Abstract
The identification of new antibacterial targets is urgently needed to address multidrug resistant and latent tuberculosis infection. Sulfur metabolic pathways are essential for survival and the expression of virulence in many pathogenic bacteria, including Mycobacterium tuberculosis. In addition, microbial sulfur metabolic pathways are largely absent in humans and therefore, represent unique targets for therapeutic intervention. In this review, we summarize our current understanding of the enzymes associated with the production of sulfated and reduced sulfur-containing metabolites in Mycobacteria. Small molecule inhibitors of these catalysts represent valuable chemical tools that can be used to investigate the role of sulfur metabolism throughout the Mycobacterial lifecycle and may also represent new leads for drug development. In this light, we also summarize recent progress in the development of inhibitors of sulfur metabolism enzymes.
Keywords: Tuberculosis, mycobacteria, sulfur metabolism, enzymes, thiols, sulfation, drug design, inhibitors
MYCOBACTERIUM TUBERCULOSIS
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), is one of the most lethal infectious agents affecting humans [1-3]. The disease infects almost two billion people or one-third of the world’s population, and accounts for an estimated 2 million deaths per year. The majority of people afflicted with TB live in developing countries, where lethal synergy with HIV infection also fuels the TB pandemic.
M. tuberculosis infection is difficult to treat, requiring 6-9 months of chemotherapy with a cocktail of four antibiotics – isoniazid, rifampin, pyrazinamide and ethambutol [4,5]. In large part, the lengthy drug therapy is necessary because mycobacteria exist as a metabolically diverse population within the human host [5]. Some bacteria will be actively dividing, rendering them susceptible to antibiotic treatment. However, less active subpopulations of also exist in stationary phase or as dormant bacteria [6,7]. Since TB drugs target biological processes required for bacterial growth (e.g., cell wall biosynthesis), they are far less effective at killing the persistent population [5,8,9].
In addition to toxic side effects, the lengthy treatment regime results in poor patient compliance and drug resistant strains are beginning to emerge [10]. The World Health Organization estimates that up to 50 million persons worldwide are infected with multidrug resistant strains of M. tuberculosis (MDR-TB) [11]. This number continues to grow as 300,000 new MDR-TB cases are diagnosed each year with 79 percent of individuals showing resistance to three or more frontline drugs [11]. Taken together, the growing problem of MDR-TB and the lack of drugs that effectively target persistent bacteria, stress the urgent need for identification of new antimicrobial targets [12,13].
Many fundamental aspects of mycobacterial metabolism and pathogenesis are poorly understood, in part because of the technical difficulties inherent to studying M. tuberculosis. The organism must be manipulated in a biosafety level 3 laboratory, and the slow growth rate (3 weeks for colonies, up to 1 year for completion of animal models) imposes limitations on apparent research productivity. However, the availability of complete mycobacterial genome sequences [14-17] and the maturation of methods for disrupting mycobacterial genes [18-20] have provided tools that can accelerate the discovery of potential drug targets and elucidate metabolic pathways that are essential for mycobacterial survival.
OVERVIEW OF TB INFECTION
M. tuberculosis infection is a complex process that initiates with aerosol inhalation to the host lung [6,21,22]. Therein, the mycobacteria are phagocytosed by alveolar macrophages. Upon entry into a macrophage, the TB bacilli interfere with normal phagosomal maturation, preventing fusion with lysosomes [23]. The ability of M. tuberculosis to side-step lysosomal degradation allows the bacilli to take up residence an endosomal environment and multiply within the host cell. In response to the infection, macrophages produce pro-inflammatory signals – cytokines and chemokines – that recruit T-cells and neutrophils to the infected tissue [22,24-27]. These cells encircle the infected macrophage, walling it off from the surrounding tissue in a structure called a granuloma [28-31]. Within the context of the granuloma, T-cells can proliferate in response to specific mycobacterial antigens and some may leave the granuloma to reenter the circulation; thus, the granuloma is a dynamic structure [32]. Activation of the immune response and induction of lung inflammation is part of the M. tuberculosis lifecycle [6,21,22]. The lung tissue damage caused by activated immune cells induces coughing and provides an exit strategy for the bacteria to spread to another host.
Less than 10% of infected individuals will develop active TB infection. In the rest, mycobacteria residing within granulomas enter into a persistent or “latent” state characterized by a lack of cell division and a change in basic metabolism [6,7,33,34]. These latent mycobacteria are difficult to eradicate since they are not reliant on machinery targeted by conventional antibiotics [5]. By unknown mechanisms, the infection can be reactivated after many years or decades to produce active, infectious TB. This event is often associated with compromised immune function due to coinfection with HIV, drug use, or aging. Hence, effective treatment of TB will require efficacy against persistent M. tuberculosis, or at the least a better understanding of the mechanisms underlying immune cell activation, bacterial adaptation and survival within the granuloma [5,13,31].
SULFUR AND MYCOBACTERIAL SURVIVAL
To complete its lifecycle, M. tuberculosis must survive within the hostile, nutrient-poor and oxidizing environment of the host macrophage [7,30,35]. At the same time, M. tuberculosis must activate sufficient immune effector functions to induce granuloma formation in the lung [21,22]. This complex interplay between mycobacteria and the host immune system likely requires several host-pathogen interaction mechanisms and, once the granuloma has been formed, induction of metabolic pathways that allow the organism to persist. At present time, the metabolic requirements of mycobacteria in the context of the granuloma are not fully understood. However, genes involved in the metabolism of sulfur have consistently been identified as up-regulated in response to oxidative stress, nutrient starvation and dormancy adaptation (culture conditions that model aspects of mycobacterial life in the granuloma) and during macrophage infection [36-45].
Sulfur is an essential element for life and plays a central role in numerous microbial metabolic processes [46]. In its reduced form, sulfur is used in the biosynthesis of the amino acids cysteine and methionine. Cysteine is incorporated into biomolecules such as proteins, coenzymes, and mycothiol (the mycobacterial equivalent of glutathione) [See Fig. (1)]. Found in all actinomycetes, mycothiol regulates cellular redox status and is essential for M. tuberculosis survival [47]. Another reduced sulfur-containing metabolite, coenzyme A (CoA), is heavily utilized for lipid metabolism (a process that is central to mycobacterial cell wall maintenance and remodeling) [48]. In its oxidized form, sulfur is present as a sulfuryl moiety (−SO3−) that can modify hydroxyls and amines in proteins, polysaccharides and lipids [See Fig. (2)] [49,50]. Extracellular presentation of sulfated metabolites plays important regulatory roles in cell-cell and host-pathogen communication [50]. Hence, acquisition and metabolism of sulfur is essential for mycobacterial virulence and survival.
Fig. (1).
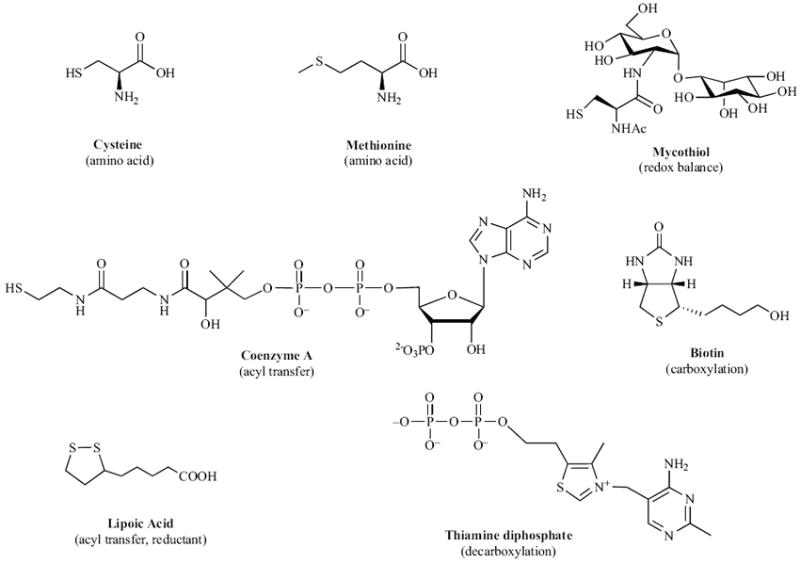
Reduced sulfur-containing metabolites in mycobacteria.
Fig. (2).
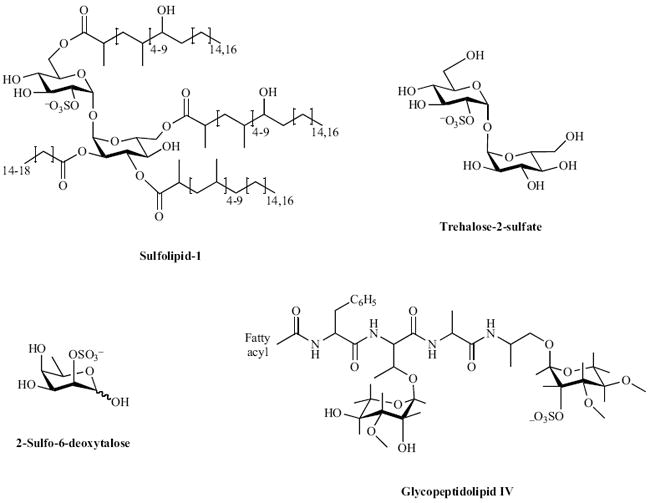
Sulfated metabolites in mycobacteria.
The identification of new antibacterial targets is essential to address MDR- and latent-TB infection [13,51]. Toward this end, mycobacterial sulfur metabolism represents a promising new area for anti-TB therapy [50,52,53]. Numerous studies have validated amino acid biosynthetic pathways and downstream metabolites as antimicrobial targets [54-57] and sulfur metabolic pathways are required for the expression of virulence in many pathogenic bacteria [58-62]. In particular, mutants in mycobacterial sulfur metabolism genes are severely impaired in their ability to persist and cause disease [41,43,61,63-66]. Furthermore, most microbial sulfur metabolic pathways are absent in humans and therefore, represent unique targets for therapeutic intervention. In this review, we focus on the enzymes associated with the production of sulfated and reduced sulfur-containing metabolites in Mycobacteria. Small molecule inhibitors of these catalysts represent valuable chemical tools that can be used to investigate the role of sulfur metabolism in M. tuberculosis survival and may also represent new leads for drug development. In this light, we also highlight major efforts toward inhibitor discovery of mycobacterial sulfur metabolic pathways.
SULFATE ASSIMILATION IN MYCOBACTERIA
Sulfate assimilation begins with the active transport of inorganic sulfate (SO42-) across the mycobacterial cell membrane by the CysTWA SubI ABC transporter complex [see Fig. (3)] [67,68]. Once sulfate is imported, it is activated by ATP sulfurylase (encoded by cysND) via adenylation to produce adenosine-5’-phosphosulfate (APS) [39,53,69]. In mycobacteria, APS lies at a metabolic branch point [53]. For sulfation of biomolecules such as proteins, lipids and polysaccharides, APS is phosphorylated at the 3’-hydroxyl by APS kinase (encoded by cysC) to form 3’-phosphoadenosine-5’-phosphosulfate (PAPS), the universal sulfate donor for sulfotransferases (STs) [53,69,70]. Transfer of −SO3− to hydroxyl or amino functionalities of biomolecules plays important roles in regulation of cell-cell communication and metabolism [50]. Alternatively, for production of reduced sulfur-containing metabolites, the sulfate moiety in APS is reduced to sulfite (SO32-) by APS reductase (gene product of cysH) [53,61,71,72]. Sulfite is further reduced to sulfide (S2-) by sulfite reductase (encoded by nirA) [73] and is the form of sulfur that is used for the biosynthesis of sulfur-containing metabolites including cysteine, methionine, coenzymes, and mycothiol [46,47,49]. Each branch of sulfate assimilation is discussed in terms of the available genetic and biochemical data below.
Fig. (3).
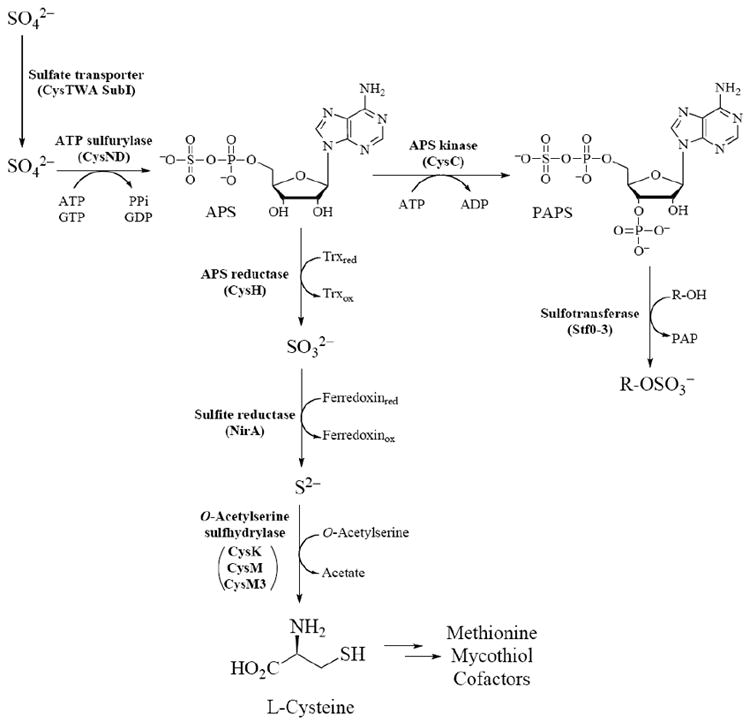
The sulfate assimilation pathway in mycobacteria.
Sulfate Import and Activation
Present at 300-500 μM, inorganic sulfate is the fourth most abundant anion in human plasma [74]. Sulfate transporters have been identified in all major human tissues investigated to date, and of particular relevance to the intracellular lifestyle of M. tuberculosis, the existence of endosomal-associated transporters has also been demonstrated [74]. The genes encoding the CysTWA SubI ABC transporter complex in mycobacteria have been identified by homology to Escherichia coli and Salmonella typhimurium [68], are essential [41], robustly up-regulated during oxidative stress [37], dormancy adaptation [36], and expressed in macrophages [44]. Consistent with this annotation, cysA or subI mutants (ΔcysA or ΔsubI, respectively) in M. bovis bacillus Calmette-Guérin (BCG) – an attenuated, vaccine strain of M. bovis – are compromised in their ability to transport sulfate [68,75]. When grown in media supplemented with casamino acids, the rate of sulfate transport in ΔcysA is ~1.1% relative to wild-type M. bovis BCG [68]. The minor amount of transport is not enough to meet bacterial sulfur requirements and hence, these sulfate transport mutants are auxotrophic for reduced sulfur.
