Abstract
In our previous study to identify the RNA internal loops that bind an aminoglycoside derivative, we determined that 6′-N-5-hexynoate kanamycin A prefers to bind 1×1 nucleotide internal loops containing C•A mismatches. In this present study, the molecular recognition between a variety of RNAs that are mutated around the C•A loop and the ligand was investigated. Studies show that both loop nucleotides and loop closing pairs affect binding affinity. Most interestingly, it was shown that there is a correlation between the thermodynamic stability of the C•A internal loops and ligand affinity. Specifically, C•A loops that had relatively high or low stability bound the ligand most weakly whereas loops with intermediate stability bound the ligand most tightly. In contrast, there is no correlation between the likelihood that a loop forms a C-A+ pair at lower pH and ligand affinity. It was also found that a 1×1 nucleotide C•A loop that bound to the ligand with the highest affinity is identical to the consensus site in RNAs that are edited by adenosine deaminases acting on RNA type 2 (ADAR2). These studies provide a detailed investigation of factors affecting small molecule recognition of internal loops containing C•A mismatches, which are present in a variety of RNAs that cause disease.
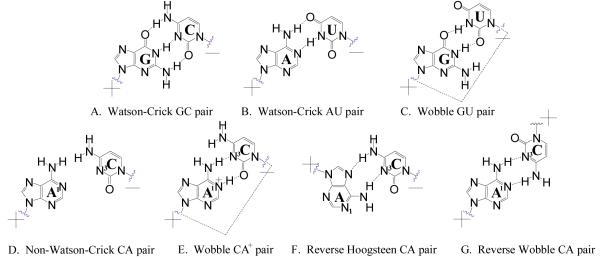
RNA has diverse structures that are important for biological function (1-7). Unlike DNA, RNA is single stranded and has secondary and tertiary structures derived from non-canonical pairing interactions. One example of a non-canonical pair is C•A which can form a variety of structures such as a non-Watson-Crick pair, a wobble C-A+ pair, a reverse Hoogsteen pair and a reverse wobble pair (Figure 1 D-G). These structures are energetically different (8). The least stable loops are the non-Watson-Crick (Figure 1D) and the reverse Hoogsteen (Figure 1F) pairs in which no hydrogen bonds form at the Watson-Crick face. The reverse wobble pair (Figure 1G) is found in parallel duplexes (9). C-A+ pairs form when A is protonated, leading to a wobble geometry (Figure 1E) (8, 10). The structure is similar to the geometry of wobble GU pairs in an anti-parallel duplex (11). Thus, C-A+ pairs fit into a helix similarly to wobble GU pairs or Watson-Crick pairs (9, 11, 12).
Figure 1.
Canonical base pairs (A-C) and four possible CA pairs (D-G) in RNA. Plus and minus signs represent the direction of strands (+/− represent anti-parallel strands and +/+ represent parallel strands).
The function of RNA is often dictated by the presence of C•A pairs. For example, a C•A loop is the preferred substrate of adenosine deaminase acting on RNA type 2 (ADAR2) (13). ADARs modify the A in C•A loop to an inosine (13-15). In Hepatitis Delta Virus (HDV), ADAR is utilized to diversify the genome, for example changing a stop codon to a tryptophan codon (16). Global ADAR substrate analysis reveals that editing is context dependent: the A in 1×1 nucleotide C•A loops is edited more efficiently in 5′-WAK-3′ (where W is A or U and K is G or U) than A’s in other contexts (17). A variety of inherited diseases are also due to defects in ADAR editing of RNA transcripts. These include editing of the 2C-subtype of the serotonin receptor, which has been implicated in contributing to Prader-Willi syndrome (18, 19). A variety of other central nervous system disorders are also caused by malfunctions in RNA editing (20). Since CA loops are often the preferred editing site(s) in RNA, small molecules that target editing sites could provide lead ligands to treat diseases caused by mis-editing by ADAR. Additionally, mutations in mitochondrial tRNAs that cause neuromuscular disorders are often due to insertion of CA 1×1 internal loops.(21-24) Thus, many diseases are due to insertion or altered processing of RNAs with 1×1 nucleotide CA internal loops.
Information on the RNA secondary structures that bind small molecules has been previously used to target RNAs that cause disease. For example, information on the preferred binding sites of kanamycin and Hoechst 33258 derivatives have been used to rationally design cell-permeable, modularly assembled ligands targeting the RNAs that cause myotonic dystrophy types 1 and 2, spinocerebellar ataxia type 3, and Huntington’s disease (25-28). It is likely that a similar approach to those mentioned above could be used to target diseases that are caused by CA loops in RNA.
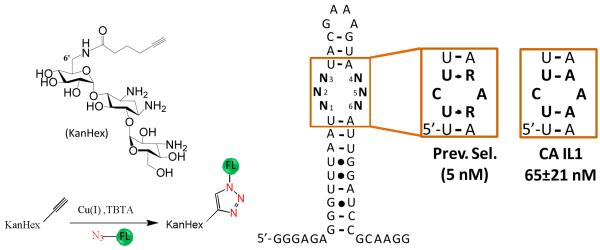
Previously, it was identified that RNAs with C•A loops were one of the preferred targets of 6′-N-5-hexynoate kanamycin A (KanHex) by using a selection-based approach (Figure 2) (29, 30). Selected RNA-KanHex interactions occurred with dissociation constant ranging from 5 to 12 nM at pH 7.0. No loops were selected that were closed by two GC pairs indicating that the thermodynamic stability of the loop could be an important determinant in high affinity binding. Various studies have that the identity and orientation of loop closing pairs affect loop stability.(31-34) We are unaware of any other studies that investigate how ligand affinity is correlated to loop stability.
Figure 2.
The 3×3 internal loop library (ILL) selection with 6′-N-5-hexynoate kanamycin A (KanHex) using a microarray platform (30). Prev. Sel. indicates the consensus sequence and the corresponding dissociation constant from a previous selection. CA ILX indicates an internal loop containing a C across from an A; the associated value is the measured Kd from this study.
