisman GA. International Union of Pharmacology. Update of the P2Y G protein-coupled nucleotide receptors: from molecular mechanisms and pathophysiology to therapy. Pharmacol Rev. 2006;58:281–341. doi: 10.1124/pr.58.3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gachet C. Regulation of platelet functions by P2 receptors. Annu Rev Pharmacol Toxicol. 2006;46:277–300. doi: 10.1146/annurev.pharmtox.46.120604.141207. [DOI] [PubMed] [Google Scholar]
- 4.Burnstock G. Purinergic signalling and disorders of the central nervous system. Nat Rev Drug Discovery. 2008;7:575–590. doi: 10.1038/nrd2605. [DOI] [PubMed] [Google Scholar]
- 5.Idzko M, Hammad H, van Nimwegen M, Kool M, Willart MA, Muskens F, Hoogsteden HC, Luttmann W, Ferrari D, Di Virgilio F, Virchow JC, Lambrecht BN. Extracellular ATP triggers and maintains asthmatic airway inflammation by activating dendritic cells. Nat Med. 2007;13:913–919. doi: 10.1038/nm1617. [DOI] [PubMed] [Google Scholar]
- 6.Shin A, Toy T, Rothenfusser S, Robson N, Vorac J, Dauer M, Stuplich M, Endres S, Cebon J, Maraskovsky E, Schnurr M. P2Y receptor signaling regulates phenotype and IFN-alpha secretion of human plasmacytoid dendritic cells. Blood. 2008;111:3062–3069. doi: 10.1182/blood-2007-02-071910. [DOI] [PubMed] [Google Scholar]
- 7.Malin SA, Davis BM, Richard KH, Reynolds IJ, Albers KM, Molliver DC. Thermal nociception and TRPV1 function are attenuated in mice lacking the nucleotide receptor P2Y2. Pain. 2008;138:484–496. doi: 10.1016/j.pain.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lugo-Garcia L, Filhol R, Lajoix AD, Gross R, Petit P, Vignon J. Expression of purinergic P2Y receptor subtypes by INS-1 insulinoma beta-cells: a molecular and binding characterization. Eur J Pharmacol. 2007;568:54–60. doi: 10.1016/j.ejphar.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Lazarowski ER, Tarran R, Grubb BR, van Heusden CA, Okada S, Boucher RC. Nucleotide release provides a mechanism for airway surface liquid homeostasis. J Biol Chem. 2004;279:36855–36864. doi: 10.1074/jbc.M405367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Müller T, Bayer H, Myrtek D, Ferrari D, Sorichter S, Ziegenhagen MW, Zissel G, Virchow JC, Luttmann W, Norgauer J, Di Virgilio F, Idzko M. The P2Y14 receptor of airway epithelial cells coupling to intracellular Ca2+ and IL-8 secretion. Am J Respir Cell Mol Biol. 2005;33:601–609. doi: 10.1165/rcmb.2005-0181OC. [DOI] [PubMed] [Google Scholar]
- 11.Chambers JK, Macdonald LE, Sarau HM, Ames RS, Freeman K, Foley JJ, Zhu Y, McLaughlin MM, Murdock P, McMillan L, Trill J, Swift A, Aiyar N, Taylor P, Vawter L, Naheed S, Szekeres P, Hervieu G, Scott C, Watson JM, Murphy AJ, Duzic E, Klein C, Bergsma DJ, Wilson S, Livi GP. A G protein-coupled receptor for UDP-glucose. J Biol Chem. 2000;275:10767–10771. doi: 10.1074/jbc.275.15.10767. [DOI] [PubMed] [Google Scholar]