Interestingly, no significant difference in the number of viable bacilli was observed in the organs of mice infected with ΔcysA and wild-type M. bovis BCG up to 63 days post-infection [68]. These data indicate that M. bovis BCG may scavenge sufficient amounts of reduced sulfur from the host for survival. However, an important question raised from the findings of this study is whether the sulfur requirements for an attenuated M. bovis strain reflect those of M. tuberculosis known to elicit a more potent host immune response [21,22,32]. It is also possible that the mycobacterial genome encodes for an additional sulfate transporter which is not expressed under culture conditions, but is specifically up-regulated during infection [52]. In support of this hypothesis, mRNA array analysis shows significant up-regulation of hypothetical protein 1739c (annotated as a putative high affinity sulfate transporter) during M. tuberculosis infection of macrophages in response to nitric oxide [38] or hypoxia [45]. Additional studies will be required to confirm the function of the putative sulfate transporter and its relevance to sulfate acquisition in vivo.
Once sulfate is transported to the cytosol, ATP sulfurylase (encoded by cysD) catalyzes the first committed step in sulfate assimilation [see Fig. (3)] [39,69]. In this reaction, the adenylyl moiety of adenosine 5’-triphosphate (ATP) is coupled to sulfate. The product that results, APS, contains a unique high-energy phosphoric-sulfuric acid anhydride bond – the biologically activated form of sulfate [49]. Formation of APS is energetically unfavorable (Keq of 10-7 – 10-8 near physiological conditions) [49] and in prokaryotes, the hydrolysis of guanosine 5’-triphosphate (GTP) is coupled to sulfurylation of ATP to surmount this energetic hurdle [76]. The GTPase (encoded by cysN) forms a heterodimer with ATP sulfurylase (CysD) and synthesis of APS synthesis is driven 1.1 × 106-fold further during GTP hydrolysis [69]. Notably, eukaryotic ATP sulfurylases do not bear any sequence or structural similarity to their prokaryotic counterparts, nor do they employ a GTPase for PAPS biosynthesis [77]. These mechanistic and structural differences, in particular the unique G protein subunit, could be exploited to develop small molecule inhibitors of bacterial sulfate activation [52].
The final step in PAPS biosynthesis is catalyzed by APS kinase (encoded by cysC) [53,70]. In this reaction, ATP is utilized to phosphorylate the 3’-hydroxyl of APS. Depending on the organism, APS kinase can be encoded as a separate protein or as a fusion with ATP sulfurylase, without significant variation in catalytic mechanism [53,78]. Most eukaryotes (including humans) encode for ATP sulfurylase (CysD) and APS kinase (CysC) on a single polypeptide. In M. tuberculosis, however, APS kinase (Cys C) is genetically fused to the GTPase subunit (CysN) of ATP sulfurylase [53]. The APS kinase domain of M. tuberculosis CysNC was identified through sequence homology and confirmed by genetic complementation [53]. In a subsequent report, a mutant strain of M. tuberculosis that removes the APS kinase domain of the bifunctional cysNC gene was constructed [70]. As expected, the cysC knockout (ΔcysC) was able to grow on sulfate as a sole sulfur source (indicating a functional ATP sulfurylase), but was unable to synthesize PAPS [70].
Fusion of APS kinase to the GTPase domain of ATP sulfurylase raised the interesting possibility of substrate channeling between subunits [52,78]. In this scenario, the final product PAPS, and not the APS intermediate, would be released into solution. Leyh and colleagues have recently tested this hypothesis for M. tuberculosis ATP sulfurylase [78]. Although PAPS synthesis is 5,800 times more efficient than APS synthesis [69], these studies demonstrate that APS is not channeled from the M. tuberculosis adenylyl-transferase to the APS kinase domain [78], consistent with the domain arrangement proposed from a recent crystal structure of the CysNC complex [77].
Collectively, CysNC and D proteins form a multifunctional enzyme complex ~300 KDa (consistent with a trimer of CysNC•D heterodimers), referred to as the sulfate-activating complex (SAC) [39,69]. In M. tuberculosis, expression of the SAC operon is induced by conditions likely to be encountered by pathogenic mycobacteria within the macrophage, including sulfur limitation, oxidative stress, and is repressed by cysteine [37,39]. The SAC operon is also up-regulated during stationary phase growth, an in vitro model of persistent M. tuberculosis infection [36]. M. tuberculosis SAC gene expression is also augmented within the intracellular environment of the macrophage [44,79]. Taken together, these data are consistent with increased activity of sulfate-activating enzymes and flux through the sulfate assimilation pathway during mycobacterial infection.
Sulfotransferases and Sulfation
Sulfotransferases (STs), the enzymes that install sulfate esters, transfer sulfate from PAPS (produced by the SAC) to a hydroxyl or, less frequently, to an amide moiety on glycoproteins, glycolipids and metabolites [see Fig. (3)] [50]. Sulfated metabolites are abundant in higher eukaryotes, particularly mammals, where they function primarily in cell-cell communication. For example, sulfated glycoproteins mediate interactions of leukocytes with endothelial cells at sites of chronic inflammation, sulfated peptides such as hirudin and cholecystokinin act as hormones, and sulfated glycolipids are involved in neuronal development [80,81]. In contrast, reports of sulfated metabolites in prokaryotes have been rare. In 1992, Long and colleagues reported the first functionally characterized sulfated metabolite from the prokaryotic world – the nodulation factor NodRm-1 from Sinorhizobium meliloti [82]. This sulfated glycolipid is secreted from the bacterium and acts on host plant cell receptors thereby initiating symbiotic infection [83].
Among pathogenic bacteria, only one family has been reported to produce sulfated metabolites – the Mycobacteria. More than 40 years ago, Goren and coworkers isolated an abundant sulfated glycolipid from the M. tuberculosis cell wall and characterized the structure shown in Fig. (2) [84-86]. Termed sulfolipid-1 or SL-1, this compound has only been observed in the tuberculosis complex; it is absent from non-pathogenic mycobacteria such as M. smegmatis. Comprising a trehalose-2-sulfate (T2S) core modified with four fatty acyl groups, SL-1 accounts for almost 1% of the dry weight of M. tuberculosis. Early studies found a correlation between the abundance of SL-1 and the virulence of different clinical M. tuberculosis isolates [87,88] and its location in the outer envelope has prompted speculation that it may be involved in host-pathogen interactions [89]. The exact function of SL-1, however, remains elusive (see [52] and references therein). Nonetheless, the biosynthetic pathway for SL-1 has recently been elucidated [52,90-92] and a comprehensive study of mutants in SL-1 biosynthesis should help clarify the role of this sulfated glycolipid in the mycobacterial lifecycle.
In addition to SL-1, other novel sulfated metabolites have been identified in M. tuberculosis using an innovative metabolomic approach that combines genetic engineering, metabolic labeling with a stable sulfur isotope (34SO42-) together with mass spectrometry analysis [70] [see Fig. (2)]. Structurally distinct sulfated metabolites have also been identified in several other mycobacterial species, including M. smegmatis, M. fortuitum, and the HIV-associated opportunistic pathogen M. avium [see Fig. (2)] [70,93-95]. Interestingly, in M. avium a sulfated cell wall glycopeptidolipid was recently found to be up-regulated in HIV patients with acquired drug resistance [93]. Significant work remains to fully characterize and elucidate the biological significance of sulfated metabolites found in mycobacteria. A major step toward this objective is to define the biosynthetic pathways of mycobacterial sulfated metabolites, including the STs responsible for installing the sulfuryl moiety.
In 2002, an analysis of mycobacterial genomes reported by Mougous and colleagues revealed a large family of open reading frames with homology to human carbohydrate sulfotransferases [50]. The predicted proteins shared regions of sequence homology associated with binding to their common substrate, PAPS. Presently, four such genes have been identified in M. tuberculosis (annotated as stf0-3) and the M. avium genome encodes nine putative STs (stf0, 1, 4-10) [52]. To date, of the 11 predicted STs found in mycobacterial genomes, genetic and biochemical studies have only been reported for Stf0 and Stf3.
Stf0 is present in a number of other pathogenic bacteria and initiates the biosynthesis of SL-1 by sulfating the disaccharide, trehalose, to form T2S [see Fig. (2)] [92]. A knockout mutant of stf0 has been reported in M. tuberculosis [96]. This study demonstrates that Stf0 is not required for survival in liquid culture, hinting toward a specific role in host infection. The structure of stf0 in complex with trehalose has recently been reported and has revealed several interesting features [92]. In the presence of trehalose, Stf0 forms a dimer both in solution and in the crystal structure. Moreover, Stf0-bound trehalose participates in the dimer interface, with hydroxyl groups from a glucose residue bound in one monomer forming interactions with the other monomer. Residues involved in substrate binding and dimerization have been identified, along with a possible general base (i.e., Glu36) that may facilitate nucleophilic attack of the 2’-hydroxyl group on PAPS. A panel of synthetic glucose and trehalose analogs has also been tested for binding revealing that any modification to the parent disaccharide compromises substrate sulfation [92]. Finally, a kinetic study of the enzyme using MS has also been reported [97]. The results address the order of substrates binding and are consistent with a random sequential mechanism involving a ternary complex with both PAPS [or 3’-phosphoadenosine-5’-phosphate, (PAP)] and trehalose (or T2S) bound in the active site.
Stf3 may play a regulatory role in M. tuberculosis virulence [98]. In a mouse model of TB infection, a mutant strain in which Stf3 was disrupted (Δstf3) was unable to produce a sulfated molecule termed, “S881”. Interestingly, when compared to wild-type M. tuberculosis, Δstf3 exhibited a hypervirulent phenotype. No relatives of the remaining stf family members are found in any other prokaryotic genomes, suggesting that they are unique to mycobacteria. Substrates for the majority of mycobacterial STs remain to be elucidated.
ST INHIBITOR DISCOVERY
Although the roles of sulfated metabolites in the mycobacterial lifecycle remain under investigation ([52] and references therein), the analogy to sulfation in higher eukaryotes is compelling. The challenges to defining their role in mycobacterial infection and survival are two-fold: (1) the collection of sulfated metabolites must be identified and structurally characterized; and, (2) the biosynthetic pathway of the sulfated metabolites must be elucidated. In addition to traditional genetic approaches, small molecule inhibitors of STs in mycobacteria would also be useful tools to dissect their physiological roles. In addition, since STs play critical biological roles in higher eukaryotes and are implicated in several disease states, they also represent promising therapeutic targets [80,99]. Since prokaryotic STs have not been discovered until relatively recently, the majority of research and inhibitor discovery has focused primarily on eukaryotic STs. Nonetheless, these studies can serve as a platform for mycobacterial ST inhibitor design and we highlight the most fruitful efforts to date.
There are two classes of STs - cytosolic and Golgi-resident enzymes [50,80,100]. In general, cytosolic STs sulfonate small molecules such as hormones and bioamines while membrane-bound STs prefer larger substrates such as proteins and carbohydrates. STs have also been further classified according to their functional role into estrogen STs (EST), heparin STs, tyrosyl protein ST (TPST), N-Acetyl glucosamine 6-O-ST and carbohydrate STs. The first crystal structure to be elucidated was that of murine estrogen sulfotransferase (mEST) in 1997 [101] and since then, structures of nine other STs have been characterized. These include cytosolic STs such as Phenol ST (SULT1A1) [102], catecholamine ST (SULT1A3) [103], mycobacterial Stf0 [92] and Golgi-resident STs (GSTs) such as heparan N-deacetylase-N-ST-1 (NDST-1) [104]. Structures of STs in complex with PAPS or PAP reveal a conserved nature of the cofactor binding site, suggesting that STs share a similar mechanisms of sulfuryl transfer. The catalytic site of each ST must also accommodate diverse substrates and these differences in specificity are reflected in the substrate-binding site of each ST [99].
Bisubstrate Analogs
To investigate molecules that inhibit both the PAPS- and substrate–binding domains of STs simultaneously, synthetic bisubstrate analogs have been employed [105,106]. Compounds were designed to incorporate elements from the cofactor, PAPS and the substrate, providing specificity via critical interactions within both binding pockets of the enzyme [107]. Inhibitor potency is achieved from the entropic advantage of linking structures that mimic each substrate. On screening a 447 member 3’-phosphoadenosine library, several bisubstrate-based compounds were identified (1) [105], (2) [106] as inhibitors of EST (see Table 1). The activities of these compounds were comparable to some of the other compounds known to be inhibitors of EST including polychlorinated biphenols (3), discovered by testing a large number of hydroxylated polychlorinated biphenyl metabolites [108] and dietary agents like Quercetin (4), identified from a study investigating the inhibitory effects of natural flavonoids on EST activity (see Table 1) [109].
Table 1.
Sulfotransferase Inhibitors
| No. | Ref. | Structure | Activity[a] (IC50) | Type of Inhibition |
|---|---|---|---|---|
| 1 | [105] |
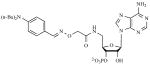
|
>80% inhibition at 200 μM | No data |
| 2 | [106] |
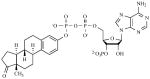
|
10 nM | Competitive with PAP |
| 3 | [108] |
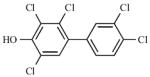
|
0.15 – 0.25 nM | Non-competitive |
| 4 | [109] |
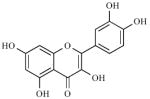
|
0.1 μM | Competitive |
| 5 | [113] |
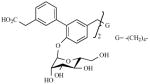
|
70 μM | Non-competitive |
| 6 | [114] |
|
20 μM | Competitive |
| 7 | [117] |
|
96 nM (Ki) | Competitive |
| 8 | [99] |
|
50 μM | No data |
| 9 | [99] |
|
30 μM | No data |
| 10 | [119] |
|
30 μM | Specific, reversible |
| 11 | [119] |
|
40 μM | Specific, irreversible |
Enzymes assayed: 1-4: EST; 5 : P-Selectin ; 6: GST; 7: β-AST-IV; 8: NodH; 9: GST-3; 10-11: TPST-2.
Similar substrate-emulating approaches have also been used to design inhibitors for E-, P- and L-selectins, all prime targets for anti-inflammatory drug discovery [110]. GSTs are involved in biosynthesis of the L-selectin ligand, 6’-sulfo sialyl Lewis X [111]. The sulfonation of sialyl Lewis X motif by GST leads to a strong interaction with receptors on L-selectin cell adhesion molecules resulting in a potent anti-inflammatory response. A “glycomimetic” strategy was used to design inhibitors for these STs. In this approach the inhibitors retained structural and functional aspects of the natural ligands, but were designed to be synthetically more feasible [112]. One selectin antagonist (5), was identified using this strategy and it is currently under clinical trials (see Table 1) [113].