Herein, the molecular recognition of KanHex by 1×1 nucleotide C•A loops is reported. Specifically, the features in the C•A loops that provide the optimal KanHex binding site were investigated by mutating and deleting loop nucleotides and mutating loop closing pairs. The stabilities of some loops at pH 7.0 and 5.5 were also determined using optical melting. Importantly, there is a correlation between loop stability at pH 7.0 and ligand affinity. That is, loops with intermediate stabilities bind the ligand with the highest affinity. Loops that are relatively more or less stable bind more weakly. In contrast, there is no correlation between the formation of C-A+ pairs at lower pH and affinity. It was also found that the consensus RNA loop binder for this ligand is similar to the consensus ADAR2 editing site (13). Thus, there could be similarities in molecular recognition of CA loops by KanHex and the RNA-binding domain of the ADAR2 protein. These studies could enable the design of compounds that bind specifically to RNAs edited by ADAR2 and modulate editing. Such compounds have not been reported in the literature.
MATERIALS & METHODS
Aminoglycoside Derivatives
KanHex and the fluorescein labeled derivatives of all aminoglycosides were synthesized from the parent aminoglycoside as previously described (29, 30).
DNA Templates and PCR Amplification of DNA Templates Encoding Selected RNAs
All DNA templates were purchased from Integrated DNA Technologies, Inc. (IDT) and used without further purification unless noted otherwise. The DNA templates encoding the mutated RNAs were PCR-amplified in 50 μL of 1X PCR buffer (10 mM Tris-HCl, pH 9.0, 50 mM KCl, 0.1% Triton X), 4.25 mM MgCl2, 0.33 mM dNTPs, 2 μM each primer (5′-CCTTGCGGATCCAAT; and 5′-GGCCGGATCCTAATACGACTCACTATAGGGAGAGGGTTTAAT), and 0.2 μL of Taq DNA polymerase. The DNA was amplified by 25 cycles of 95 °C for 30 s, 50 °C for 30 s, and 72 °C for 1 min. All PCR reactions were analyzed by gel electrophoresis on a 5% agarose gel stained with ethidium bromide.
RNA Transcription and Purification
RNA oligonucleotides were transcribed using an RNAMaxx Transcription kit (Stratagene) according to the manufacturer’s protocol with 12.5 μL of the amplified DNA from the PCR reaction described above. After transcription, 1 unit of DNase I (Invitrogen) was added, and the sample was incubated for an additional 30 min at 37 °C. The transcribed RNAs were then purified by gel electrophoresis on a denaturing 15% polyacrylamide gel. The RNAs were visualized by UV shadowing and extracted into 300 mM NaCl by tumbling overnight at 4 °C. The resulting solution was concentrated with 2-butanol and ethanol precipitated. The RNAs were dissolved in 150 μL of nanopure water, and the concentrations were determined by their absorbances at 260 nm and the corresponding extinction coefficients. Extinction coefficients were determined using HyTher version 1.0 (35, 36). The parameters used by HyTher are based on information about the extinction coefficients of nearest neighbors in RNA (37).
Fluorescence Binding Assays
Dissociation constants were determined using an in solution, fluorescence-based assay. RNA was folded in HBI (8 mM Na2PO4, pH 7.0, 185 mM NaCl, 0.1 mM EDTA) and 40 μg/mL BSA at 60 °C for 5 min. The solution was allowed to cool slowly to room temperature. Then, fluorescently labeled KanHex was added to a final concentration of 50 nM. Serial dilutions (1:2) were completed in 1X HBII (HBI supplemented with 40 μg/mL BSA and 50 nM fluorescently labeled KanHex). The solutions were incubated for 30 min at room temperature and then transferred to wells of a black 96-well plate. Fluorescence intensity was measured on a Bio-Tek FLX-800 plate reader. The change in fluorescence intensity as a function of RNA concentration was fit to equation 1 (38):
| (eq 1) |
where I is the observed florescence intensity, I0 is the fluorescence intensity in the absence of RNA, Δε is the difference between the fluorescence intensity in the absence of RNA and the fluorescence intensity in the presence of infinite RNA concentration, [FL]0 is the concentration of the fluorescently labeled KanHex, [RNA]0 is the concentration of the selected internal loop or control RNA, and Kt is the dissociation constant.
Optical Melting Experiments and the Determination of Thermodynamic Parameters
Single stranded oligonucleotides were purchased from Integrated DNA Technology (IDT) and purified by preparative thin layer chromatography using 55:35:10 (v:v:v) 1-propanol : ammonium hydroxide : water as the mobile phase. The product was identified by UV shadowing and was extracted from the silica by tumbling in NANOpure water for 3 h. All non-self complementary duplexes were formed by mixing the corresponding oligonucleotides in an equimolar ratio.
Melting experiments were completed in 8 mM Na2HPO4 (pH 5.5 or 7.0), 180 mM NaCl, and 0.1 mM Na2EDTA using a Beckman Coulter DU800 UV-Vis spectrometer with an attached peltier heater. Melting curves of absorbance versus temperature were acquired at 260 nm with heating rate of 1 °C/min from 12 °C to 89 °C. Melting curves were fit to a two-state model using the MeltWin program (http://www.meltwin.com) (39). The program fits the shape of each curve with sloping base lines and temperature-independent ΔHo and ΔSo using a nonlinear least-squares algorithm (39-41). Thermodynamic parameters for the non-self complementary duplexes were also obtained by plotting the inverse of the melting temperature of oligonucleotides duplex (TM−1) versus . The curves were then fit to Equation 2 (42):
| (eq. 2) |
where R is the gas constant, 1.987 cal K−1 mol−1.
In order to determine if a loop of interest likely forms a C-A+ pair at lower pH’s, the stabilities of the duplexes were compared at pH 7.0 and 5.5. The difference in free energies, ΔΔG037, pH, was calculated according to Equation 3 (based on the ΔG037 values calculated from Equation 2):
| (eq. 3) |
Determination of Loop Stability
The stabilities of the CA internal loops, , were calculated using to Equation 4 (43):
| (eq. 4) |
where is the free energy for formation of a duplex containing a CA internal loop as calculated from Equation 2, is the free energy for formation of the duplex without the CA internal loop as calculated from Equation 2, and is the free energy for the nearest neighbor that was disrupted by insertion of the internal loops (40, 44).