- 12.Abbracchio MP, Boeynaems JM, Barnard EA, Boyer JL, Kennedy C, Miras-Portugal MT, King BF, Gachet C, Jacobson KA, Weisman GA, Burnstock G. Characterization of the UDP-glucose receptor (re-named here the P2Y14 receptor) adds diversity to the P2Y receptor family. Trends Pharmacol Sci. 2003;24:52–55. doi: 10.1016/S0165-6147(02)00038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scrivens M, Dickenson JM. Functional expression of P2Y14 receptor in human neutrophils. Eur J Pharmacol. 2006;543:166–173. doi: 10.1016/j.ejphar.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 14.Bassil AK, Bourdu S, Townson KA, Wheeldon A, Jarvie EM, Zebda N, Abuin A, Grau E, Livi GP, Punter L, Latcham J, Grimes AM, Hurp DP, Downham KM, Sanger GJ, Winchester WJ, Morrison AD, Moore GBT. UDP-glucose modulates gastric function through P2Y14 receptor-dependent and -independent mechanisms. Am J Physiol Gastrointest Liver Physiol. 2009;296:G923–G930. doi: 10.1152/ajpgi.90363.2008. [DOI] [PubMed] [Google Scholar]
- 15.Ko H, Das A, Carter RL, Fricks IP, Zhou Y, Ivanov AA, Melman A, Joshi BV, Kováč P, Hajduch J, Kirk KL, Harden TK, Jacobson KA. Molecular recognition in the P2Y14 receptor: Probing the structurally permissive terminal sugar moiety of UDP-glucose. Bioorg Med Chem. doi: 10.1016/j.bmc. 2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko H, Fricks I, Ivanov AA, Harden TK, Jacobson KA. Structure activity relationship of uridine 5′-diphosphoglucose (UDP-Glucose) analogues as agonists of the human P2Y14 receptor. J Med Chem. 2007;50:2030–2039. doi: 10.1021/jm061222w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov AA, Fricks I, Harden TK, Jacobson KA. Molecular dynamics simulation of the P2Y14 receptor. Ligand docking and identification of a putative binding site of the distal hexose moiety. Bioorg Med Chem Lett. 2007;17:761–766. doi: 10.1016/j.bmcl.2006.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fricks I, Maddiletti S, Carter R, Lazarowski ER, Nicholas RA, Jacobson KA, Harden TK. UDP is a competitive antagonist at the human P2Y14 receptor and a full agonist at the rat P2Y14 receptor. J Pharm Exp Therap. 2008;325:588–594. doi: 10.1124/jpet.108.136309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomalia DA, Reyna LA, Svenson S. Dendrimers as multi-purpose nanodevices for oncology drug delivery and diagnostic imaging. Cellular delivery of therapeutic macromolecules. Biochem Soc Trans. 2007;35:61–67. doi: 10.1042/BST0350061. [DOI] [PubMed] [Google Scholar]
- 20.Lee CC, MacKay JA, Fréchet JMJ, Szoka FC. Designing dendrimers for biological applications. Nat Biotechnol. 2005;23:1517–1526. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 21.Zeng F, Zimmerman SC. Dendrimers in supramolecular chemistry: from molecular recognition to self-assembly. Chem Rev. 1997;97:1681–1712. doi: 10.1021/cr9603892. [DOI] [PubMed] [Google Scholar]
- 22.Gillies ER, Fréchet JMJ. Dendrimers and dendritic polymers in drug delivery. Drug Discovery Today. 2005;10:35–43. doi: 10.1016/S1359-6446(04)03276-3. [DOI] [PubMed] [Google Scholar]
- 23.D'Emanuele A, Attwood D. Dendrimer-drug interactions. Adv Drug Delivery Rev. 2005;57:2147–2162. doi: 10.1016/j.addr.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Patri AK, Majoros IJ, Baker JR. Dendritic polymer macromolecular carriers for drug delivery. Curr Opin Chem Biol. 2002;6:466–471. doi: 10.1016/s1367-5931(02)00347-2. [DOI] [PubMed] [Google Scholar]
- 25.Woller EK, Walter ED, Morgan JR, Singel DJ, Cloninger MJ. Altering the strength of lectin binding interactions and controlling the amount of lectin clustering using mannose/hydroxyl-functionalized dendrimers. J Am Chem Soc. 2003;125:8820–8826. doi: 10.1021/ja0352496. [DOI] [PubMed] [Google Scholar]