Kinase-Derived Inhibitors
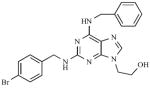
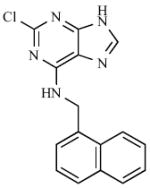
The “kinase inhibitor” approach exploits the similarity between reactions catalyzed by STs and kinases. Since STs and kinases use adenosine-based donor nucleotides to transfer an anionic moiety onto their respective substrates (PAPS for STs and ATP for kinases), it was proposed that ATP derivatives might also function as ST inhibitors [80,114]. Furthermore, the hydrophobic adenine binding pockets of EST [101,115] and heparin N-sulfotransferase [104] are similar to those of several kinases. A 2,6,9-trisubstituted purine library [116], originally designed to target cyclin dependent kinase 2, was tested for inhibitory activity with carbohydrate STs. Of the 139 compounds screened, the six most potent purines exhibited half maximal inhibitory concentrations (IC50s) that ranged from 20 – 40 μM (6) [114], with five of them having a common benzyl substituent at N6 (see Table 1). Though these inhibitors showed selectivity for carbohydrate STs, achieving selectivity over kinases remains a challenge. A high throughput screen of 35,000 purine and pyrimidine analogs has also identified a potent inhibitor of β–arylsulfotransferases (β-AST-IV) (7) [117] (see Table 1).
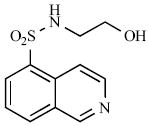
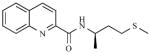
A second class of kinase inhibitors, isoquinoline sulfonamides, has also been tested for inhibitory activity against a panel of STs consisting of EST, NodH, GST-2 [118]. Isoquinoline sulfonamide inhibitors were developed after a crystal structure of cyclic adenosine-5’-phosphate (cAMP) dependent protein kinase in complex with isoquinoline showed that the heterocycle moiety was bound in the subsite occupied by the adenine ring of ATP. Of the 100 isoquinoline and quinoline derivatives screened, the most active compounds inhibited single enzyme selectively with modest IC50 values in the range of 30 – 100 μM (8, 9) [99,119] (see Table 1).
Combinatorial Target-Guided Ligand Assembly
In this strategy, a library of ligands or ‘monomers’ carry a common chemical handle to facilitate their combinatorial assembly [119]. In the first round, monomers were screened against the ST target at concentrations of 1 mM or higher. Compounds that demonstrated inhibitory activity were then used to construct a library of ‘dimers’ via an oxime linkage, and were screened for inhibitors. This approach resulted in the identification of two of the first known inhibitors of Golgi-resident tyrosyl protein ST-2 (TPST-2) (10, 11) [119] (see Table 1).
ST inhibitors identified in the studies above are a promising start in drug discovery efforts. However, to date the majority of ST inhibitor compounds possess fairly modest IC50s, are not “drug-like”, or suffer from a lack of specificity. Recent advances in structure-based drug design and high-throughput screening should greatly facilitate the discovery of new inhibitors for STs and other sulfonucleotide-binding enzymes.
OXIDATIVE ANTIMICROBIAL ACTIVITY
In order to replicate and persist in its human host, M. tuberculosis must survive within the hostile environment of the macrophage, where bactericidal oxidants - superoxide (O2·−) and nitric oxide (NO·) – are generated in response to infection [35]. Two enzymes, nicotinamide adenine dinucleotide phosphate-oxidase (NADPH oxidase) and inducible nitric oxide synthase (NOS2), are largely responsible for production of these reactive oxygen and nitrogen intermediates (termed ROI and RNI, respectively) [120,121].
NADPH oxidase is a membrane protein that generates O2·− by transferring electrons from NADPH inside the cell across the phagosomal membrane; the electrons are coupled to molecular oxygen to produce O2·− [122]. Subsequently, O2·− can accept an electron spontaneously or be reduced by superoxide dismutase (SOD) to form hydrogen peroxide (H2O2) [123]. In turn, H2O2 can oxidize cellular targets or be converted into the highly damaging hydroxyl radical (OH·) through the iron-catalyzed Fenton-Haber-Weiss reaction [124]. In the NOS2 reaction, the guanidino nitrogen of arginine undergoes a five-electron oxidation via a N-ω-hydroxy-L-arginine (NOHLA) intermediate to yield ·NO [125]. The combination of the two oxidant-generating systems can also exert a synergistic effect in bacterial killing as macrophages can generate O2·− simultaneously with ·NO, yielding the more reactive peroxynitrite (ONOO−) [126]. A consequence of NADPH and NOS2 enzymatic activities and the resulting “oxidative burst” is that phagocytosed bacteria are killed by oxidative damage to a range of protein and DNA targets [35,125,127].
In mice, activation of macrophages induces production of NOS2 and phagosomal NADPH oxidase, via ligation of toll-like receptors (TLRs), or via stimulation by the cytokines IFN-γ or TNF-α [128,129]. In mouse models of TB, numerous studies have demonstrated that NOS2 plays an essential role in controlling persistent infection. Macrophages can inhibit mycobacterial growth via NOS2-generated RNI, inhibition of NOS2 during persistent infection leads to reactivation of disease, and NOS2 gene-disrupted mice are extremely susceptible to TB infection [128,129]. More recently, a proteomics study has identified proteins in M. tuberculosis that are targeted by RNI stress [130]. Notably, many essential metabolic and antioxidant defense enzymes are among those proteins found modified for RNI.
While good evidence exists for ROI-mediated bacterial killing of other bacterial, fungal and parasitic pathogens, their bactericidal effect on mycobacteria has been less clear. Studies demonstrate that M. tuberculosis resists killing by ROI in vitro and that mice with defects in p47 or gp91 subunits of phagocyte NADPH oxidase (Phox) are also relatively resistant to TB infection [128,131]. However, NADPH oxidase is highly active during the persistent phase of M. tuberculosis infection in mice [132]. This observation suggests that M. tuberculosis must possess extremely effective detoxification pathways to counter ROI stress. Consistent with this hypothesis, mice deficient in the KatG catalase-peroxidase survived better in pg91phox-deficient mice [132]. More recently, it was shown that macrophages deficient in early stages of Phox assembly exhibited reduced bacterial killing, correlating with decreased production of ROI [133]. Taken together, these observations indicate that survival of M. tuberculosis within macrophages depends upon the ability of the bacterial to counter oxidative assault.
Mycobacteria produce enzymes such as SOD, peroxidases, catalase, and nitrosothiol reductase to help counteract the effect of ROI/RNI and promote intracellular survival and persistence in the host [35,134-136]. In addition to enzymatic detoxification of ROI and RNI, reduced sulfur-containing metabolites are an essential component of bacterial antioxidant defense systems [137-141]. Specifically in mycobacteria, low molecular-weight thiols such as mycothiol [see Fig. (1)], play a central role in maintaining a reducing cellular environment [47,137]. Proper redox homeostasis is essential for normal cellular function and to mitigate the effects of oxidative stress. Hence, the metabolic route used for the production of reduced sulfur-containing metabolites [see Fig. (3)] is predicted to be important for mycobacterial survival [52,53,61]. Consistent with this hypothesis, expression of mycobacterial genes involved in reductive sulfate assimilation are induced by oxidative stress and within the environment of the macrophage [36-45].
Sulfate Reduction
APS reductase (encoded by cysH) catalyzes the first committed step in the biosynthesis of reduced sulfur compounds [see Fig. (3)]. In this reaction, APS is reduced to SO32- and adenosine-5’-phosphate (AMP) [142]. Thioredoxin (Trx), a 12.7 kDa protein with a redox active disulfide bond, supplies the reducing potential necessary for this two-electron reduction [143]. The SO32- product of this reaction is reduced further to S2-, which is used for the biosynthesis of reduced sulfur-containing metabolites, such as cysteine, methionine, CoA, iron-sulfur clusters and mycothiol [46,67] [see Fig (1)]. Consistent with its important metabolic role, APS reductase was identified in a screen for essential genes in M. bovis BCG [41] and cysH is actively expressed during the dormant phase of M. tuberculosis and in the environment of the macrophage [36,44].
Humans do not reduce sulfate for de novo cysteine biosynthesis and therefore, do not have a CysH equivalent. Thus, APS reductase may be an attractive drug target if the enzyme is required for bacterial survival or virulence in vivo [52,53,61,72]. To test this hypothesis, Senaratne and coworkers generated an M. tuberculosis mutant strain lacking CysH (ΔcysH) [61]. As predicted, the mutant strain was auxotrophic for cysteine and could only be grown in media supplemented with this amino acid, methionine or glutathione (from which cysteine can be generated catabolically). The cysH mutant exhibited attenuated virulence in BALB/c and C57BL/6 immunocompetent mice. Growth kinetics in the lungs, spleen and liver of mice infected with ΔcysH or wild-type M. tuberculosis were also quantified. Strikingly, the number of colony-forming units recovered from the ΔcysH mutant mirrored those of wild-type M. tuberculosis during the acute stage of infection [up to 16 days post-infection (pi)]. However, the number of viable bacteria in the mutant became significantly less (i.e., by 3 orders of magnitude) coincident with the emergence of adaptive TH1- mediated immunity and the induction of persistence in the mouse (between 16 and 42 days pi) [144]. In addition, ΔcysH was most compromised in the liver, where the host’s oxidative antimicrobial response is thought to play an especially important role in antimicrobial defense. Since the replication of ΔcysH in mouse tissues during the first 16 days post infection was identical to that of wild-type, these data suggest that mouse tissues can provide M. tuberculosis with sufficient reduced sulfur-containing amino acids (e.g., cysteine and methionine), for initial growth (see discussion below) [52,61,68,145]. Hence, APS reductase activity appears to be dispensable during the acute phase of infection, but indispensable in the later, persistence phase where access to or supply of reduced sulfur-containing nutrients becomes limiting [61].
As discussed above, NOS2 plays a vital role in controlling persistent M. tuberculosis infection in mice [6,146,147]. In order to test the role of APS reductase in protecting the bacteria against the effects of NOS2, NOS2-/- mice were infected with wild-type and ΔcysH M. tuberculosis [61]. In contrast to the observation made in wild-type mice, ΔcysH did not lose viability after the first 21 days pi in NOS-/- mice; all mice succumbed to infection within 26 to 31 days. Thus, ΔcysH is significantly more virulent when NOS2 is absent. Taken together, these studies indicate that APS reductase plays a central role in protecting M. tuberculosis against the effects of reactive nitrogen species produced by NOS2 and is critical for bacterial survival in the persistence phase of infection in mice [61]. Furthermore, a follow-up study demonstrates that immunization of mice with ΔcysH generates protection equivalent to that of the BCG vaccine in mice infected with M. tuberculosis [148].
Attenuation of ΔcysH in a mouse model of M. tuberculosis infection and the importance of APS reductase in mycobacterial persistence further motivated investigation of the molecular details of the reaction catalyzed by APS reductase [61]. Biochemical, spectroscopic, mass spectrometry and structural investigation of APS reductase support a two-step mechanism, in which APS undergoes nucleophilic attack by an absolutely conserved cysteine to form an enzyme S-sulfocysteine intermediate, E-Cys-Sγ−SO3− [61,71,72,149,150]. In a subsequent step, SO32− is released in a Trx-dependent reaction. During the catalytic cycle, nucleophilic attack at Sγ atom of the S-sulfocysteine intermediate results in the transient formation of a mixed disulfide between Trx and APS reductase, with concomitant release of sulfite. The structure of this complex has recently been reported and reveals a unique protein-protein interface as a potential candidate for disruption for small molecule or peptide inhibitors [151].
In addition to the conserved catalytic cysteine, the primary sequence of APS reductase is also distinguished by the presence of a conserved iron-sulfur cluster motif, -CysCys-X~80-CysXXCys- [53,71]. Biochemical studies demonstrate that the four cysteines in this motif coordinate a [4Fe-4S] cluster, and that this cofactor is essential for catalysis [71,72]. The first structure of an assimilatory APS reductase was recently reported, with its [4Fe-4S] cluster intact and APS bound in the active site [149]. Consistent with prior biochemical observations, the structure revealed that APS binds in close proximity to the iron-sulfur center. Hence, compounds that target the metal site may represent promising approaches toward rational inhibitor design. This approach is actively being explored, as well as inhibitors that target the Trx-APS reductase interface and will be reported in due course [152].
The final step in sulfate reduction, the six electron reduction of SO32- to S2-, is catalyzed by sulfite reductase (encoded by nirA) [see Fig (3)] [73]. Like cysH, nirA is an essential gene [41] and is active during the dormant phase of M. tuberculosis [36,44]. The sulfite reductase in M. tuberculosis belongs to the family of ferredoxin-dependent sulfite/nitrite reductases [73]. These enzymes contain a [4Fe-4S] center and a siroheme. In this reaction, the external electron donor (likely ferredoxin) binds transiently to sulfite reductase and transfers electrons to the [4Fe-4S] center, one at a time. Subsequently, sulfite reduction is accomplished by transferring electrons from the cluster to the siroheme, which coordinates the sulfite substrate. In 2005, Schnell and coworkers reported the structure of M. tuberculosis NirA [73]. Interestingly, the structure depicts a covalent bond between the side chains of residues Tyr69 and Cys161 adjacent to the siroheme in the active site of sulfite reductase. Site-directed mutagenesis of either residue impairs catalytic activity, though their involvement in the mechanism of sulfite reduction is presently unknown [73].
Cysteine Biosynthesis
De novo cysteine biosynthesis in mycobacterium occurs via condensation of S2- with O-acetyl-L-serine (synthesized by cysE, a serine acetyl transferase) [46,67] [see Fig. (3)]. The M. tuberculosis genome contains three O-acetylserine sulfhydrylase genes, cysM, cysK and cysM3 that can catalyze this reaction. Notably, cysE and cysM are essential for survival in a mouse model of M. tuberculosis infection or in primary macrophages, respectively [40,43]; cysM is also up-regulated under oxidative stress conditions [37].
In 2005, Burns and colleagues presented in vitro evidence for an additional pathway to make cysteine from sulfide [see Fig. (4)] [153]. In this pathway, a sulfide carrier protein, CysO, is converted into a thiocarboxylate by MoeZ (Rv3206), and then alkylated by O-acetyl serine in a reaction catalyzed by CysM. Subsequently, an S–N acyl rearrangement takes places to afford CysO-cysteine which is hydrolyzed by Mec+ (Rv1334) to release cysteine and regenerate CysO. An appealing feature of this pathway is that a protein-bound thiocarboxylate would be much more stable to oxidative species in the macrophage, relative to free sulfide [153]. Analysis of mRNA expression demonstrates that each of these genes is up-regulated during exposure to toxic oxidants [37].