RESULTS & DISCUSSION
We previously reported a 2-Dimensional Combinatorial Screening (2DCS) selection in which the RNAs from a 3×3 nucleotide internal loop library that bind 6′-N-5-hexynoate kanamycin A (KanHex) were identified (Figure 2) (30). It was determined that the RNA-binding ligand prefers symmetric internal loops that display a cytosine across from an adenine (C•A) closed by UA or UG base pairs (Figure 2) and binds with a stoichiometry of 1:1.(30) Furthermore, 62.5% of selected loops contain C across from A whereas only 33% of the 4,096-membered library contains this motif (30). This represents a significant bias in the selection (p-value = 0.0124; or there is less than a 1.24% chance the loops were selected randomly). The consensus loop that has the highest affinity was a 1×1 nucleotide loop containing a CA mismatch closed by two UA base pairs (5 nM) (30). In addition, no selected internal loop was closed by two CG/GC base pairs. These results suggest that both CA loops and their closing pairs are important for high affinity binding to KanHex. Thus, we studied these factors by determining the affinities of loops with mutated or deleted nucleotides and mutated closing pairs.
The identity and position of loop nucleotides are important for binding KanHex
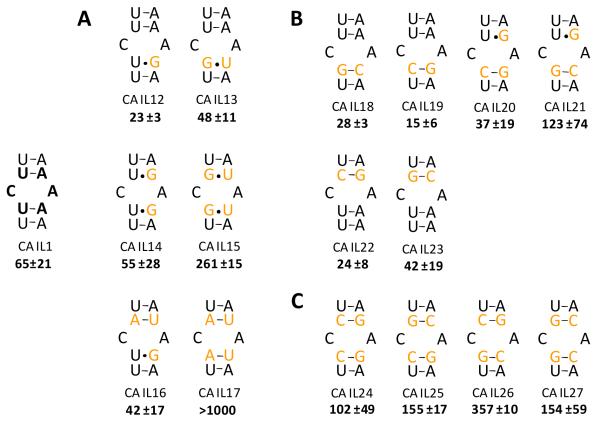
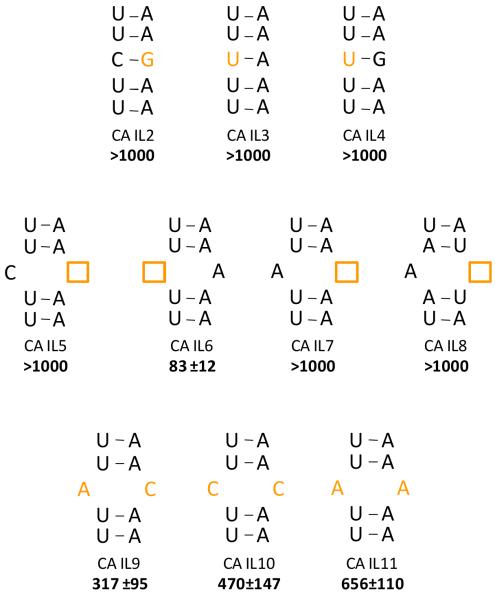
The various internal loops and their closing base pairs in this study (Figures 2-4) were inserted into the same RNA cassette used in the 2DCS selection experiment (Figure 2). The starting construct contains a CA mismatch closed by two UA base pairs (CA IL1, Figure 2). CA IL1 binds KanHex with a dissociation constant of 65 nM. The loop was then systematically mutated in order to determine the effect of loop nucleotides and loop closing base pairs on molecular recognition (Figure 3). The first set of mutations generated fully paired RNAs, CA IL2, IL3 and IL4 (Figure 3). There was no change in fluorescence intensity when up to 1 μM of CA IL2, IL3 or IL4 was added to 50 nM of KanHex, indicating that the fully paired RNAs do not bind the ligand or bind very weakly.
Figure 4.
The RNA constructs used to determine how loop closing pairs affect binding affinity to KanHex. Nucleotides are derived from the boxed region in Figure 2. Dissociation constants are reported in nanomolar.
Figure 3.
The RNA constructs used to determine how the identity and position of loop nucleotides affect binding to KanHex. Nucleotides are derived from the boxed region in Figure 2. Dissociation constants are reported in nanomolar.
In order to determine if the C and/or the A in the loop are important for the molecular recognition of KanHex, one of the nucleotides or the other was deleted. Deletion of the A5 generates a C2 bulge (CA IL5, Figure 3) and binding was abolished (Kd > 1 μM). Interestingly, deletion of the C2 residue, which creates an A5 bulge (CA IL6, Figure 3) resulted in a dissociation constant of 83 nM. Mutating C2 to A2 results in a 1-nucleotide bulge in which the A is on the other side of the helix, or CA IL7 (Figure 3). CA IL7 binds weakly to KanHex with a Kd > 1 μM. Switching the orientation of both closing base pairs in CA IL7 to 5′-AU-3′ (generating CA IL8) does not rescue binding affinity. CC (CA IL10) and AA (CA IL11) mismatches were also studied. They bind KanHex about 8- and 10-fold more weakly, respectively, than CA IL1. Interestingly, a previous study showed that kanamycin A bound to 1×1 nucleotide CC loops present in the 5′ untranslated region (UTR) of thymidylate synthase messenger RNA with weak dissociation constants (>2 μM) (45). In summary, the identity of the loop nucleotides, particularly A5, is important for high affinity binding to KanHex.
Loop closing pairs affect KanHex binding affinity
To investigate the importance of C•A loop closing base pairs, they were systematically altered. Of the 36 possible CA mismatches with different closing base pairs, 16 were studied (Figure 4). Series A contains RNAs in which the closing base pairs are mutated to afford U•G or G•U wobble base pairs; series B contains RNAs in which the stability of closing base pair is increased by incorporating one CG or GC base pair; and series C RNAs have two CG or GC closing base pairs.
