Abstract
Genome-wide association studies have identified genetic markers in kallikrein-related peptidase 3 (KLK3) associated with prostate cancer. However, some of these markers are also associated with prostate-specific antigen (PSA) levels, so it is unclear whether the polymorphisms are causal or if the association with risk is solely due to detection bias through PSA screening. PSA is a biologically active serine protease, cleaving insulin-like growth factor-binding protein. We examined the association of single-nucleotide polymorphisms (SNPs) in KLK3 with prostate cancer risk, disease-specific survival and pre-diagnostic PSA levels in a case–control study nested within the Physicians’ Health Study, which began in 1982, with over 27 years of follow-up. We genotyped SNPs spanning the entire KLK3 locus to capture common variation at high resolution. Six polymorphisms were significantly associated with prostate cancer incidence (P < 0.05); the odds ratios per minor allele ranged from 0.88 to 0.73. For four of these, the odds ratios were lower when restricting to cases diagnosed in the pre-PSA screening era (before 1989). The four alleles significantly associated with lower PSA levels were also associated with lower prostate cancer risk. KLK3 variants were not significantly associated with stage at diagnosis, risk of lethal cancer or survival. Our results suggest that detection bias due to the association of KLK3 variants with PSA levels cannot completely explain the association with prostate cancer risk. Understanding the mechanism by which genetic variation in KLK3 affects prostate cancer risk has important implications for study of the biological role of PSA in prostate tumorigenesis.
Introduction
Prostate-specific antigen (PSA), encoded by the kallikrein-related peptidase 3 (KLK3) gene, is a serine protease produced by epithelial cells in the prostate. PSA is produced almost exclusively by the prostate and circulating levels increase in the presence of cancer (1); this latter association led to the development of PSA as a screening biomarker and an indicator of disease progression. However, PSA is also influenced by age, prostate size, inflammation and infection.
Recent genome-wide association studies (GWAS) identified numerous genetic regions associated with prostate cancer risk (2–11), including the locus on chromosome 19 containing KLK3. The minor allele of the single-nucleotide polymorphism (SNP) rs2735839 in this region was associated with a 17–46% decreased risk of prostate cancer (P = 1.5 × 10−18); Eeles et al. (8) initially reported these results in a multistage design in which stage 1 of the study specifically excluded controls with PSA levels >0.5 ng/ml. The goal of this sampling strategy was to decrease the possibility of including controls with occult prostate cancer, but this design raised the question of whether this genetic variant was truly a prostate cancer risk factor or merely associated with the likelihood of diagnosis by virtue of its link with PSA levels. Several groups have observed an association between rs2735839 and PSA levels (12–14). Ahn et al. (12) genotyped tag SNPs across KLK3 and observed no association with prostate cancer risk in several populations; they only saw an association of rs2735839 when they artificially restricted controls to those with PSA levels <0.5 ng/ml. Eeles et al. responded by noting that although the association may be partly attributed to an association with lower PSA level, they also observed the association in the second stage of their study when controls were not selected based on PSA (15). Additional case–control studies have observed associations of KLK3 variation with prostate cancer risk (16–20) and mortality (21), though association of SNPs with PSA levels have also been observed (22,23). The reported associations of KLK3 SNPs with prostate cancer may be different across studies due to differences in the screening behavior of participants and related differences in PSA levels of cases and non-cases, the selection of controls or the play of chance.
Because some of the SNPs are clearly associated with PSA levels, their association with prostate cancer risk could solely be due to detection bias with PSA screening. However, PSA has a biological role beyond simply serving as a screening tool for prostate cancer. The association of these SNPs with PSA levels demonstrates their functionality or high correlation with functional SNPs. It is possible that these SNPs may affect PSA levels and that PSA activity is involved in biologically driving prostate cancer development or may also affect prostate cancer initiation through an independent mechanism. PSA degrades insulin-like growth factor-binding protein (IGFBP) 5 (24) and cleaves IGFBP3, which reduces its affinity for insulin-like growth factor (IGF)-1 (25); both mechanisms would increase free IGF-1 levels (26). We and others have shown that total IGF-1 levels are associated with risk of advanced prostate cancer (27). We therefore analyzed the association between SNPs in the KLK3 gene locus and prostate cancer risk and survival in the Physicians’ Health Study (PHS), considering pre-diagnostic PSA from study baseline in 1982.