Fig. (4).

Postulated alternate cysteine biosynthetic pathway [153].
Like most organisms, mycobacteria do not have large pools of free cysteine [145]. Once cysteine is produced it is rapidly utilized in protein synthesis, or for the biosynthesis of methionine and reduced sulfur containing cofactors [See Fig. (1)]. The most abundant thiol metabolite in mycobacteria (present in millimolar concentrations) is mycothiol [154]. Found in all actinomycetes, mycothiol is essential for M. tuberculosis survival and intracellular levels of this thiol are associated with changes in resistance to antibiotics and oxidative stress [47].
MYCOTHIOL
Mycothiol (MSH) or 1D-myo-inosityl 2-(N-acetyl-L-cysteinyl) amido-2-deoxy-α-D-glucopyranoside, is an unusual conjugate of N-acetylcysteine (AcCys) with 1D-myo-inosityl 2-acetamido-2-deoxy-α-D-glucopyranoside (GlcNAc-Ins) [see Fig. (5)], and is the major low-molecular mass thiol in most action-mycetes, including mycobacteria [154]. MSH is the functional equivalent of glutathione (GSH) in mycobacteria [47,155] and is associated with the protection of M. tuberculosis from toxic oxidants and antibiotics [137]. Interestingly, the thiol in MSH undergoes copper-ion catalyzed autoxidation 30-fold more slowly than cysteine and 7-fold more slowly than glutathione [156]. Thus, high concentrations of cellular MSH may increase the capacity of actinomycetes to mitigate the negative effects of oxidative stress.
Fig. (5).
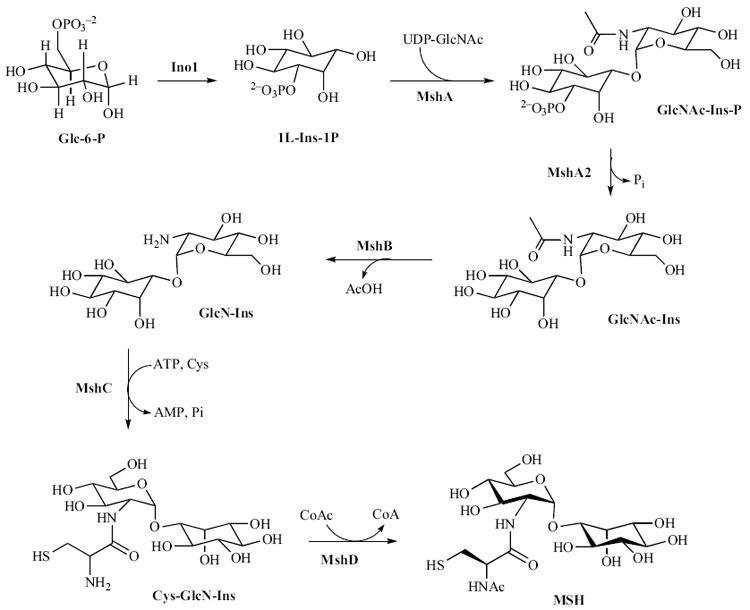
MSH biosynthetic pathway.
Apart from protection against toxic oxidants, M. tuberculosis relies upon MSH for growth in an oxygen-rich environment and for establishing the pattern of resistance to isoniazid and rifampin [137]. While previous reviews on MSH give a detailed overview of the MSH biochemistry [47] and MSH-dependent proteins [155], the purpose of this section is to highlight research avenues that would help clarify the functional role of MSH in the mycobacterial lifecycle and highlight promising drug targets in MSH metabolism.
Overview of Mycothiol Biosynthesis
Over a series of seminal papers, R. C. Fahey, G. L. Newton and Y. Av-Gay have elucidated the biosynthetic pathway of MSH [see Fig. (5)]. Production of MSH begins from the biosynthesis of 1L-myo-inositol 1-phosphate (1L-Ins-1-P), produced from glucose-6-phosphate in a reaction catalyzed by inositol-1-phosphate synthase (Ino1) [157]. From this precursor, five enzymes catalyze the conversion of 1L-Ins-1-P to MSH. In the first step, a glycosyl-transferase, MshA, catalyzes the reaction between a UDP-N-acetylglucosamine (UDP-GlcNAc) and 1L-Ins-1-P, generating UDP and 1-O-(2-acetamido-2-deoxy-α-D-glucopyranosyl)-D-myo-inositol 3-phosphate (GlcNAc-Ins-P) [157]. A phosphatase, as yet uncharacterized, but designated MshA2, dephosphorylates GlcNAc-Ins-P to produce 1-O-(2-acetamido-2-deoxy-α-D-glucopyranosyl)-D-myo-inositol (GlcNAc-Ins), the substrate for MshB [157]. In the next step, GlcNAc-Ins is deacetylated by MshB to yield 1-O-(2-amino-1-deoxy-α-D-glucopyranosyl)-D-myo-inositol (GlcN-Ins) [158]. Subsequently, MshC catalyses the ATP-dependent ligation of L-cysteine to GlcN-Ins to produce 1-O-[[(2R)-2-amino-3-mercapto-1-oxopropyl]amino]-2-deoxy-α-D-glucopyranosyl)-D-myo-inositol (Cys-GlcN-Ins) [159]. In the final step, N-acetylation of Cys-GlcN-Ins with acetyl-CoA is catalyzed by MshD to afford MSH [160]. The total chemical synthesis of MSH has also been reported [161,162].
The genes encoding the enzymes responsible for MSH biosynthesis have been identified using a variety of methods including transposon [163] and chemical mutagenesis [158,164,165]. In turn, these mutants have been utilized to determine the indispensability of the respective genes in the biosynthesis of MSH and their consequence on the viability of mycobacteria [166-168]. Significant progress in the biochemical characterization of these enzymes has also been made [158,160,163,164,167,169].
Mycothiol Biosynthetic Enzymes
The gene encoding the glycosyltransferase, MshA was first identified as a transposon mutant in M. smegmatis that did not produce measurable amounts of GlcNAc-Ins and MSH [163]. By virtue of homology, MshA belongs to the known CAZy family 4 glycosyltransferases, [163,170] which includes a number of sucrose synthases, mannosyl transferases and GlcNAc transferases. This classification strongly suggested that MshA is the glucosyl-transferase required for the biosynthesis of GlcNAc-Ins. M. smegmatis and M. tuberculosis mshA sequences were shown to be 75% identical over a 446-residue overlap. The M. tuberculosis mshA ortholog, Rv0486, complemented the mutant phenotype in M. smegmatis, thereby confirming its function. In M. smegmatis [163] and M. tuberculosis [66], mshA is essential for production of GlcNAc-Ins and therefore, for MSH synthesis. Interestingly, however, transposon mutants in mshA are viable in M. smegmatis [163], whereas in M. tuberculosis mshA is essential for growth [66]. The gene encoding the phosphatase, MshA2, remains to be identified.
MshB was the first gene identified in the MSH biosynthetic pathway [158]. The deacetylase is encoded by the M. tuberculosis open reading frame Rv1170 and was first discovered as a homolog of Rv1082, a mycothiol S-conjugate amidase (Mca). Although MshB does exhibit some amidase activity, deacetylation of GlcNAc-Ins is the preferred reaction [158]. Characterization and crystallographic studies have revealed that MshB is a Zn2+ metalloprotein and that deacetylase activity is dependent on the presence of a divalent metal cation [171,172].
Disruption of mshB results in decreased production of MSH (limited to about 5-10% of the parental M. smegmatis strain [173] and 20% that of the parental M. tuberculosis strain during log-phase growth, increasing to 100% of the wild-type MSH levels during the stationery phase [137]). Hence, MSH synthesis is not abolished in mshB mutants and, in the absence of MshB, MSH biosynthesis is accomplished via an alternative deacetylase activity that produces modest levels of GlcN-Ins [137,173]. Under culture conditions, the amount of MSH produced in mshB mutants during log phase growth is sufficient to provide MSH-dependent resistance to moderate oxidative stress. In addition, since normal quantities of MSH are produced in mshB mutants during stationary phase, it was not possible to examine the role of MSH during dormancy-like conditions in these studies.
The role of MshC involving ATP-dependent ligation of L-cysteine with GlcN-Ins was first elucidated by Bornemann and coworkers [159]. First identified in M. smegmatis [169], homologs of mshC have been identified in Streptomyces coelicolor A3, Corynebacterium striatum [154] and orthologs of M. tuberculosis MshC (Rv2130c) were also found in M. leprae [15], M. bovis [16], and in M. avium [17]. Interestingly, the enzyme encoded by mshC appears to have evolved by gene duplication of the cysteinyl-tRNA synthetase, cysS (Rv3580c) as evidenced by their similar mechanism of action [169]. In the reaction catalyzed by MshC, the 2’ amine of GlcN-Ins carries out nucleophilic attack of an activated cysteinyl-AMP intermediate to produce Cys-GlcN-Ins. Presumably, a general base removes a proton from the amino group leading to the formation of a tetrahedral intermediate, which decomposes to form the amide [169].
In M. smegmatis, chemical and transposon mutants lacking MshC activity do not produce detectible amounts of MSH [141]. In the chemical mutants, mshC was sequenced and a point mutation (Leu205Pro) was identified. This region in MshC is largely conserved among actinomycetes and hence, the Leu205Pro substitution was concluded to be responsible for the lack of MshC activity in the mutant [141]. In contrast to M. smegmatis that does not require MSH for growth, a targeted disruption of mshC in M. tuberculosis Erdman produced no viable clones possessing either the disrupted mshC gene or reduced levels of MSH. Thus, the mshC gene is required for MSH production and is essential for M. tuberculosis Erdman survival [63]. The differences in the responses of the mutants between the two strains of mycobacteria could be attributed to the fact that M. smegmatis has a larger genome (7 vs. 4.4 Mb) relative to M. tuberculosis and therefore, includes genes that facilitate its growth in the absence of MSH [174].
MshD catalyzes the final step in MSH biosynthesis. In this reaction, Cys-GlcN-Ins is acetylated using acetyl-CoA [159]. mshD was identified during the characterization of a M. smegmatis transposon mutant lacking the transacetylase activity required for MSH biosynthesis. Sequencing from the site of insertion identified the gene that encodes for mycothiol synthase or MshD. A homology search revealed a mshD ortholog in M. tuberculosis as Rv0819 and exhibits MSH synthase activity when expressed in E. coli [160]. A crystal structure of MshD from M. tuberculosis showed structural homology to the GNAT family of N-acetyl-transferases [54,175,176].
MshD mutants in M. smegmatis produce high levels of Cys-GlcN-Ins along with two other thiols, N-formyl-Cys-GlcN-Ins (fCys-GlcN-Ins) and N-succinyl-Cys-GlcN-Ins (succ-Cys-GlcN-Ins) and ~1% the amount of MSH found in the wild-type strain [174,177]. These data suggest that in the absence of mshD, mycobacteria can make use of closely related analogs of MSH such as fCys-GlcN-Ins to maintain a reducing environment in the cells [174,177]. This hypothesis is further supported by the observation that mshD transposon mutants in M. smegmatis are as resistant to peroxide-induced oxidative stress as their parental strain [177]. On the other hand, M. tuberculosis mshD mutants appear to grow poorly under other stress conditions such as low-pH media or in the absence of catalase and oleic acid [174].
MSH and Antibiotic Resistance
The formation of MSH-adducts of various anti-mycobacterial agents like cerulenin and rifamycin S [178] suggests that M. tuberculosis can use MSH in detoxification reactions [see Fig. (6)] [164]. An as yet unidentified MSH-S-transferase is believed to catalyze the formation of the MSH-drug adduct [179]. MSH-S-conjugate amidase (discussed below) then catalyzes the hydrolysis of the MSH-drug adduct to produce a mercapturic acid containing the drug moiety, which is excreted from the cell [164]. Alternatively, since the oxidation state of cell wall components could alter cell wall permeability, MSH may also confer antibiotic resistance by influencing the overall cellular redox state [137]. Experiments with mshC mutants demonstrate that MSH, and not any biosynthetic intermediate en route to MSH, is critical for antibiotic and peroxide resistence [141]. While studies with mutants lacking the various MSH biosynthetic enzymes support the idea of MSH-related resistance to antibiotics, [137,160,163,165], the exact mechanism involved in detoxification of different classes of toxins remain to be elucidated.
Fig. (6).
MSH-mediated detoxification pathways.
Mycothiol-dependent Detoxification
M. smegmatis mutants that lack genes encoding for enzymes involved in MSH biosynthesis exhibit increased sensitivity to oxidative stress and antibiotics such as streptomycin, ethionamide, rifampin, and alkylating toxins. These findings suggest that either MSH or MSH-dependent enzymes are involved in protecting mycobacteria from oxidants and toxins [141,165] and have lead to the study of enzymes that utilize MSH as a cofactor or a substrate for their activity. NAD/MSH-dependent formaldehyde dehydro-genase (MscR) was the first enzyme identified as using MSH as a cofactor [180] and is discussed at length in a recent review [155]; two other important enzymes involved in MSH metabolism and detoxification are mycothione reductase (Mtr) and Mycothiol-S-conjugate Amidase (Mca) [see Fig. (6)], discussed below.
Mycothione Reductase
To maintain a large cellular pool of reduced MSH, mycothione reductase catalyzes the reduction of oxidized MSH also known as mycothione (MSSM) [see Fig. (6)] [155]. M. tuberculosis MSH disulfide reductase (Mtr, encoded by Rv2855) was identified by homology to glutathione reductases [181,182]. Mtr is a member of the pyridine nucleotide-disulfide reductase superfamily. The reductase is a homodimeric flavoprotein disulfide isomerase and requires FAD as a cofactor [155,181]. NADPH reduces FAD, which then transfers reducing equivalents to the redox-active disulfide in Mtr to generate a stable two-electron reduced enzyme [181,182]. Subsequently, Mtr reduces the disulfide in MSSM via dithiol-disulfide interchange, with concomitant oxidation of NADPH [181,182].