In series A, the most deleterious mutation switches the orientation of both 5′-UA-3′ closing base pairs to 5′-AU-3′ (CA IL17) as no binding of KanHex is observed (Kd > 1000 nM). Similarly, when both closing base pairs were mutated to 5′-GU-3′ (CA IL15), a Kd of 261 nM was obtained. Interestingly, if one of the 5′-AU-3′ pairs in CA IL17 is replaced with a 5′-UG-3′ pair, affording CA IL16, binding is rescued (Kd=42 nM). Likewise, if one of the 5′-GU-3′ closing pairs in CA IL15 is replaced with 5′-UA-3′, then binding affinity is restored (CA IL13). Moreover, replacement of one or both 5′-UA-3′ pairs with 5′-UG-3′ pairs does not significantly affect affinity (CA IL12 and IL14). This suggests the identity and orientation of closing base pairs are important for binding to KanHex. This may be due to the ligand forming direct contacts to the closing pair or other factors such as electrostatic interactions.
Next, the loop closing pairs were systematically altered to contain one or two CG/GC closing base pairs (Series B and C, Figure 4). In Series B, only one GC or CG base pair was introduced to the RNA construct, and the highest affinity C•A internal loop was CA IL19 (15 nM). Other mutations were also well tolerated, either not affecting affinity or increasing affinity slightly as compared to the starting construct RNA. The exception is CA IL21, which contains a 5′GC and a 3′UG closing pair, and binds with a dissociation constant of ~125 nM. In contrast, when both closing pairs are mutated to GC pairs, a 2- to 5-fold decrease in affinity as compared to the starting construct RNA is observed (Series C, Figure 4).
In summary, the important features in the RNA for binding to KanHex are: 1.) a purine at position 4; 2.) the presence of at least one 5′-UA-3′, 5′-UG-3′ or 5′-CG-3′ closing base pair; and 3.) the absence of two 5′-AU-3′, 5′-GU-3′ or 5′-GC-3′ closing base pairs. Evidently, the orientation and identity of the loop closing pairs are important for tight binding of KanHex to internal loops containing CA mismatches.
Aminoglycoside selectivity for binding CA IL19
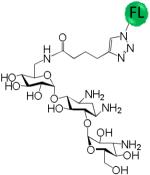
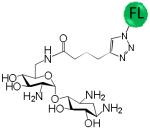
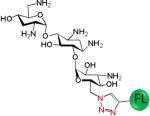
A variety of aminoglycosides are known to interact with RNA, and most often the RNA binds with highest affinity to the aminoglycoside with the greatest number of amino groups (typically neomycin B). (46, 47) Thus, there is often little aminoglycoside specificity in binding RNA targets when the aminoglycosides have a similar number of amino groups. In order to determine if the consensus C•A most specifically binds KanHex over other aminoglycosides, a series of binding assays were completed for CA IL19. The results of these studies are summarized in Table 1. The analysis shows that KanHex is the highest affinity ligand that binds CAIL19; specificities for other aminoglycosides range from 12- to >330-fold. Not surprisingly, the next highest affinity binder is the neomycin B derivative (with the largest number of amino groups (six)). The largest difference in affinity is observed between KanHex and the neamine derivative which has the smallest number of amino groups. Interestingly, KanHex and the neamine derivative have the same number of amino groups. Thus, the interaction of the KanHex to CA IL19 is due to specific interactions with the structure of the aminoglycosides and is not due to simple charge-charge interactions. Similar specificities have been observed with aminoglycosides binding to RNA internal and hairpin loops when the RNAs have been selected to bind to aminoglycosides via 2DCS. (30, 48-51) Other studies have suggested that aminoglycoside recognition is shape-dependent.(47) The results presented here suggest that high affinity binding is also sequence dependent.
Table 1.
Selectivity of CA IL 19 for binding to a series of fluorescently labeled aminoglycosides
| Aminoglycoside | Kd (nM) | Selectivity |
|---|---|---|
|
15 ± 6 | -- |
|
747 ± 23 | 50 |
|
793 ±116 | 53 |
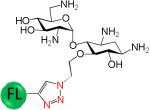
|
>5000 | >333 |
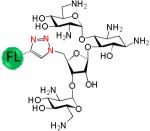
|
181 ± 21 | 12 |
Thermodynamic stability of the C•A loop is correlated with binding affinity
The fact that C•A internal loops containing two CG/GC closing base pairs bind more weakly to KanHex suggested that loop stability might play a role in binding affinity. Previous studies have shown that closing pairs affect the stability of RNA 1×1 nucleotide internal loops with loops closed by two GC pairs being the most stable (32); additional reports have elucidated other factors that affect stability (33, 34). It is also possible that some C•A loops form wobble C-A+ pairs, which would also contribute to loop stability. To address thermodynamic stability and structural differences of internal loops containing CA mismatches and their effects on binding affinity, optical melting experiments were completed. All 9-mer oligonucleotides duplexes (non-self complementary) are composed of 5′- and 3′- constant flanking regions, , and the C•A internal loops with different closing base pairs were inserted in between the flanking regions. The stabilities of the duplexes with internal loops were then compared to the corresponding fully paired duplex to calculate the free energy of loop formation (thermodynamic stability of the loop; Equation 3).
The thermodynamic stabilities of eight C•A internal loops were determined. As shown in Table 1, the range of free energy values for loop formation is in good agreement with previous reports (8, 31). The thermodynamic stability of C•A internal loops containing CG/GC closing pairs are the most stable (CA IL27, ΔG037 = 12.83 ± 0.12 kcal/mol). In contrast, C•A internal loops containing two 5′-GU-3′ closing base pairs are the least stable (CA IL15, ΔG037 = 6.79 ± 0.02 kcal/mol). Interestingly, CA IL19 is less stable than CA IL23 by 1.46 kcal/mol; the two RNAs have the same closing pairs but in different orientations. Similarly, CA IL20 and CA IL21 have the same closing pairs but in different orientations, and their stabilities differ by about 1.4 kcal/mol. Hence, the stability of C•A internal loops depends on both the orientation and the identity of the loop closing base pairs, in good agreement with previous reports.(31-34)
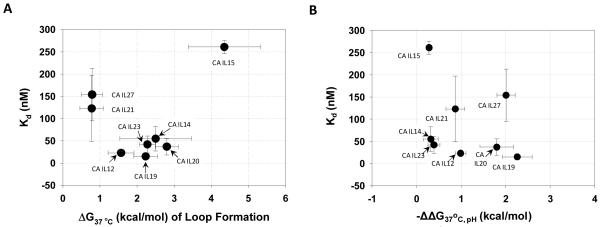
In general, the internal loops with intermediate affinity bind to KanHex with the highest affinity (Figure 5A). Less stable and more stable loops bind more weakly. The two most stable loops are CA IL21 and CA IL27. These loops bind weakly to KanHex with dissociation constants of 123 and 154 nM, respectively. CA IL15 is the least stable loop studied (destabilizing by 4.35 kcal/mol) and binds to the ligand with a Kd of 261 nM. The stabilities of the other loops studied (CA IL12, 14, 19, 20, and 23) are intermediate, ranging from 1.57 to 2.80 kcal/mol (Table 2). These loops bind with the highest affinity, having dissociation constants ranging from 15 to 55 nM (Figure 4).