Materials and methods
Study population
The PHS began as a randomized, double-blind placebo-controlled trial of aspirin and β-carotene for the prevention of cardiovascular disease and cancer among 22 071 healthy United States physicians. Men were excluded at baseline if they had any serious medical conditions including all cancers (except non-melanoma skin cancer). Blood samples were collected from 68% of the physicians at baseline in 1982–84. A detailed description of PHS has been published previously (28). Participants are followed through annual questionnaires to collect data on diet, health and lifestyle behaviors and medical history and biannually through postcards to ascertain health end points, including prostate cancer. All self-reported prostate cancer cases are verified through medical record and pathology review. Through this systematic medical record review, we abstract data on PSA at diagnosis, tumor stage and Gleason score. Death certificates and records are obtained and reviewed to determine cause of all deaths. The follow-up rate for cancer incidence is 96% and for mortality is 98%. Metastases are reported on follow-up questionnaires sent to all men diagnosed with prostate cancer and confirmed by medical record review.
For this study, we utilized a nested case–control design, with controls selected through risk-set sampling and matched to cases on age (±1 year for cases ≤55 and, if necessary, within 5 years for older cases), smoking status (never, former, current) and follow-up time. We restricted participants to self-reported Caucasians to reduce potential population stratification. In total, 1030 cases (diagnosed from 1982–2005) and 1327 controls were included for the case–control analysis. In the case-only survival analysis, we included an additional 83 prostate cancer cases diagnosed after being matched as controls in the case–control analysis (n = 1113).
Written consent was obtained from each participant at the time of initial enrollment and this investigation was approved by the Human Subjects Committee at Brigham and Women’s Hospital.
SNP selection and genotyping
We identified bona fide and suitably informative SNPs from among those represented in independently derived SNP databases prior to the completion of The International HapMap Project, namely the open access NCBI dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) and Japanese SNP (http://snp.ims.u-tokyo.ac.jp/) databases as well as the Celera SNP Reference Database (29). To ensure the selection of validated SNPs, we only chose SNPs represented in the NCBI dbSNP database that are also independently represented in at least one of the other two databases. SNPs distributed throughout the KLK3 gene locus were identified at an average resolution of approximately 2–3 SNPs/kb. The criteria for SNP selection included an expected minor allele frequency > 10% among the Caucasian men represented in the study cohort. In total, 29 SNPs were genotyped using the Sequenom technology at The Broad Institute Center for Genotyping and Analysis (http://www.broad.mit.edu/genotyping/). Six SNPs were excluded for low genotyping completion (<85%). The data completion rate was 94.2% for the 23 SNPs included in the current analyses. They span the entire KLK3 gene locus, including 5850 bp encoding the 5′ transcriptional initiation site to the 3′ polyadenylation site that defines the intron/exon organization of the ‘classical’ KLK3-201 messenger RNA transcript of 1464 bp (Ensembl Transcript ID ENST00000326003) encoding the 261 amino acid sequence of PSA, as well as additional upstream promoter sequences (4611 bp from rs925013 to the 5′ transcriptional initiation site) and downstream sequences (886 bp from the 3′ polyadenylation site to rs2569735). All known predicted or confirmed KLK3 messenger RNA variant transcripts generated by alternative splicing, exon truncation, cryptic exons and intron retention are derived from sequences within this >10 kb region (Ensembl Gene ID ENSG00000142515) (30,31).
PSA and IGF-1 measurement
PSA levels were previously measured on archived blood samples, collected in 1982; intra-batch coefficient of variation was 10.4% and inter-batch coefficient of variation was 16%, as described in Gann et al. (32). To assess the potential impact of the SNPs on IGF-1 levels, we analyzed previously assayed plasma samples for IGF as previously reported in Chan et al. (27).