Phenotypic characterization of an actinomycete mtr mutant has not been reported to date and genome-wide transposon mutagenesis has yielded conflicting results. In one study, a transposon mutant in M. tuberculosis mtr was reported to be viable [183]. In contrast, another study using high-density Himar-1 transposon mutagenesis reported that mtr is essential for M. tuberculosis survival [42]. One possible explanation for these conflicting data could be the relative importance of (or requirement for) Mtr in MSH reduction during different stages of growth. Transcriptional analysis of M. bovis BCG reveals that mtr mRNA is actively transcribed during exponential bacterial growth [166]. In the same study, mtr mRNA expression was absent in the stationery phase suggesting that Mtr might only be required to maintain the redox balance during intense periods of metabolic activity (e.g., during the growth phase) [166]. However, another study found high MSH levels throughout the growth cycle, including the stationery phase [173]. These findings suggest that, in the absence of Mtr, another thiol reductase might reduce MSSM [155]. Additional experiments will be required to clarify the importance of Mtr in MSH reduction throughout the mycobacterium lifecycle and to determine whether or not it is essential for bacterial viability.
Mycothiol-S-conjugate Amidase
In mycobacteria, mycothiol-S-conjugate Amidase (Mca) plays a major role in electrophile detoxification [see Fig. (6)] [164]. This enzyme was discovered in connection with its ability to detoxify a thiol-specific fluorescent alkylating agent, monobromobimane (mBBr), a compound commonly used for the quantitative determination of thiols. mBBr binds to MSH forming a MSH-mBBr adduct, MSmB and can be cleaved by Mca to produce glucosaminyl inositol and acetyl cysteinyl bimane, a mercapturic acid which is rapidly excreted from the cell [164]. Mca was first purified from M. smegmatis and has an ortholog in the M. tuberculosis genome, Rv1082, identified by N-terminal amino acid sequencing [164]. Studies probing the substrate specificity of Mca indicate that the enzyme specifically recognized the MSH moiety in the conjugate, but is relatively non-specific for the group attached to the sulfur in the MSH-toxin conjugate [164].
Mca and mshB exhibit an overall sequence identity of 32% [171]. Interestingly, in vitro studies indicate that MshB possesses amidase activity with MSH substrate [164]. Moreover, Mca can function as a deacetylase [157,164] and partially restored MSH production when introduced into an M. Smegmatis mshB mutant [173]. Based on the sequence identity between Mca and MshB and the crystal structure of MshB, a model for the active site of Mca has been proposed [171,172]. With the exception Lys19 in Mca replaced by Ser20 in MshB, other critical catalytic residues, including the zinc-binding site and an aspartate are perfectly conserved. The Lys to Ser alteration may play an important role in disaccharide binding [171]; a crystal structure of Mca will be important to define the MSH binding site.
Apart from the mBBr model substrate, the substrates for Mca include the MSH conjugate of cerulenin, an antibiotic that inhibits fatty acid synthetase and other antibiotic adducts. Mca homologs have been found in several antibiotic biosynthesis operons such as those for avermectin (Streptomyces avermitilis) and eythromycin (Saccharopolyspora erythrae) [47,184]. In addition, it has been demonstrated that MSH forms a conjugate with Rifamycin SV and this complex is a substrate for M. tuberculosis Mca [178]. Treatment of mca mutant and wild-type M. smegmatis strains with Rafamycin SV showed that the MSH-Rifamycin SV adduct is converted to mercapturic acid only in the wild-type [168]. Taken together, these findings demonstrate that MSH and Mca in mycobacteria work together to detoxify antibiotics [47].
Drug Targets in Mycothiol Metabolism
Mca plays a critical role in mycobacterial detoxification of antibiotics. Therefore, inhibitors of Mca could enhance the sensitivity of MSH-producing bacteria to antibiotics, establishing Mca as a promising new drug target. Toward this end, 1,500 natural product extracts and synthetic libraries were screened to identify lead compounds [185-187]. Two classes of bromotyrosine-derived natural products were competitive inhibitors of Mca [see Table 2, (1, 2)]; non-competitive inhibitors were also identified in this screen [see Table 2, (3, 4)]. These results motivated the total synthesis of a competitive inhibitor [see Table 2, (1)] that inhibits Mca with an IC50 value of 30 μM [188].
Table 2.
Mca Inhibitors
| No. | Ref | Structure | Activity (IC50) |
|---|---|---|---|
| 1 | [187] |
|
3 μM (M. tb); 2 μM (M. smeg) |
| 2 | [187] |
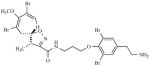
|
30 μM (M. tb & M. smeg) |
| 3 | [205] |

|
10 μM (M. tb); 0.5 μM (M. smeg) |
| 4 | [185] |
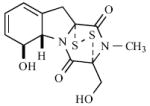
|
50 μM (M. tb & M. smeg) |
| 5 | [189] |
|
MIC = 1.5 – 15.5 μg ml-1 (M. smeg) |
| 6 | [190] |

|
7500 nmol min-1 mg-protein-1 (M. tb) |
| 7 | [191] |
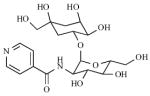
|
46 μM (M. tb) |
| 8 | [191] |

|
44 μM (M. tb) |
Recently, a series of compounds based on the structure of the natural product bromotyrosine inhibitor were synthesized and screened against mycobacteria and other gram-positive bacteria [189]. One of the lead compounds identified from this study termed, EXEG1706 [see Table 2, (5)], exhibited low minimum inhibitory concentrations (2.5 – 25 μg ml-1) for M. smegmatis, M. bovis and against methicillin-sensitive and -resistant Staphylococcus aureus, and S. aureus. However, this class of compounds was also active against mycobacterial Mca mutant strains and against gram-positive bacteria that do not produce MSH. Thus, in addition to Mca, it appears that these compounds inhibit other protein targets in vivo. Another approach used to identify Mca inhibitors has been the synthesis of MSH analogs. Synthesis of a simplified thioglycosidic analog of MSH [see Table 2, (6)] [190] and a variety of amide-functionalized MSH analogs synthesized from quinic acid led to the identification of inhibitors with modest inhibitory activities (IC50 values around 50 μM) [191] [see Table 2, (7, 8)].
In addition to Mca, other possible drug targets that could block MSH biosynthesis are the enzymes encoded by mshA and mshC (both essential genes in M. tuberculosis [63,66]). The identification of inhibitors for MshC has been initiated [157] and identification of inhibitors for another UDP-GlcNAc-dependent glycosyltransferase, MurG [192] suggests that MshA is also likely to be a drug-able target. Also, although high-density transposon mutagenesis studies have identified mshD as nonessential for the growth of M. tuberculosis in minimal culture medium [42], the survival of M. tuberculosis MshD mutants is severely compromised in activated and non-activated macrophages [40]. Thus, MshD could be a promising drug target and further analysis of this mutant in animal models of TB infection may be warranted.
OTHER SOURCES OF REDUCED SULFUR
Consistent with the requirement for sulfur in mycobacterial survival, the ability of mycobacteria to scavenge reduced sulfur from its host has been confirmed in M. bovis BCG and in M. tuberculosis [61,68,145]. Several potential sources of reduced sulfur in the human host are discussed below.
Cysteine and Cystine
Mutation of cysH in M. smegmatis and M. tuberculosis produces a cysteine auxotroph and this defect can be rescued by the addition of cysteine to the growth medium [53,61]. The finding that growth of ΔcysH mutant can be restored by the addition of cysteine suggests that cysteine or cystine (the oxidized form of cysteine) can be transported into M. smegmatis and M. tuberculosis. In addition, when [35S] cysteine is added to a growing culture of M tuberculosis, more than 70% of the radioactive sulfur taken up by the bacteria is found in methionine, also consistent with import of cysteine [145]. While genes that encode the cysteine/cystine transporter in mycobacteria have not yet been identified, cysteine and cystine uptake systems have been characterized in other prokaryotes [193,194]. In humans, cystine is the preferred form of cysteine for the synthesis of glutathione in macrophages and is present in plasma at ~25 – 35 μM [195].
Genetic screens for amino acid auxotrophs in M. bovis BCG (an attenuated version of bovine bacillus) have not isolated cysteine auxotrophs [68,75]. In the first report, only three auxotrophs were identified, one for methionine and two for leucine [75]. A subsequent study isolated two auxotrophs, both for methionine [68]. Since isolation of mycobacterial auxotrophs depends on the growth medium composition [196], it is possible that the use of casamino acids to rescue the growth of transposon mutants in these studies selected only a small subset of amino-acid auxotrophs. Consistent with this hypothesis, the approximate concentration of sulfur-containing amino acids in 1% (w/v) casamino acids is expected to be ~900 μM methionine and ~60 μM cystine.
The methionine auxotrophs identified by transposon mutagenesis in M. bovis BCG mapped to genes in the sulfate assimilation pathway, in particular to sulfate transport genes, subI [75] and cysA [68]. Since sulfate serves as the precursor for cysteine synthesis, defects in the sulfate assimilation pathway should result in cysteine auxotrophy. Surprisingly however, growth of M. bovis BCG subI or cysA mutants could not be rescued by supplementation with cysteine (in contrast to the cysH knockout in M. smegmatis and M. tuberculosis) and instead, required methionine supplementation. In addition to the inability of cysteine to rescue defects in sulfate transport, the same study also reported that wild-type M. bovis BCG grew slowly on growth media supplemented with 0.3 mM cysteine and not at all in the presence of 0.5 mM cysteine. In contrast, toxicity has not been observed in wild-type strains of M. smegmatis [53] or M. tuberculosis [61] grown in the presence 1 – 2 mM cysteine. Hence, it is possible that mutation of sulfate transport genes, subI or cysA, impact cysteine/cystine import directly or that M. bovis BCG does not transport cysteine/cystine efficiently. Further investigation into the differences between mycobacterial strains, growth media and other critical factors, such as inoculum densities, for the requirements for sulfur-containing compounds are warranted.
Methionine and Reverse Transsulfuration
The interesting observation that defects in sulfate transport or its reduction could be rescued by methionine supplementation suggested that a functional reverse transsulfuration pathway [see Fig. (7)], used to produce cysteine from methionine, was present in mycobacteria [53,68,145]. Indeed, the existence of this pathway has recently been confirmed [145]. Although a methionine transporter has not yet been identified in mycobacteria, an apparent Km of 80 μM for the transporter has been estimated in M. bovis BCG [68] and the estimated concentration of methionine in humans is ~25 μM [197].
Fig. (7).
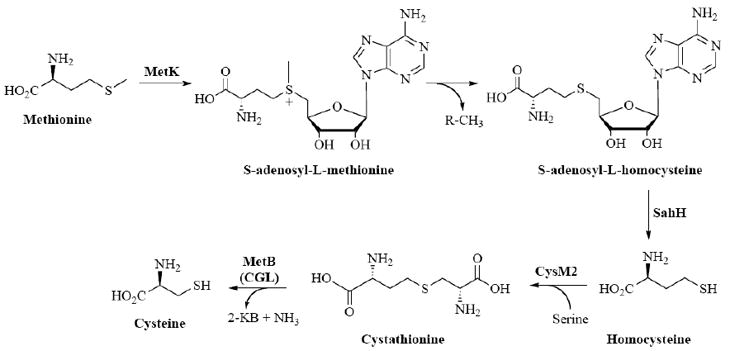
Reverse transsulfurylation pathway in mycobacteria.
Once methionine is transported into the bacteria, three enzymatic steps are required for conversion into homocysteine [see Fig. (7)] [67]. Subsequently, CysM2 (Rv1077) converts homocysteine to cystathionine. Interestingly, in M. tuberculosis, Rv1079 (annotated as metB) encodes a bifunctional cysteine γ-lyase (CGL)-cystathionine γ-synthase (CGS) enzyme [145]. Hence, conversion of cystathionine to cysteine (the final step in reverse transsulfuration) can be catalyzed by the CGL activity encoded by Rv1079 [145]. Alternatively, the CGL activity of Rv1079 can also support transsulfuration (conversion of cysteine to methionine) in M. tuberculosis by converting cysteine to cystathione. Cystathione is then transformed into methionine by two subsequent reactions: MetC (Rv3340) converts cystathionine into homocysteine and MetE/MetH (Rv1133c/Rv2124c) catalyzes methylation of homocysteine to produce methionine.
Interestingly, mutation of metB in M. tuberculosis results in a prototrophic methionine mutant [198]. In other words, MetB is not absolutely required for methionine production. This finding can be explained by the action of MetZ (Rv0391), an O-succinylhomoserine sulfurylase which bypasses the requirement for MetB and MetC by condensing S2- with O-succinylhomoserine to produce homocysteine directly. In mice, a ΔmetB strain was somewhat attenuated [198]. However, no differences in bacterial load in the lungs, liver or spleen, were observed between the metB mutant and wild-type M. tuberculosis in immunocompetent mice up to 80 days post-infection [198]. This growth phenotype contrasts that of cysH mutants in M. tuberculosis where viability decreases significantly, specifically during the persistence phase of infection [61].
Glutathione
In mycobacteria, a large amount of the reduced sulfur in cells is used to make mycothiol, the dominant low molecular weight thiol used to maintain redox equilibrium and scavenge reactive oxygen species in the cell [154]. Similarly, GSH – a tripeptide, γ-glutamyl-cysteinylglycine found in many prokaryotes and eukaryotes – is also present at high intracellular levels [155] and may provide a source of reduced sulfur for mycobacteria in the host. Estimates of GSH concentration in human cells and macrophages range from 1 – 7 mM [199,200]. An analog of GSH, nitrosoglutathione (GSNO), is bactericidal in M. bovis [201] and M. tuberculosis [202]. Use of GSNO has facilitated identification and characterization of the ABC transporter dipeptide permease (Dpp, Rv3663 – Rv3666) responsible for GSH catabolism and utilization [201,202]. Interestingly, GSH is not transported into mycobacterial cells as the tripeptide, but rather as the dipeptide, Cys-Gly [202]. Hence, import of GSH involves proteolysis by a γ-glutamyl transpeptidase (GgtA, Rv0773c) and subsequent transport via Dpp. Consistent with the proposed route of GSH catabolism and import, mutants in the transpeptidase or the permease are resistant to the toxic effects of GSNO [202]. In culture, it has been reported that GSH exhibits bacteriostatic activity at a concentration of 5 mM [203]. This effect appears to be mediated intracellularly since mutations in the dpp or ggtA relieve this phenomenon [203]. It has been noted that M. smegmatis [204] does not share a bacteriostatic effect of reduced glutathione at 5 mM. Further experiments are needed to elucidate the intracellular processing of the dipeptide and mechanism of its apparent toxicity.