Figure 5.
A, Correlation of affinity of C•A internal loops for KanHex and the thermodynamic stabilities of the loops. The sequences for the loops are shown in Table 1. B, The affinity of C•A internal loops with KanHex as a function of thermodynamic stability at different pH’s. ΔΔG037,pH is calculated by taking the difference between the ΔG037,pH=5.5 and ΔG037,pH=7.0. There is no correlation between affinity and the likelihood that a loop forms a C-A+ pair at lower pH.
Table 2.
Thermodynamic stability of duplex formation of RNAs with and without AC mismatches at pH 7.0 or pH 5.5.
| Duplexes with Loops | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Average of Two-State Melt Curve Fits |
Linear Fit to 1/TM vs ln(CT/4) Plots |
||||||||||
| −ΔH0 (kcal/mol) |
−ΔS0 (cal/K•mol) |
−ΔG037 (kcal/mol) |
−TM (0C) |
−ΔH0 (kcal/mol) |
−ΔS0 (cal/K•mol) |
−ΔG037 (kcal/mol) |
−TM (0C) |
ΔGo37, loop (kcal/mol) |
ΔΔGpH (kcal/mol) |
||
| CA IL12 | 5′-CCGAAGCCG-3′ 3′-GGCUCUGGC-5′ |
86.47±10.95 | 251.39±35.08 | 8.50±0.08 | 44.2 | 99.48±5.69 | 292.90±18.09 | 8.63±0.09 | 43.7 | 1.57±0.35 | 0.86 |
| 90.75±9.27 | 262.23±29.29 | 9.42±0.20 | 47.2 | 95.23±3.61 | 276.44±11.35 | 9.49±0.09 | 47.0 | ||||
|
| |||||||||||
| CA IL14 | 5′-CCGGAGCCG-3′ 3′-GGCUCUGGC-5′ |
79.23±8.40 | 227.69±26.24 | 8.61±0.128 | 45.4 | 79.22±0.94 | 227.70±2.98 | 8.60±0.02 | 45.3 | 2.50±0.96 | 0.14 |
| 67.62±11.75 | 191.30±37.51 | 8.29±0.17 | 45.3 | 75.87±7.40 | 217.35±23.41 | 8.46±0.22 | 45.1 | ||||
|
| |||||||||||
| CA IL15 | 5′-CCGUAUCCG-3′ 3′-GGCGCGGGC-5′ |
74.33±29.18 | 217.14±94.63 | 6.99±0.33 | 38.9 | 109.47±3.35 | 331.05±10.86 | 6.79±0.02 | 37.7 | 4.35±0.97 | 0.28 |
| 57.84±14.51 | 164.18±47.52 | 6.92±0.40 | 39.1 | 119.90±4.21 | 365.57±13.66 | 6.51±0.035 | 37.0 | ||||
|
| |||||||||||
| CA IL19 | 5′-CCGAAGCCG-3′ 3′-GGCUCCGGC-5′ |
83.08±13.72 | 231.99±41.95 | 11.13±0.72 | 55.2 | 79.85±5.27 | 222.11±16.16 | 10.97±0.27 | 55.2 | 2.23±0.31 | 2.26 |
| 90.24±12.15 | 250.78±36.94 | 12.46±0.70 | 58.8 | 103.27±3.45 | 290.32±10.47 | 13.23±0.20 | 58.5 | ||||
|
| |||||||||||
| CA IL20 | 5′-CCGGAGCCG-3′ 3′-GGCUCCGGC-5′ |
82.50±21.80 | 231.10±66.92 | 10.82±1.01 | 54.0 | 65.69±1.55 | 179.24±4.77 | 10.09±0.07 | 54.8 | 2.80±0.31 | 1.80 |
| 82.38±17.92 | 228.84±54.69 | 11.40±0.958 | 56.5 | 91.52±6.72 | 256.73±20.53 | 11.89±0.37 | 56.3 | ||||
|
| |||||||||||
| CA IL21 | 5′-CCGGACCCG-3′ 3′-GGCUCGGGC-5′ |
87.21±14.16 | 142.12±43.51 | 12.12±0.87 | 58.2 | 89.14±3.15 | 248.01±9.58 | 12.22±0.18 | 58.1 | 0.78±0.32 | 0.87 |
| 93.05±11.44 | 264.20±34.55 | 13.11±0.72 | 60.1 | 94.92±1.76 | 263.84±5.31 | 13.09±0.11 | 60.0 | ||||
|
| |||||||||||
| CA IL23 | 5′-CCGCAACCG-3′ 3′-GGCGCUGGC-5′ |
99.35±12.45 | 283.05±38.39 | 11.56±0.55 | 53.6 | 119.34±2.47 | 344.72±7.93 | 12.43±0.11 | 53.1 | 2.28±0.21 | 0.39 |
| 81.95±11.80 | 227.58±35.95 | 11.36±0.65 | 56.4 | 95.40±1.65 | 268.76±5.05 | 12.04±0.08 | 53.6 | ||||
|
| |||||||||||
| CA IL27 | 5′-CCGCACCCG-3′ 3′-GGCGCGGGC-5′ |
75.71±7.75 | 203.58±23.23 | 12.57±0.53 | 63.9 | 79.10±1.69 | 213.67±5.08 | 12.83±0.12 | 63.8 | 0.79±0.29 | 2.01 |
| 116.26±18.0 | 320.74±53.22 | 16.78±1.47 | 67.0 | 93.10±2.05 | 252.34±6.06 | 14.84±0.16 | 67.4 | ||||
| Duplexes without Loops | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Average of Two-State Melt Curve Fits |
Linear Fit to 1/TM vs ln(CT/4) Plots |
||||||||
| −ΔH0 (kcal/mol) |
−ΔS0 (cal/K•mol) |
−ΔG037 (kcal/mol) |
−TM (0C) |
−ΔH0 (kcal/mol) |
−ΔS0 (cal/K•mol) |
−ΔG037 (kcal/mol) |
−TM (0C) |
||
| 5′-CCGCCG-3′ 3′-GGCGGC-5′ |
57.52±8.88 | 152.79±27.01 | 10.13±0.51 | 57.7 | 56.84±1.13 | 150.53±4.46 | 10.15±0.06 | 58.1 | |