- 26.Woller EK, Cloninger MJ. The lectin-binding properties of six generations of mannose-functionalized dendrimers. Org Lett. 2002;4:7–10. doi: 10.1021/ol016568+. [DOI] [PubMed] [Google Scholar]
- 27.Jayaraman N, Nepogodiev SA, Stoddart JF. Synthetic carbohydrate-containing dendrimers. Chem –Eur J. 1997;3:1193–1199. [Google Scholar]
- 28.Page D, Roy R. Synthesis and biological properties of mannosylated starburst poly(amidoamine) dendrimers. Bioconjugate Chem. 1997;8:714–723. doi: 10.1021/bc970126u. [DOI] [PubMed] [Google Scholar]
- 29.Kieburg C, Lindhorst TK. Glycodendrimer synthesis without using protecting groups. Tetrahedron Lett. 1997;38:3885–3888. [Google Scholar]
- 30.Mammen M, Choi SK, Whitesides GM. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew Chem, Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 31.Stiriba SE, Frey H, Haag R. Dendritic polymers in biomedical applications: from potential to clinical use in diagnostics and therapy. Angew Chem, Int Ed. 2002;41:1329–1334. doi: 10.1002/1521-3773(20020415)41:8<1329::aid-anie1329>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 32.Krause W, Hackmann-Schlichter N, Maier FK, Müller R. Dendrimers in diagnostics. Top Curr Chem. 2000;210:261–308. [Google Scholar]
- 33.Wathier M, Jung PJ, Carnahan MA, Kim T, Grinstaff MW. Dendritic macromers as in situ polymerizing biomaterials for securing cataract incisions. J Am Chem Soc. 2004;126:12744–12745. doi: 10.1021/ja045870l. [DOI] [PubMed] [Google Scholar]
- 34.Grinstaff MW. Biodendrimers: New Polymeric Biomaterials for Tissue Engineering. Chem –Eur J. 2002;8:2838–2846. [PubMed] [Google Scholar]
- 35.Lee H, Baker JR, Larson RG. Molecular dynamics studies of the size, shape, and internal structure of 0% and 90% acetylated fifth-generation polyamidoamine dendrimers in water and methanol. J Phys Chem B. 2006;110:4014–4019. doi: 10.1021/jp056148s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maiti PK, Çağin T, Wang G, Goddard WA. Structure of PAMAM dendrimers: generations 1 through 11. Macromolecules. 2004;37:6236–6254. [Google Scholar]
- 37.Lee I, Athey BD, Wetzel AW, Meixner W, Baker JR. Structural molecular dynamics studies on polyamidoamine dendrimers for a therapeutic application: effects of pH and generation. Macromolecules. 2002;35:4510–4520. [Google Scholar]
- 38.Esfand R, Tomalia DA. Poly(amidoamine) (PAMAM) dendrimers: from biomimicry to drug delivery and biomedical applications. Drug Discovery Today. 2001;6:427–436. doi: 10.1016/s1359-6446(01)01757-3. [DOI] [PubMed] [Google Scholar]
- 39.Kim Y, Hechler B, Klutz A, Gachet C, Jacobson KA. Toward multivalent signaling across G protein-coupled receptors from poly(amidoamine) dendrimers. Bioconjugate Chem. 2008;19:406–411. doi: 10.1021/bc700327u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim Y, Klutz AM, Hechler B, Gao ZG, Gachet C, Jacobson KA. Application of the functionalized congener approach to dendrimer-based signaling agents acting through A2A adenosine receptors. Purinergic Signalling. 2008;5:39–50. doi: 10.1007/s11302-008-9113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klutz AM, Gao ZG, Lloyd J, Shainberg A, Jacobson KA. Enhanced A3 adenosine receptor selectivity of multivalent nucleoside-dendrimer conjugates. J Nanobiotechnol. 2008;6:12. doi: 10.1186/1477-3155-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haugland RP. Handbook of Fluorescent Probes and Research Products. 9th. Molecular Probes, Invitrogen; 2002. pp. 20–35. [Google Scholar]