OUTLOOK
The emergence of antibiotic resistance and the problem of mycobacterial persistence in M. tuberculosis urgently stress the need for new target identification. Toward this end, mycobacterial sulfur metabolic pathways represent a promising new area for anti-TB therapy. In the last several years excellent progress has been made, leading to the identification and validation of several potential drug targets in sulfate assimilation and MSH metabolism. At the same time, many aspects of mycobacterial sulfur metabolism remain poorly understood and represent exciting areas of new or continued investigation. Significant work remains to validate additional targets, improve inhibitor potency for existing targets and to further define the roles that sulfated and many reduced sulfur-containing metabolites play in mycobacterial virulence and persistence. Finally, a wide variety of microbes including Pseudomonas aeruginosa, Bacillus anthracis, and Yersinia pestis also rely on unique sulfur metabolic pathways for their own survival. Hence, in the fight against multidrug resistant microbes, investigation of microbial sulfur metabolism in mycobacteria and other pathogens should be fertile scientific ground in the years to come.
Acknowledgments
We wish to acknowledge the many colleagues who have contributed to the ideas put forth in this review. Financial support from the Chemical Biology Ph.D. program and the Life Sciences Institute at the University of Michigan is gratefully acknowledged.
ABBREVIATIONS
- TB
Tuberculosis
- MDR
Multidrug resistant
- CoA
Coenzyme A
- −SO3−
Sulfuryl moiety
- SO42-
Sulfate
- APS
Adenosine-5’-phosphosulfate
- PAPS
3’-Phosphoadenosine-5’-phosphosulfate
- SO32-
Sulfite
- S2-
Sulfide
- BCG
Bacillus Calmette-Guérin
- ATP
Adenosine 5’-triphosphate
- GTP
Guanosine 5’-triphosphate
- SAC
Sulfate-activating complex
- ST
Sulfotransferase
- SL-1
Sulfolipid-1
- T2S
Trehalose-2-sulfate
- PAP
3’-Phosphoadenosine-5’-phosphate
- EST
Estrogen ST
- TPST
Tyrosyl protein ST
- GST
Golgi-resident ST
- β-AST
β–arylsulfotransferases
- cAMP
Cyclic adenosine-5’-phosphate
- IC50
Half maximal inhibitory concentration
- O2·−
Superoxide
- NO·
Nitric oxide
- NADPH oxidase
Nicotinamide adenine dinucleotide phosphate-oxidase
- NOS2
Inducible nitric oxide synthase
- ROI
Reactive oxygen intermediate
- RNI
Reactive nitrogen intermediate
- SOD
Superoxide dismutase
- H2O2
Hydrogen peroxide
- OH·
Hydroxyl radical
- NOHLA
N-ω-hydroxy-L-arginine
- ONOO−
Peroxynitrite
- TLR
Toll-like receptor
- Phox
Phagocyte NADPH oxidase
- AMP
Adenosine-5’-phosphate
- Trx
Thioredoxin
- Cys-Sγ−SO3−
S-sulfocysteine
- [4Fe-4S]
Four iron-four sulfur cluster
- MSH
Mycothiol
- AcCys
N-acetylcysteine
- GlcNAc-Ins
1D-myo-inosityl 2-acetamido-2-deoxy-α-D-glucopyranoside
- GSH
Glutathione
- 1L-Ins-1-P
1L-myo-inositol 1-phosphate
- UDP-GlcNAc
UDP-N-acetylglucosamine
- GlcNAc-Ins-P
1-O-(2-acetamido-2-deoxy-α-D-glucopyranosyl)-D-myo-inositol 3-phosphate
- GlcNAc-Ins
1-O-(2-acetamido-2-deoxy-α-D-glucopyranosyl)-D-myo-inositol
- GlcN-Ins
1D -myo-inosityl-2-amino-2-deoxy-α-D-glucopyranoside
- Cys-GlcN-Ins
1-O-[[(2R)-2-amino-3-mercapto-1-oxopropyl]amino]-2-deoxy-α-D-glucopyranosyl)-D-myo-inositol
- fCys-GlcN-Ins
N-formyl-Cys-GlcN-Ins
- succ-Cys-GlcN-Ins
N-succinyl-Cys-GlcN-Ins
- MSSM
Mycothione
- mBBr
Monobromobimane
- CGL
Cysteine γ-lyase
- CGS
Cystathionine γ-synthase
- GSNO
Nitrosoglutathione
References
- 1.Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, Raviglione MC, Dye C. Arch Intern Med. 2003;163:1009. doi: 10.1001/archinte.163.9.1009. [DOI] [PubMed] [Google Scholar]
- 2.Dye C, Scheele S, Dolin P, Pathania V, Raviglione MC. JAMA. 1999;282:677. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 3.Kochi A. Bull World Health Organ. 2001;79:71. [PMC free article] [PubMed] [Google Scholar]
- 4.Blumberg HM, Burman WJ, Chaisson RE, Daley CL, Etkind SC, Friedman LN, Fujiwara P, Grzemska M, Hopewell PC, Iseman MD, Jasmer RM, Koppaka V, Menzies RI, O’Brien RJ, Reves RR, Reichman LB, Simone PM, Starke JR, Vernon AA. Am J Respir Crit Care Med. 2003;167:603. doi: 10.1164/rccm.167.4.603. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y. Annu Rev Pharmacol Toxicol. 2005;45:529. doi: 10.1146/annurev.pharmtox.45.120403.100120. [DOI] [PubMed] [Google Scholar]
- 6.Flynn JL, Chan J. Infect Immun. 2001;69:4195. doi: 10.1128/IAI.69.7.4195-4201.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y. Front Biosci. 2004;9:1111. [Google Scholar]
- 8.Mitchison DA. Chest. 1979;76:771. doi: 10.1378/chest.76.6_supplement.771. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y. Front Biosci. 2004;9:975. doi: 10.2741/1289. [DOI] [PubMed] [Google Scholar]
- 10.Espinal MA. Tuberculosis (Edinb) 2003;83:44. doi: 10.1016/s1472-9792(02)00058-6. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization. 2003 http://www.who.int/gtb/
- 12.Duncan K. Tuberculosis (Edinb) 2003;83:201. doi: 10.1016/s1472-9792(02)00076-8. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, P-M K, Denkins S. Drug Disc Today. 2006;11:21. [Google Scholar]
- 14.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Nature. 1998;393:537. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 15.Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honore N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies RM, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutherford KM, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward JR, Barrell BG. Nature. 2001;409:1007. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 16.Garnier T, Eiglmeier K, Camus JC, Medina N, Mansoor H, Pryor M, Duthoy S, Grondin S, Lacroix C, Monsempe C, Simon S, Harris B, Atkin R, Doggett J, Mayes R, Keating L, Wheeler PR, Parkhill J, Barrell BG, Cole ST, Gordon SV, Hewinson RG. Proc Natl Acad Sci USA. 2003;100:7877. doi: 10.1073/pnas.1130426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Bannantine JP, Zhang Q, Amonsin A, May BJ, Alt D, Banerji N, Kanjilal S, Kapur V. Proc Natl Acad Sci USA. 2005;102:12344. doi: 10.1073/pnas.0505662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glickman MS, Cahill SM, Jacobs WR., Jr J Biol Chem. 2001;276:2228. doi: 10.1074/jbc.C000652200. [DOI] [PubMed] [Google Scholar]
- 19.Parish T, Stoker NG. Microbiology. 2000;146(Pt 8):1969. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- 20.van Kessel JC, Hatfull GF. Nat Methods. 2007;4:147. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 21.Flynn JL, Chan J. Annu Rev Immunol. 2001;19:93. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 22.Houben EN, Nguyen L, Pieters J. Curr Opin Microbiol. 2006;9:76. doi: 10.1016/j.mib.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 23.Vergne I, Chua J, Lee HH, Lucas M, Belisle J, Deretic V. Proc Natl Acad Sci USA. 2005;102:4033. doi: 10.1073/pnas.0409716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giacomini E, Iona E, Ferroni L, Miettinen M, Fattorini L, Orefici G, Julkunen I, Coccia EM. J Immunol. 2001;166:7033. doi: 10.4049/jimmunol.166.12.7033. [DOI] [PubMed] [Google Scholar]
- 25.Henderson RA, Watkins SC, Flynn JL. J Immunol. 1997;159:635. [PubMed] [Google Scholar]
- 26.Salgame P. Curr Opin Immunol. 2005;17:374. doi: 10.1016/j.coi.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Uehira K, Amakawa R, Ito T, Tajima K, Naitoh S, Ozaki Y, Shimizu T, Yamaguchi K, Uemura Y, Kitajima H, Yonezu S, Fukuhara S. Clin Immunol. 2002;105:296. doi: 10.1006/clim.2002.5287. [DOI] [PubMed] [Google Scholar]
- 28.Cosma CL, Humbert O, Ramakrishnan L. Nat Immunol. 2004;5:828. doi: 10.1038/ni1091. [DOI] [PubMed] [Google Scholar]
- 29.Russell DG. Nat Rev Microbiol. 2007;5:39. doi: 10.1038/nrmicro1538. [DOI] [PubMed] [Google Scholar]
- 30.Saunders BM, Cooper AM. Immunol Cell Biol. 2000;78:334. doi: 10.1046/j.1440-1711.2000.00933.x. [DOI] [PubMed] [Google Scholar]
- 31.Ulrichs T, Kaufmann SH. J Pathol. 2006;208:261. doi: 10.1002/path.1906. [DOI] [PubMed] [Google Scholar]
- 32.Lazarevic V, Nolt D, Flynn JL. J Immunol. 2005;175:1107. doi: 10.4049/jimmunol.175.2.1107. [DOI] [PubMed] [Google Scholar]
- 33.Boshoff HI, Barry CE., 3rd Nat Rev Microbiol. 2005;3:70. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 34.Tufariello JM, Chan J, Flynn JL. Lancet Infect Dis. 2003;3:578. doi: 10.1016/s1473-3099(03)00741-2. [DOI] [PubMed] [Google Scholar]
- 35.Nathan C, Shiloh MU. Proc Natl Acad Sci USA. 2000;97:8841. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hampshire T, Soneji S, Bacon J, James BW, Hinds J, Laing K, Stabler RA, Marsh PD, Butcher PD. Tuberculosis (Edinb) 2004;84:228. doi: 10.1016/j.tube.2003.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manganelli R, Voskuil MI, Schoolnik GK, Dubnau E, Gomez M, Smith I. Mol Microbiol. 2002;45:365. doi: 10.1046/j.1365-2958.2002.03005.x. [DOI] [PubMed] [Google Scholar]
- 38.Ohno H, Zhu G, Mohan VP, Chu D, Kohno S, Jacobs WR, Jr, Chan J. Cell Microbiol. 2003;5:637. doi: 10.1046/j.1462-5822.2003.00307.x. [DOI] [PubMed] [Google Scholar]
- 39.Pinto R, Tang QX, Britton WJ, Leyh TS, Triccas JA. Microbiology. 2004;150:1681. doi: 10.1099/mic.0.26894-0. [DOI] [PubMed] [Google Scholar]
- 40.Rengarajan J, Bloom BR, Rubin EJ. Proc Natl Acad Sci USA. 2005;102:8327. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sassetti CM, Boyd DH, Rubin EJ. Proc Natl Acad Sci USA. 2001;98:12712. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sassetti CM, Boyd DH, Rubin EJ. Mol Microbiol. 2003;48:77. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 43.Sassetti CM, Rubin EJ. Proc Natl Acad Sci USA. 2003;100:12989. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. J Exp Med. 2003;198:693. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Proc Natl Acad Sci USA. 2001;98:7534. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kredich NM. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. 2. Niedhardt FC, editor. Vol. 1. ASM Press; Washington, D. C.: 1996. pp. 514–527. [Google Scholar]
- 47.Newton GL, Fahey RC. Arch Microbiol. 2002;178:388. doi: 10.1007/s00203-002-0469-4. [DOI] [PubMed] [Google Scholar]
- 48.Takayama K, Wang C, Besra GS. Clin Microbiol Rev. 2005;18:81. doi: 10.1128/CMR.18.1.81-101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leyh TS. Crit Rev Biochem Mol Biol. 1993:28–515. doi: 10.3109/10409239309085137. [DOI] [PubMed] [Google Scholar]
- 50.Mougous JD, Green RE, Williams SJ, Brenner SE, Bertozzi CR. Chem Biol. 2002;9:767. doi: 10.1016/s1074-5521(02)00175-8. [DOI] [PubMed] [Google Scholar]
- 51.Nathan C. Nature. 2004;431:899. doi: 10.1038/431899a. [DOI] [PubMed] [Google Scholar]