| 58.44±10.67 | 155.43±32.62 | 10.23±0.57 | 58.0 | 53.38±1.18 | 139.75±3.62 | 10.04±0.06 | 58.8 | ||
|
| |||||||||
| Duplex 12 |
5′-CCGAGCCG-3′ 3′-GGCUUGGC-5′ |
80.12±5.31 | 223.72±16.37 | 10.73±0.23 | 54.2 | 80.42±2.42 | 224.64±7.44 | 10.75±0.11 | 54.2 |
|
| |||||||||
| Duplex 14 |
5′-CCGGGCCG-3′ 3′-GGCUUGGC-5′ |
71.76±1.81 | 199.3±5.73 | 9.84±0.18 | 52.0 | 92.42±2.20 | 263.70±6.83 | 10.63±0.08 | 51.4 |
|
| |||||||||
| Duplex 15 |
5′-CCGUUCCG-3′ 3′-GGCGGGGC-5′ |
110.48±9.29 | 320.39±29.02 | 11.11±0.31 | 50.4 | 98.18±3.89 | 282.15±12.07 | 10.67±0.14 | 50.6 |
|
| |||||||||
| Duplex 19 |
5′-CCGAGCCG-3′ 3′-GGCUCGGC-5′ |
91.02±6.25 | 247.13±18.41 | 14.38±0.54 | 66.3 | 102.31±1.86 | 280.58±5.50 | 15.28±0.15 | 66.0 |
|
| |||||||||
| Duplex 20 |
5′-CCGGGCCG-3′ 3′-GGCUCGGC-5′ |
98.78±3.88 | 271.44±11.64 | 14.59±0.27 | 64.6 | 104.45±2.29 | 288.38±6.84 | 15.00±0.17 | 64.4 |
|
| |||||||||
| Duplex 21 |
5′-CCGGCCCG-3′ 3′-GGCUGGGC-5′ |
95.85±8.87 | 262.87±26.38 | 14.32±0.67 | 64.4 | 111.31±0.89 | 308.88±2.65 | 15.51±0.07 | 64.2 |
|
| |||||||||
| Duplex 23 |
5′-CCGCACCG-3′ 3′-GGCGUGGC-5′ |
114.69±2.51 | 316±7.42 | 16.42±0.21 | 66.3 | 119.47±2.07 | 330.96±6.14 | 16.82±0.17 | 66.2 |
|
| |||||||||
| Duplex 27 |
5′-CCGCCCCG-3′ 3′-GGCGGGGC-5′ |
91.56±6.41 | 241.02±18.78 | 16.81±0.58 | 76.2 | 92.39±2.84 | 243.45±8.33 | 16.88±0.25 | 76.1 |
One of the structures that a CA mismatch can form is a wobble C-A+ pair (Figure 1E). The pKa for adenine N1 is 3.5; thus it is usually deprotonated at physiological pH. However, large pKa shifts have been observed for adenine residues in the leadzyme (52), the hairpin ribozyme (53), the Hepatitis Delta Virus ribozyme (54), and the Varkud satellite ribozyme (55). Previous studies have also shown that the protonation state of adenine in an C•A mismatch is context- and sequence-dependent (8, 10). Since the affinity of the RNA internal loop-ligand complex is correlated to loop stability, we also determined if affinity was correlated with formation of C-A+ pairs at lower pH’s.
The thermodynamic parameters for the eight CA internal loops that were studied at pH 7.0 were determined at pH 5.5 (Table 2, red). Based on these data, CA IL19 has the largest change in stability, ΔΔG037,pH, while CA IL15 has the smallest ΔΔG037,pH This suggests that C•A mismatch in CA IL19 but not in CA IL15 is likely a C-A+ wobble pair at pH 5.5. However, there is no correlation between the affinity of C•A internal loops for KanHex and ΔΔG037,pH (Figure 5B) and therefore the likelihood that the loop forms a C-A+ pair at lower pH’s. For example, CA IL27 (Kd = 154 nM) has a similar ΔΔG037,pH as CA IL19 (Kd = 15 nM) but binds weakly to KanHex. In contrast, CA IL14 and CA IL23 have similar ΔΔG037,pH’s as CA IL15 (Kd = 261 nM) but bind to KanHex with Kd’s of 55±28 nM and 42±19 nM, respectively.
Without high resolution structures, it is difficult to determine if the presence of KanHex affects the protonation state of the CA mismatches or vice versa (56). Binding assays completed at lower pH’s cannot give insight into this matter since lowering the pH also changes the protonation state of the ligand; both affect binding affinity. Likewise, optical melting experiments completed in the presence of the ligand cannot deconvolute thermodynamic contributions due to binding of ligand and a change in protonation state of the C-A+ mismatch if one occurs. Since the highest affinity CA mismatches are those with intermediate stability, it is likely that formation of a C-A+ pair would decrease affinity.