- 43.Kobayashi H, Brechbiel MW. Dendrimer-based nanosized MRI contrast agents. Curr Pharm Biotechnol. 2004;5:539–549. doi: 10.2174/1389201043376571. [DOI] [PubMed] [Google Scholar]
- 44.Longmire M, Choyke PL, Kobayashi H. Dendrimer-based contrast agents for molecular imaging. Curr Topics Med Chem. 2008;8:1180–1186. doi: 10.2174/156802608785849021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim Y, Klutz AM, Jacobson KA. Systematic investigation of polyamidoamine dendrimers surface-modified with poly(ethylene glycol) for drug delivery applications: Synthesis, characterization, and evaluation of cytotoxicity. Bioconjugate Chem. 2008;19:1660–1672. doi: 10.1021/bc700483s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fricks IP, Carter RL, Lazarowski ER, Harden TK. Gi-dependent cell signaling responses of the human P2Y14-receptor in model cell systems. J Pharmacol Exp Therap. 2009 doi: 10.1124/jpet.109.150730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, IJzerman AP, Stevens RC. The 2.6 Å crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ivanov AA, Jacobson KA. Molecular modeling of a PAMAM-CGS21680 dendrimer bound to an A2A adenosine receptor homodimer. Bioorg Med Chem Lett. 2008;18:4312–4315. doi: 10.1016/j.bmcl.2008.06.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gandia J, Galino J, Amaral O, Soriano A, Lluís C, Franco R, Ciruela F. Detection of higher-order G protein-coupled receptor oligomers by a combined BRET-BiFC technique. FEBS Lett. 2008;582:2979–2984. doi: 10.1016/j.febslet.2008.07.045. [DOI] [PubMed] [Google Scholar]
- 50.Jacobson KA. Functionalized congener approach to the design of ligands for G protein-coupled receptors (GPCRs) Bioconjugate Chem. 2009 doi: 10.1021/bc9000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lazarowski ER, Shea DA, Boucher RC, Harden TK. Release of cellular UDP-glucose as a potential extracellular signaling molecule. Mol Pharmacol. 2003;63:1190–1197. doi: 10.1124/mol.63.5.1190. [DOI] [PubMed] [Google Scholar]
- 52.Cordeaux Y, Briddon SJ, Alexander SP, Kellam B, Hill SJ. Agonist occupied A3 adenosine receptors exist within heterogeneous microdomains of individual living cells. FASEB J. 2008;22:850–860. doi: 10.1096/fj.07-8180com. [DOI] [PubMed] [Google Scholar]
- 53.Kitchens KM, El-Sayed MEH, Ghandehari H. Transepithelial and endothelial transport of poly (amidoamine) dendrimers. Adv Drug Delivery Rev. 2005;57:2163–2176. doi: 10.1016/j.addr.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 54.Duncan R, Izzo L. Dendrimer biocompatibility and toxicity. Adv Drug Delivery Rev. 2005;57:2215–2237. doi: 10.1016/j.addr.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 55.Harden TK, Scheer AG, Smith MM. Differential modification of the interaction of cardiac muscarinic cholinergic and beta-adrenergic receptors with a guanine nucleotide binding component(s) Mol Pharmacol. 1982;21:570–580. [PubMed] [Google Scholar]
- 56.Nourse A, Millar DB, Minton AP. Physicochemical characterization of generation 5 polyamidoamine dendrimers. Biopolymers. 2000;53:316–328. doi: 10.1002/(SICI)1097-0282(20000405)53:4<316::AID-BIP4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