- 52.Schelle MW, Bertozzi CR. Chembiochem. 2006;7:1516. doi: 10.1002/cbic.200600224. [DOI] [PubMed] [Google Scholar]
- 53.Williams SJ, Senaratne RH, Mougous JD, Riley LW, Bertozzi CR. J Biol Chem. 2002;277:32606. doi: 10.1074/jbc.M204613200. [DOI] [PubMed] [Google Scholar]
- 54.Aoki Y, Kondoh M, Nakamura M, Fujii T, Yamazaki T, Shimada H, Arisawa M. J Antibiot (Tokyo) 1994;47:909. doi: 10.7164/antibiotics.47.909. [DOI] [PubMed] [Google Scholar]
- 55.Ejim LJ, Blanchard JE, Koteva KP, Sumerfield R, Elowe NH, Chechetto JD, Brown ED, Junop MS, Wright GD. J Med Chem. 2007;50:755. doi: 10.1021/jm061132r. [DOI] [PubMed] [Google Scholar]
- 56.Jacques SL, Mirza IA, Ejim L, Koteva K, Hughes DW, Green K, Kinach R, Honek JF, Lai HK, Berghuis AM, Wright GD. Chem Biol. 2003;10:989. doi: 10.1016/j.chembiol.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 57.Kugler M, Loeffler W, Rapp C, Kern A, Jung G. Arch Microbiol. 1990;153:276. doi: 10.1007/BF00249082. [DOI] [PubMed] [Google Scholar]
- 58.Bogdan JA, Nazario-Larrieu J, Sarwar J, Alexander P, Blake MS. Infect Immun. 2001;69:6823. doi: 10.1128/IAI.69.11.6823-6830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ejim LJ, D’Costa VM, Elowe NH, Loredo-Osti JC, Malo D, Wright GD. Infect Immun. 2004;72:3310. doi: 10.1128/IAI.72.6.3310-3314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lestrate P, Delrue RM, Danese I, Didembourg C, Taminiau B, Mertens P, De Bolle X, Tibor A, Tang CM, Letesson JJ. Mol Microbiol. 2000;38:543. doi: 10.1046/j.1365-2958.2000.02150.x. [DOI] [PubMed] [Google Scholar]
- 61.Senaratne RH, De Silva AD, Williams SJ, Mougous JD, Reader JR, Zhang T, Chan S, Sidders B, Lee DH, Chan J, Bertozzi CR, Riley LW. Mol Microbiol. 2006;59:1744. doi: 10.1111/j.1365-2958.2006.05075.x. [DOI] [PubMed] [Google Scholar]
- 62.Yang Z, Pascon RC, Alspaugh A, Cox GM, McCusker JH. Microbiology. 2002;148:2617. doi: 10.1099/00221287-148-8-2617. [DOI] [PubMed] [Google Scholar]
- 63.Sareen D, Newton GL, Fahey RC, Buchmeier NA. J Bacteriol. 2003;185:6736. doi: 10.1128/JB.185.22.6736-6740.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huet G, Daffe M, Saves I. J Bacteriol. 2005;187:6137. doi: 10.1128/JB.187.17.6137-6146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huet G, Castaing JP, Fournier D, Daffe M, Saves I. J Bacteriol. 2006;188:3412. doi: 10.1128/JB.188.9.3412-3414.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buchmeier N, Fahey RC. FEMS Microbiol Lett. 2006;264:74. doi: 10.1111/j.1574-6968.2006.00441.x. [DOI] [PubMed] [Google Scholar]
- 67.Wheeler PRB, J S. In: Tuberculosis and the Tubercle Bacillus. Cole ST, editor. ASM Press; Washington, D. C.: 2005. pp. 309–339. [Google Scholar]
- 68.Wooff E, Michell SL, Gordon SV, Chambers MA, Bardarov S, Jacobs WR, Jr, Hewinson RG, Wheeler PR. Mol Microbiol. 2002;43:653. doi: 10.1046/j.1365-2958.2002.02771.x. [DOI] [PubMed] [Google Scholar]
- 69.Sun M, Andreassi JL, 2nd, Liu S, Pinto R, Triccas JA, Leyh TS. J Biol Chem. 2005;280:7861. doi: 10.1074/jbc.M409613200. [DOI] [PubMed] [Google Scholar]
- 70.Mougous JD, Leavell MD, Senaratne RH, Leigh CD, Williams SJ, Riley LW, Leary JA, Bertozzi CR. Proc Natl Acad Sci USA. 2002;99:17037. doi: 10.1073/pnas.252514899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carroll KS, Gao H, Chen H, Leary JA, Bertozzi CR. Biochemistry. 2005;44:14647. doi: 10.1021/bi051344a. [DOI] [PubMed] [Google Scholar]
- 72.Carroll KS, Gao H, Chen H, Stout CD, Leary JA, Bertozzi CR. PLoS Biol. 2005;3:e250. doi: 10.1371/journal.pbio.0030250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schnell R, Sandalova T, Hellman U, Lindqvist Y, Schneider G. J Biol Chem. 2005;280:27319. doi: 10.1074/jbc.M502560200. [DOI] [PubMed] [Google Scholar]
- 74.Markovich D. Physiol Rev. 2001;81:1499. doi: 10.1152/physrev.2001.81.4.1499. [DOI] [PubMed] [Google Scholar]
- 75.McAdam RA, Weisbrod TR, Martin J, Scuderi JD, Brown AM, Cirillo JD, Bloom BR, Jacobs WR., Jr Infect Immun. 1995;63:1004. doi: 10.1128/iai.63.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu C, Suo Y, Leyh TS. Biochemistry. 1994;33:7309. doi: 10.1021/bi00189a036. [DOI] [PubMed] [Google Scholar]
- 77.Mougous JD, Lee DH, Hubbard SC, Schelle MW, Vocadlo DJ, Berger JM, Bertozzi CR. Mol Cell. 2006;21:109. doi: 10.1016/j.molcel.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 78.Sun M, Leyh TS. Biochemistry. 2006;45:11304. doi: 10.1021/bi060421e. [DOI] [PubMed] [Google Scholar]
- 79.Triccas JA, Berthet FX, Pelicic V, Gicquel B. Microbiology. 1999;145(Pt 10):2923. doi: 10.1099/00221287-145-10-2923. [DOI] [PubMed] [Google Scholar]
- 80.Armstrong JAB, C R. Curr Opin Drug Disc. 2000;3:502. [PubMed] [Google Scholar]
- 81.Fukuda M, Hiraoka N, Akama TO, Fukuda MN. J Biol Chem. 2001;276:47747. doi: 10.1074/jbc.R100049200. [DOI] [PubMed] [Google Scholar]
- 82.Schwedock JS, Long SR. Genetics. 1992;132:899. doi: 10.1093/genetics/132.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roche P, Debelle F, Maillet F, Lerouge P, Faucher C, Truchet G, Denarie J, Prome JC. Cell. 1991;67:1131. doi: 10.1016/0092-8674(91)90290-f. [DOI] [PubMed] [Google Scholar]
- 84.Goren MB. Biochim Biophys Acta. 1970;210:127. doi: 10.1016/0005-2760(70)90068-8. [DOI] [PubMed] [Google Scholar]
- 85.Goren MB. Biochim Biophys Acta. 1970;210:116. doi: 10.1016/0005-2760(70)90067-6. [DOI] [PubMed] [Google Scholar]
- 86.Goren MB, Brokl O, Das BC, Lederer E. Biochemistry. 1971;10:72. doi: 10.1021/bi00777a012. [DOI] [PubMed] [Google Scholar]
- 87.Gangadharam PR, Cohn ML, Middlebrook G. Tubercle. 1963;44:452. doi: 10.1016/s0041-3879(63)80087-2. [DOI] [PubMed] [Google Scholar]
- 88.Goren MB, Brokl O, Schaefer WB. Infect Immun. 1974;9:142. doi: 10.1128/iai.9.1.142-149.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Daffe M, Draper P. Adv Microb Physiol. 1998;39:131. doi: 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- 90.Bhatt K, Gurcha SS, Bhatt A, Besra GS, Jacobs WR., Jr Microbiology. 2007;153:513. doi: 10.1099/mic.0.2006/003103-0. [DOI] [PubMed] [Google Scholar]
- 91.Converse SE, Mougous JD, Leavell MD, Leary JA, Bertozzi CR, Cox JS. Proc Natl Acad Sci USA. 2003;100:6121. doi: 10.1073/pnas.1030024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mougous JD, Petzold CJ, Senaratne RH, Lee DH, Akey DL, Lin FL, Munchel SE, Pratt MR, Riley LW, Leary JA, Berger JM, Bertozzi CR. Nat Struct Mol Biol. 2004;11:721. doi: 10.1038/nsmb802. [DOI] [PubMed] [Google Scholar]
- 93.Khoo KH, Jarboe E, Barker A, Torrelles J, Kuo CW, Chatterjee D. J Biol Chem. 1999;274:9778. doi: 10.1074/jbc.274.14.9778. [DOI] [PubMed] [Google Scholar]
- 94.Lopez JA, Ludwig EH, McCarthy BJ. J Biol Chem. 1992;267:10055. [PubMed] [Google Scholar]
- 95.McCarthy C. Infect Immun. 1976;14:1241. doi: 10.1128/iai.14.5.1241-1252.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Desmarais D, Jablonski PE, Fedarko NS, Roberts MF. J Bacteriol. 1997;179:3146. doi: 10.1128/jb.179.10.3146-3153.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pi N, Hoang MB, Gao H, Mougous JD, Bertozzi CR, Leary JA. Anal Biochem. 2005;341:94. doi: 10.1016/j.ab.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 98.Mougous JD, Senaratne RH, Petzold CJ, Jain M, Lee DH, Schelle MW, Leavell MD, Cox JS, Leary JA, Riley LW, Bertozzi CR. Proc Natl Acad Sci USA. 2006;103:4258. doi: 10.1073/pnas.0510861103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rath VL, Verdugo D, Hemmerich S. Drug Discov Today. 2004;9:1003. doi: 10.1016/S1359-6446(04)03273-8. [DOI] [PubMed] [Google Scholar]
- 100.Nimmagadda D, Cherala G, Ghatta S. Indian J Exp Biol. 2006;44:171. [PubMed] [Google Scholar]
- 101.Kakuta Y, Pedersen LG, Carter CW, Negishi M, Pedersen LC. Nat Struct Biol. 1997;4:904. doi: 10.1038/nsb1197-904. [DOI] [PubMed] [Google Scholar]
- 102.Gamage NU, Duggleby RG, Barnett AC, Tresillian M, Latham CF, Liyou NE, McManus ME, Martin JL. J Biol Chem. 2003;278:7655. doi: 10.1074/jbc.M207246200. [DOI] [PubMed] [Google Scholar]
- 103.Bidwell LM, McManus ME, Gaedigk A, Kakuta Y, Negishi M, Pedersen L, Martin JL. J Mol Biol. 1999;293:521. doi: 10.1006/jmbi.1999.3153. [DOI] [PubMed] [Google Scholar]
- 104.Kakuta Y, Sueyoshi T, Negishi M, Pedersen LC. J Biol Chem. 1999;274:10673. doi: 10.1074/jbc.274.16.10673. [DOI] [PubMed] [Google Scholar]
- 105.Armstrong JI, Ge X, Verdugo DE, Winans KA, Leary JA, Bertozzi CR. Org Lett. 2001;3:2657. doi: 10.1021/ol0162217. [DOI] [PubMed] [Google Scholar]
- 106.Armstrong JI, Verdugo DE, Bertozzi CR. J Org Chem. 2003;68:170. doi: 10.1021/jo0260443. [DOI] [PubMed] [Google Scholar]
- 107.Radzicka A, Wolfenden R. Methods Enzymol. 1995;249:284. doi: 10.1016/0076-6879(95)49039-6. [DOI] [PubMed] [Google Scholar]
- 108.Kester MH, Bulduk S, Tibboel D, Meinl W, Glatt H, Falany CN, Coughtrie MW, Bergman A, Safe SH, Kuiper GG, Schuur AG, Brouwer A, Visser TJ. Endocrinology. 2000;141:1897. doi: 10.1210/endo.141.5.7530. [DOI] [PubMed] [Google Scholar]
- 109.Otake Y, Nolan AL, Walle UK, Walle T. J Steroid Biochem Mol Biol. 2000;73:265. doi: 10.1016/s0960-0760(00)00073-x. [DOI] [PubMed] [Google Scholar]
- 110.Gonzalez-Amaro R, Sanchez-Madrid F. Crit Rev Immunol. 1999;19:389. [PubMed] [Google Scholar]
- 111.Bistrup A, Bhakta S, Lee JK, Belov YY, Gunn MD, Zuo FR, Huang CC, Kannagi R, Rosen SD, Hemmerich S. J Cell Biol. 1999;145:899. doi: 10.1083/jcb.145.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wong C-H. J Am Chem Soc. 1997;119:8152. doi: 10.1021/ja964290o. [DOI] [PubMed] [Google Scholar]
- 113.Kogan TP, Dupre B, Bui H, McAbee KL, Kassir JM, Scott IL, Hu X, Vanderslice P, Beck PJ, Dixon RA. J Med Chem. 1998;41:1099. doi: 10.1021/jm9704917. [DOI] [PubMed] [Google Scholar]
- 114.Armstrong JI, Portley AR, Chang YT, Nierengarten DM, Cook BN, Bowman KG, Bishop A, Gray NS, Shokat KM, Schultz PG, Bertozzi CR. Angew Chem Int Ed Engl. 2000;39:1303. doi: 10.1002/(sici)1521-3773(20000403)39:7<1303::aid-anie1303>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 115.Kakuta Y, Petrotchenko EV, Pedersen LC, Negishi M. J Biol Chem. 1998;273:27325. doi: 10.1074/jbc.273.42.27325. [DOI] [PubMed] [Google Scholar]
- 116.Chang YT, Gray NS, Rosania GR, Sutherlin DP, Kwon S, Norman TC, Sarohia R, Leost M, Meijer L, Schultz PG. Chem Biol. 1999;6:361. doi: 10.1016/S1074-5521(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 117.Chapman E, Ding S, Schultz PG, Wong CH. J Am Chem Soc. 2002;124:14524. doi: 10.1021/ja021086u. [DOI] [PubMed] [Google Scholar]
- 118.Verdugo GE, Wong C-H. Wiley-VCH. 2003;2:781–802.