SUMMARY
The molecular recognition of KanHex by C•A internal loops was investigated by systematically mutating the loop nucleotides and closing base pairs and studying their affinities. Results revealed that the identity and orientation of the closing pairs affects binding of KanHex as do the identity and position of loop nucleotides. Interestingly, loops with intermediate thermodynamic stability bind most tightly to KanHex while those that are relatively more or less stable bind more weakly. Affinity is not correlated with the likelihood that a loop forms a C-A+ wobble pair at lower pH’s.
ACKNOWLEDGMENTS
We thank Jessica Childs-Disney for assistance with manuscript preparation. M.D.D. dedicates this work to Kaitlyn Disney.
This work was funded by the National Institutes of Health, R01-GM079235.
ABBREVIATIONS
- ADAR
adenosine deaminase acting on RNA
- A-site
aminoacyl-tRNA site
- BSA
bovine serum albumin
- DMSO
dimethyl sulfoxide
- dNTP
deoxyribonucleotide triphosphate
- HDV
Hepatitis Delta Virus
- HEPES
N-(2-hydroxyethyl)piperazine-N’-2-ethanesulfonic acid
- IL
internal loop
- KanHex
6′-N-5-hexynoate kanamycin A
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- RNA-PSP
RNA Privileged Space Predictor
- rRNA
ribosomal RNA
- RT-PCR
reverse transcriptase-polymerase chain reaction
- SDS
sodium dodecyl sulfate
- TBTA
tris(benzyltriazolylmethyl)amine
- Tris-HCl
tris(hydroxymethyl)aminomethane-hydrochloride
REFERENCES
- 1.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 2.Zaug AJ, Cech TR. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986;231:470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- 3.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 4.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci U S A. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 6.Gallego J, Varani G. Targeting RNA with small-molecule drugs: therapeutic promise and chemical challenges. Acc Chem Res. 2001;34:836–843. doi: 10.1021/ar000118k. [DOI] [PubMed] [Google Scholar]
- 7.von Ahsen U, Noller HF. Footprinting the sites of interaction of antibiotics with catalytic group I intron RNA. Science. 1993;260:1500–1503. doi: 10.1126/science.8502993. [DOI] [PubMed] [Google Scholar]
- 8.Chen G, Kennedy SD, Turner DH. A CA(+) pair adjacent to a sheared GA or AA pair stabilizes size-symmetric RNA internal loops. Biochemistry. 2009;48:5738–5752. doi: 10.1021/bi8019405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang SB, Hung LW, Chi YI, Holbrook EL, Carter RJ, Holbrook SR. Structure of an RNA internal loop consisting of tandem C-A+ base pairs. Biochemistry. 1998;37:11726–11731. doi: 10.1021/bi980758j. [DOI] [PubMed] [Google Scholar]
- 11.Pan B, Mitra SN, Sundaralingam M. Structure of a 16-mer RNA duplex r(GCAGACUUAAAUCUGC)2 with wobble C.A+ mismatches. J Mol Biol. 1998;283:977–984. doi: 10.1006/jmbi.1998.2140. [DOI] [PubMed] [Google Scholar]
- 12.Varani G, McClain WH. The G × U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong SK, Sato S, Lazinski DW. Substrate recognition by ADAR1 and ADAR2. RNA. 2001;7:846–858. doi: 10.1017/s135583820101007x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bass BL. RNA editing and hypermutation by adenosine deamination. Trends Biochem Sci. 1997;22:157–162. doi: 10.1016/s0968-0004(97)01035-9. [DOI] [PubMed] [Google Scholar]
- 15.Das AK, Carmichael GG. ADAR editing wobbles the microRNA world. ACS Chem Biol. 2007;2:217–220. doi: 10.1021/cb700064h. [DOI] [PubMed] [Google Scholar]
- 16.Casey JL. Hepatits Delta Virus. Springer-Verlag; Berlin: 2006. RNA editing in hepatitis delta virus; pp. 67–89. [DOI] [PubMed] [Google Scholar]
- 17.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 18.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB, Niswender CM, Copeland SC, Herrick-Davis K. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 19.Morabito MV, Abbas AI, Hood JL, Kesterson RA, Jacobs MM, Kump DS, Hachey DL, Roth BL, Emeson RB. Mice with altered serotonin 2C receptor RNA editing display characteristics of Prader-Willi syndrome. Neurobiol Dis. 2010;39:169–180. doi: 10.1016/j.nbd.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morabito MV, Emeson RB. RNA editing as a therapeutic target for CNS disorders. Neuropsychopharmacology. 2009;34:246. doi: 10.1038/npp.2008.159. [DOI] [PubMed] [Google Scholar]
- 21.Wittenhagen LM, Kelley SO, Wittenhagen LM, Roy MD, Kelley SO. Dimerization of a pathogenic human mitochondrial tRNA. Nat Struct Biol. 2002;9:586–590. doi: 10.1038/nsb820. [DOI] [PubMed] [Google Scholar]
- 22.Roy MD, Wittenhagen LM, Kelley SO. Structural probing of a pathogenic tRNA dimer. Rna. 2005;11:254–260. doi: 10.1261/rna.7143305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wittenhagen LM, Kelley SO. Impact of disease-related mitochondrial mutations on tRNA structure and function. Trends Biochem Sci. 2003;28:605–611. doi: 10.1016/j.tibs.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Zifa E, Giannouli S, Theotokis P, Stamatis C, Mamuris Z, Stathopoulos C, Ling J, Roy H, Qin D, Rubio MA, Alfonzo JD, Fredrick K, Ibba M, Wani AA, Ahanger SH, Bapat SA, Rangrez AY, Hingankar N, Suresh CG, Barnabas S, Patole MS, Shouche YS. Mitochondrial tRNA mutations: clinical and functional perturbations. RNA Biol. 2007;4:38–66. doi: 10.4161/rna.4.1.4548. [DOI] [PubMed] [Google Scholar]
- 25.Disney MD, Lee MM, Pushechnikov A, Childs-Disney JL. The role of flexibility in the rational design of modularly assembled ligands targeting the RNAs that cause the myotonic dystrophies. Chembiochem. 2010;11:375–382. doi: 10.1002/cbic.200900716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, Disney MD. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J Am Chem Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee MM, Childs-Disney JL, Pushechnikov A, French JM, Sobczak K, Thornton CA, Disney MD. Controlling the specificity of modularly assembled small molecules for RNA via ligand module spacing: targeting the RNAs that cause myotonic muscular dystrophy. J Am Chem Soc. 2009;131:17464–17472. doi: 10.1021/ja906877y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee MM, Pushechnikov A, Disney MD. Rational and modular design of potent ligands targeting the RNA that causes myotonic dystrophy 2. ACS Chem Biol. 2009;4:345–355. doi: 10.1021/cb900025w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Disney MD, Childs-Disney JL. Using selection to identify and chemical microarray to study the RNA internal loops recognized by 6′-N-acylated kanamycin A. Chembiochem. 2007;8:649–656. doi: 10.1002/cbic.200600569. [DOI] [PubMed] [Google Scholar]
- 30.Childs-Disney JL, Wu M, Pushechnikov A, Aminova O, Disney MD. A small molecule microarray platform to select RNA internal loop-ligand interactions. ACS Chem Biol. 2007;2:745–754. doi: 10.1021/cb700174r. [DOI] [PubMed] [Google Scholar]
- 31.Kierzek R, Burkard ME, Turner DH. Thermodynamics of single mismatches in RNA duplexes. Biochemistry. 1999;38:14214–14223. doi: 10.1021/bi991186l. [DOI] [PubMed] [Google Scholar]
- 32.Davis AR, Znosko BM. Thermodynamic characterization of single mismatches found in naturally occurring RNA. Biochemistry. 2007;46:13425–13436. doi: 10.1021/bi701311c. [DOI] [PubMed] [Google Scholar]
- 33.Davis AR, Kirkpatrick CC, Znosko BM. Structural characterization of naturally occurring RNA single mismatches. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq793. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis AR, Znosko BM. Positional and neighboring base pair effects on the thermodynamic stability of RNA single mismatches. Biochemistry. 2010 doi: 10.1021/bi100146z. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peyret N, Seneviratne PA, Allawi HT, SantaLucia J., Jr. Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 36.SantaLucia J., Jr. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci U S A. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puglisi JD, Tinoco I., Jr. Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Rando RR. Specific binding of aminoglycoside antibiotics to RNA. Chem Biol. 1995;2:281–290. doi: 10.1016/1074-5521(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 39.McDowell JA, Turner DH. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996;35:14077–14089. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- 40.Xia T, SantaLucia J, Jr., Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 41.Petersheim M, Turner DH. Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry. 1983;22:256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- 42.Borer PN, Dengler B, Tinoco I, Jr., Uhlenbeck OC. Stability of ribonucleic acid double-stranded helices. J Mol Biol. 1974;86:843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- 43.Gralla J, Crothers DM. Free energy of imperfect nucleic acid helices. 3. Small internal loops resulting from mismatches. J Mol Biol. 1973;78:301–319. doi: 10.1016/0022-2836(73)90118-6. [DOI] [PubMed] [Google Scholar]
- 44.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 45.Tok JB, Cho J, Rando RR. Aminoglycoside antibiotics are able to specifically bind the 5′-untranslated region of thymidylate synthase messenger RNA. Biochemistry. 1999;38:199–206. doi: 10.1021/bi9819428. [DOI] [PubMed] [Google Scholar]
- 46.Wong CH, Hendrix M, Priestley ES, Greenberg WA. Specificity of aminoglycoside antibiotics for the A-site of the decoding region of ribosomal RNA. Chem Biol. 1998;5:397–406. doi: 10.1016/s1074-5521(98)90073-4. [DOI] [PubMed] [Google Scholar]
- 47.Tor Y, Hermann T, Westhof E. Deciphering RNA recognition: aminoglycoside binding to the hammerhead ribozyme. Chem Biol. 1998;5:R277–283. doi: 10.1016/s1074-5521(98)90286-1. [DOI] [PubMed] [Google Scholar]
- 48.Aminova O, Paul DJ, Childs-Disney JL, Disney MD. Two-dimensional combinatorial screening identifies specific 6′-acylated kanamycin A- and 6′-acylated neamine-RNA hairpin interactions. Biochemistry. 2008;47:12670–12679. doi: 10.1021/bi8012615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Disney MD, Labuda LP, Paul DJ, Poplawski SG, Pushechnikov A, Tran T, Velagapudi SP, Wu M, Childs-Disney JL. Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J Am Chem Soc. 2008;130:11185–11194. doi: 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- 50.Paul DJ, Seedhouse SJ, Disney MD. Two-dimensional combinatorial screening and the RNA Privileged Space Predictor program efficiently identify aminoglycoside-RNA hairpin loop interactions. Nucleic Acids Res. 2009;37:5894–5907. doi: 10.1093/nar/gkp594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tran T, Disney MD. Two-dimensional combinatorial screening of a bacterial rRNA A-site-like motif library: defining privileged asymmetric internal loops that bind aminoglycosides. Biochemistry. 2010;49:1833–1842. doi: 10.1021/bi901998m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Legault P, Pardi A. Unusual dynamics and pKa shift at the active site of a lead-dependent ribozyme. J Am Chem Soc. 1997;119:6621–6628. [Google Scholar]
- 53.Guo M, Spitale RC, Volpini R, Krucinska J, Cristalli G, Carey PR, Wedekind JE. Direct Raman measurement of an elevated base pKa in the active site of a small ribozyme in a precatalytic conformation. J Am Chem Soc. 2009;131:12908–12909. doi: 10.1021/ja9060883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakano S, Chadalavada DM, Bevilacqua PC. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 55.Wilson TJ, Li NS, Lu J, Frederiksen JK, Piccirilli JA, Lilley DM. Nucleobase-mediated general acid-base catalysis in the Varkud satellite ribozyme. Proc Natl Acad Sci U S A. 2010;107:11751–11756. doi: 10.1073/pnas.1004255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaul M, Barbieri CM, Kerrigan JE, Pilch DS. Coupling of drug protonation to the specific binding of aminoglycosides to the A site of 16 S rRNA: elucidation of the number of drug amino groups involved and their identities. J Mol Biol. 2003;326:1373–1387. doi: 10.1016/s0022-2836(02)01452-3. [DOI] [PubMed] [Google Scholar]