- 119.Kehoe JW, Maly DJ, Verdugo DE, Armstrong JI, Cook BN, Ouyang YB, Moore KL, Ellman JA, Bertozzi CR. Bioorg Med Chem Lett. 2002;12:329. doi: 10.1016/s0960-894x(01)00744-2. [DOI] [PubMed] [Google Scholar]
- 120.Huang PL, Dawson TM, Bredt DS, Snyder SH, Fishman MC. Cell. 1993;75:1273. doi: 10.1016/0092-8674(93)90615-w. [DOI] [PubMed] [Google Scholar]
- 121.Pollock JD, Williams DA, Gifford MA, Li LL, Du X, Fisherman J, Orkin SH, Doerschuk CM, Dinauer MC. Nat Genet. 1995;9:202. doi: 10.1038/ng0295-202. [DOI] [PubMed] [Google Scholar]
- 122.Ushio-Fukai M. Sci STKE. 2006;2006:re8. doi: 10.1126/stke.3492006re8. [DOI] [PubMed] [Google Scholar]
- 123.McCord JM, Fridovich I. J Biol Chem. 1969;244:6049. [PubMed] [Google Scholar]
- 124.Hampton MB, Kettle AJ, Winterbourn CC. Blood. 1998;92:3007. [PubMed] [Google Scholar]
- 125.Chan JF, J L. In: Nitric Oxide and Infection. Fang FC, editor. Kluwer Academic and Plenum Publishers; New York: 1999. pp. 281–310. [Google Scholar]
- 126.Padmaja S, Huie RE. Biochem Biophys Res Commun. 1993;195:539. doi: 10.1006/bbrc.1993.2079. [DOI] [PubMed] [Google Scholar]
- 127.Schlosser-Silverman E, Elgrably-Weiss M, Rosenshine I, Kohen R, Altuvia S. J Bacteriol. 2000;182:5225. doi: 10.1128/jb.182.18.5225-5230.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chan J, Xing Y, Magliozzo RS, Bloom BR. J Exp Med. 1992;175:1111. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Thoma-Uszynski S, Stenger S, Takeuchi O, Ochoa MT, Engele M, Sieling PA, Barnes PF, Rollinghoff M, Bolcskei PL, Wagner M, Akira S, Norgard MV, Belisle JT, Godowski PJ, Bloom BR, Modlin RL. Science. 2001;291:1544. doi: 10.1126/science.291.5508.1544. [DOI] [PubMed] [Google Scholar]
- 130.Rhee KY, Erdjument-Bromage H, Tempst P, Nathan CF. Proc Natl Acad Sci USA. 2005;102:467. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cooper AM, Segal BH, Frank AA, Holland SM, Orme IM. Infect Immun. 2000;68:1231. doi: 10.1128/iai.68.3.1231-1234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. Mol Microbiol. 2004;52:1291. doi: 10.1111/j.1365-2958.2004.04078.x. [DOI] [PubMed] [Google Scholar]
- 133.Daniel DS, Dai G, Singh CR, Lindsey DR, Smith AK, Dhandayuthapani S, Hunter RL, Jr, Jagannath C. J Immunol. 2006;177:4688. doi: 10.4049/jimmunol.177.7.4688. [DOI] [PubMed] [Google Scholar]
- 134.Dosanjh NS, Rawat M, Chung JH, Av-Gay Y. FEMS Microbiol Lett. 2005;249:87. doi: 10.1016/j.femsle.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 135.Jaeger T, Budde H, Flohe L, Menge U, Singh M, Trujillo M, Radi R. Arch Biochem Biophys. 2004;423:182. doi: 10.1016/j.abb.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 136.Jaeger T, Flohe L. Biofactors. 2006;27:109. doi: 10.1002/biof.5520270110. [DOI] [PubMed] [Google Scholar]
- 137.Buchmeier NA, Newton GL, Koledin T, Fahey RC. Mol Microbiol. 2003;47:1723. doi: 10.1046/j.1365-2958.2003.03416.x. [DOI] [PubMed] [Google Scholar]
- 138.Haramaki N, Han D, Handelman GJ, Tritschler HJ, Packer L. Free Radic Biol Med. 1997;22:535. doi: 10.1016/s0891-5849(96)00400-5. [DOI] [PubMed] [Google Scholar]
- 139.Kwon YW, Masutani H, Nakamura H, Ishii Y, Yodoi J. Biol Chem. 2003;384:991. doi: 10.1515/BC.2003.111. [DOI] [PubMed] [Google Scholar]
- 140.Muller S. Mol Microbiol. 2004;53:1291. doi: 10.1111/j.1365-2958.2004.04257.x. [DOI] [PubMed] [Google Scholar]
- 141.Rawat M, Newton GL, Ko M, Martinez GJ, Fahey RC, Av-Gay Y. Antimicrob Agents Chemother. 2002;46:3348. doi: 10.1128/AAC.46.11.3348-3355.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lampreia JP, A S, Moura JJG. Methods Enzymol. 1994;243:241. [Google Scholar]
- 143.Gonzalez Porque P, Baldesten A, Reichard P. J Biol Chem. 1970;245:2371. [PubMed] [Google Scholar]
- 144.Shi L, Jung YJ, Tyagi S, Gennaro ML, North RJ. Proc Natl Acad Sci USA. 2003;100:241. doi: 10.1073/pnas.0136863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wheeler PR, Coldham NG, Keating L, Gordon SV, Wooff EE, Parish T, Hewinson RG. J Biol Chem. 2005;280:8069. doi: 10.1074/jbc.M412540200. [DOI] [PubMed] [Google Scholar]
- 146.MacMicking J, Xie QW, Nathan C. Annu Rev Immunol. 1997;15:323. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 147.MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Proc Natl Acad Sci USA. 1997;94:5243. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Senaratne RH, Mougous JD, Reader JR, Williams SJ, Zhang T, Bertozzi CR, Riley LW. J Med Microbiol. 2007;56:454. doi: 10.1099/jmm.0.46983-0. [DOI] [PubMed] [Google Scholar]
- 149.Chartron J, Carroll KS, Shiau C, Gao H, Leary JA, Bertozzi CR, Stout CD. J Mol Biol. 2006;364:152. doi: 10.1016/j.jmb.2006.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gao H, Leary J, Carroll KS, Bertozzi CR, Chen H. J Am Soc Mass Spectrom. 2007;18:167. doi: 10.1016/j.jasms.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chartron J, Shiau C, Stout CD, Carroll KS. Biochemistry. 2007;46:3942. doi: 10.1021/bi700130e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Carroll KS. 2007 personal communication. [Google Scholar]
- 153.Burns KE, Baumgart S, Dorrestein PC, Zhai H, McLafferty FW, Begley TP. J Am Chem Soc. 2005;127:11602. doi: 10.1021/ja053476x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Newton GL, Arnold K, Price MS, Sherrill C, Delcardayre SB, Aharonowitz Y, Cohen G, Davies J, Fahey RC, Davis C. J Bacteriol. 1996;178:1990. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rawat M, Av-Gay Y. FEMS Microbiol Rev. :2007. doi: 10.1111/j.1574-6976.2006.00062.x. [DOI] [PubMed] [Google Scholar]
- 156.Newton GL, Bewley CA, Dwyer TJ, Horn R, Aharonowitz Y, Cohen G, Davies J, Faulkner DJ, Fahey RC. Eur J Biochem. 1995;230:821. doi: 10.1111/j.1432-1033.1995.0821h.x. [DOI] [PubMed] [Google Scholar]
- 157.Newton GL, Ko M, Ta P, Av-Gay Y, Fahey RC. Protein Expr Purif. 2006;47:542. doi: 10.1016/j.pep.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 158.Newton GL, Av-Gay Y, Fahey RC. J Bacteriol. 2000;182:6958. doi: 10.1128/jb.182.24.6958-6963.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bornemann C, Jardine MA, Spies HS, Steenkamp DJ. Biochem J. 1997;325(Pt 3):623. doi: 10.1042/bj3250623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Koledin T, Newton GL, Fahey RC. Arch Microbiol. 2002;178:331. doi: 10.1007/s00203-002-0462-y. [DOI] [PubMed] [Google Scholar]
- 161.Jardine MA, Spies HS, Nkambule CM, Gammon DW, Steenkamp DJ. Bioorg Med Chem. 2002;10:875. doi: 10.1016/s0968-0896(01)00383-2. [DOI] [PubMed] [Google Scholar]
- 162.Lee S, Rosazza JP. Org Lett. 2004;6:365. doi: 10.1021/ol0362008. [DOI] [PubMed] [Google Scholar]
- 163.Newton GL, Koledin T, Gorovitz B, Rawat M, Fahey RC, Av-Gay Y. J Bacteriol. 2003;185:3476. doi: 10.1128/JB.185.11.3476-3479.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Newton GL, Av-Gay Y, Fahey RC. Biochemistry. 2000;39:10739. doi: 10.1021/bi000356n. [DOI] [PubMed] [Google Scholar]
- 165.Newton GL, Unson MD, Anderberg SJ, Aguilera JA, Oh NN, delCardayre SB, Av-Gay Y, Fahey RC. Biochem Biophys Res Commun. 1999;255:239. doi: 10.1006/bbrc.1999.0156. [DOI] [PubMed] [Google Scholar]
- 166.Hayward D, Wiid I, van Helden P. IUBMB Life. 2004;56:131. doi: 10.1080/15216540410001674012. [DOI] [PubMed] [Google Scholar]
- 167.Newton GL, Ta P, Bzymek KP, Fahey RC. J Biol Chem. 2006;281:33910. doi: 10.1074/jbc.M604724200. [DOI] [PubMed] [Google Scholar]
- 168.Rawat M, Uppal M, Newton G, Steffek M, Fahey RC, Av-Gay Y. J Bacteriol. 2004;186:6050. doi: 10.1128/JB.186.18.6050-6058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sareen D, Steffek M, Newton GL, Fahey RC. Biochemistry. 2002;41:6885. doi: 10.1021/bi012212u. [DOI] [PubMed] [Google Scholar]
- 170.Campbell JA, Davies GJ, Bulone V, Henrissat B. Biochem J. 1997;326(Pt 3):929. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Maynes JT, Garen C, Cherney MM, Newton G, Arad D, Av-Gay Y, Fahey RC, James MN. J Biol Chem. 2003;278:47166. doi: 10.1074/jbc.M308914200. [DOI] [PubMed] [Google Scholar]
- 172.McCarthy AA, Peterson NA, Knijff R, Baker EN. J Mol Biol. 2004;335:1131. doi: 10.1016/j.jmb.2003.11.034. [DOI] [PubMed] [Google Scholar]
- 173.Rawat M, Kovacevic S, Billman-Jacobe H, Av-Gay Y. Microbiology. 2003;149:1341. doi: 10.1099/mic.0.26084-0. [DOI] [PubMed] [Google Scholar]
- 174.Buchmeier NA, Newton GL, Fahey RC. J Bacteriol. 2006;188:6245. doi: 10.1128/JB.00393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Vetting MW, Roderick SL, Yu M, Blanchard JS. Protein Sci. 2003;12:1954. doi: 10.1110/ps.03153703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vetting MW, Yu M, Rendle PM, Blanchard JS. J Biol Chem. 2006;281:2795. doi: 10.1074/jbc.M510798200. [DOI] [PubMed] [Google Scholar]
- 177.Newton GL, Ta P, Fahey RC. J Bacteriol. 2005;187:7309. doi: 10.1128/JB.187.21.7309-7316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Steffek M, Newton GL, Av-Gay Y, Fahey RC. Biochemistry. 2003;42:12067. doi: 10.1021/bi030080u. [DOI] [PubMed] [Google Scholar]
- 179.Hand CE, Honek JF. J Nat Prod. 2005;68:293. doi: 10.1021/np049685x. [DOI] [PubMed] [Google Scholar]
- 180.Misset-Smits M, van Ophem PW, Sakuda S, Duine JA. FEBS Lett. 1997;409:221. doi: 10.1016/s0014-5793(97)00510-3. [DOI] [PubMed] [Google Scholar]
- 181.Patel MP, Blanchard JS. Biochemistry. 1999;38:11827. doi: 10.1021/bi991025h. [DOI] [PubMed] [Google Scholar]
- 182.Patel MP, Blanchard JS. Biochemistry. 2001;40:5119. doi: 10.1021/bi0029144. [DOI] [PubMed] [Google Scholar]
- 183.McAdam RA, Quan S, Smith DA, Bardarov S, Betts JC, Cook FC, Hooker EU, Lewis AP, Woollard P, Everett MJ, Lukey PT, Bancroft GJ, Jacobs WR, Jr, Duncan K. Microbiology. 2002;148:2975. doi: 10.1099/00221287-148-10-2975. [DOI] [PubMed] [Google Scholar]
- 184.Lombo F, Velasco A, Castro A, de la Calle F, Brana AF, Sanchez-Puelles JM, Mendez C, Salas JA. Chembiochem. 2006;7:366. doi: 10.1002/cbic.200500325. [DOI] [PubMed] [Google Scholar]
- 185.Nicholas GM, Eckman LL, Newton GL, Fahey RC, Ray S, Bewley CA. Bioorg Med Chem. 2003;11:601. doi: 10.1016/s0968-0896(02)00345-0. [DOI] [PubMed] [Google Scholar]
- 186.Nicholas GM, Eckman LL, Ray S, Hughes RO, Pfefferkorn JA, Barluenga S, Nicolaou KC, Bewley CA. Bioorg Med Chem Lett. 2002;12:2487. doi: 10.1016/s0960-894x(02)00385-2. [DOI] [PubMed] [Google Scholar]
- 187.Nicholas GM, Newton GL, Fahey RC, Bewley CA. Org Lett. 2001;3:1543. doi: 10.1021/ol015845+. [DOI] [PubMed] [Google Scholar]
- 188.Fetterolf B, Bewley CA. Bioorg Med Chem Lett. 2004;14:3785. doi: 10.1016/j.bmcl.2004.04.095. [DOI] [PubMed] [Google Scholar]
- 189.Pick N, Rawat M, Arad D, Lan J, Fan J, Kende AS, Av-Gay Y. J Med Microbiol. 2006;55:407. doi: 10.1099/jmm.0.46319-0. [DOI] [PubMed] [Google Scholar]
- 190.Knapp S, Gonzalez S, Myers DS, Eckman LL, Bewley CA. Org Lett. 2002;4:4337. doi: 10.1021/ol0269796. [DOI] [PubMed] [Google Scholar]
- 191.Metaferia BB, Ray S, Smith JA, Bewley CA. Bioorg Med Chem Lett. 2007;17:444. doi: 10.1016/j.bmcl.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 192.Hu Y, Helm JS, Chen L, Ginsberg C, Gross B, Kraybill B, Tiyanont K, Fang X, Wu T, Walker S. Chem Biol. 2004;11:703. doi: 10.1016/j.chembiol.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 193.Burguiere P, Auger S, Hullo MF, Danchin A, Martin-Verstraete I. J Bacteriol. 2004;186:4875. doi: 10.1128/JB.186.15.4875-4884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Muller A, Thomas GH, Horler R, Brannigan JA, Blagova E, Levdikov VM, Fogg MJ, Wilson KS, Wilkinson AJ. Mol Microbiol. 2005;57:143. doi: 10.1111/j.1365-2958.2005.04691.x. [DOI] [PubMed] [Google Scholar]
- 195.Wang XF, Cynader MS. J Neurochem. 2000;74:1434. doi: 10.1046/j.1471-4159.2000.0741434.x. [DOI] [PubMed] [Google Scholar]
- 196.Pavelka MS, Jr, Jacobs WR., Jr J Bacteriol. 1999;181:4780. doi: 10.1128/jb.181.16.4780-4789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wheeler PRR, C . In: Tuberculosis Pathogenesis, Protection and Control. Bloom BR, editor. ASM Press; Washington, D. C.: 1994. p. 353. [Google Scholar]
- 198.Smith DA, Parish T, Stoker NG, Bancroft GJ. Infect Immun. 2001;69:1142. doi: 10.1128/IAI.69.2.1142-1150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Eckert KG, Elbers FR, Eyer P. Biochem Pharmacol. 1989;38:3253. doi: 10.1016/0006-2952(89)90622-9. [DOI] [PubMed] [Google Scholar]
- 200.Roederer M, Staal FJ, Osada H, Herzenberg LA, Herzenberg LA. Int Immunol. 1991;3:933. doi: 10.1093/intimm/3.9.933. [DOI] [PubMed] [Google Scholar]
- 201.Green RM, Seth A, Connell ND. Infect Immun. 2000;68:429. doi: 10.1128/iai.68.2.429-436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Dayaram YK, Talaue MT, Connell ND, Venketaraman V. J Bacteriol. 2006;188:1364. doi: 10.1128/JB.188.4.1364-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Venketaraman V, Dayaram YK, Talaue MT, Connell ND. Infect Immun. 2005;73:1886. doi: 10.1128/IAI.73.3.1886-1889.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Muse W. 2007 personal communication. [Google Scholar]
- 205.Nicholas GM, Hong TW, Molinski TF, Lerch ML, Cancilla MT, Lebrilla CB. J Nat Prod. 1999;62:1678. doi: 10.1021/np990190v. [DOI] [PubMed] [Google Scholar]