Statistical analysis
Genotypes were coded in a codominant fashion to estimate separate odds ratios for heterozygotes and homozygote carriers of the minor allele and in an additive fashion (assuming a multiplicative increase in the odds of disease per minor allele carried) to calculate a one degree of freedom test of association. Using a nested case–control design, we assessed the risk of incident prostate cancer using unconditional logistic regression models, adjusting for the matching factors (age, smoking status and duration of follow-up). We corrected for multiple testing, considering the linkage disequilibrium (LD) between SNPs, using the method described in Conneely and Boehnke (33), using the package P-ACT in the R statistical software. We additionally performed logistic regression comparing cases diagnosed before 1989 to controls (‘pre-1989 risk’) and cases diagnosed after 1991 to controls (‘post-1991 risk’). Among prostate cancer cases, we performed an analysis of time to lethal prostate cancer outcome using Cox regression models adjusting for age at diagnosis. Lethal prostate cancer was defined as death due to prostate cancer (n = 168) or the development of bony metastases (n = 11); follow-up began at the time of prostate cancer diagnosis and individuals were censored at the time of death from another cause or the end of follow-up (5 March 2010).
We used analysis of variance to compare the genotypes (three categories) to baseline (log-transformed) PSA levels and reported the P-value for trend. This analysis was performed for all individuals who never developed prostate cancer during follow-up (non-cases). In prostate cancer cases, the same method was used to compare genotypes with reported PSA levels at diagnosis. We also used analysis of variance to examine the association of genotypes with total IGF-1 levels in cases and non-cases. We limited the IGF-1 analysis to SNPs that were significantly associated with risk or PSA levels (P < 0.05). Analyses were performed with SAS version 9.1 statistical software; all P-values are two sided.
Results
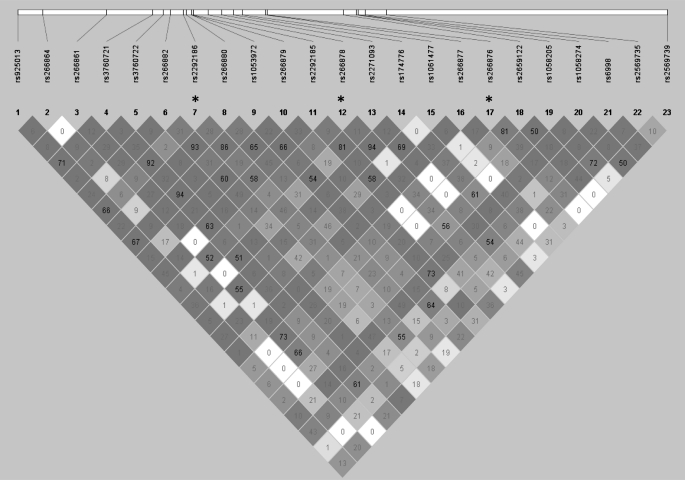
Characteristics of the baseline and clinical characteristics of the PHS cases and controls are presented in Table I. We genotyped 29 SNPs dispersed across the entire KLK3 gene; six SNPs were excluded for low genotyping completion (<85%), so 23 were included in the analysis (Figure 1). Three of the SNPs violated Hardy–Weinberg Equilibrium in the control subjects (rs2292186, rs266876, rs266878; P < 0.0001); results for these SNPs were included but should be interpreted with caution.
Table I.
Characteristics of prostate cancer cases and controls in the PHS
| Case–control incidence analysis | Cases (n = 1030) | Controls (n = 1327) |
| Age at study onset, mean ± SD | 59.0 ± 8.3 | 59.3 ± 8.0 |
| Gleason score, n (%) | ||
| 2–6 | 478 (52.3) | |
| 7 | 296 (32.4) | |
| 8–10 | 140 (15.3) | |
| Clinical stage, n (%) | ||
| T1, T2 | 849 (86.9) | |
| T3, T4, N1, M1 | 128 (13.1) | |
| PSA at baseline (μg/l), median (10th−90th)a | 3.0 (1.0–16.1) | 1.1 (0.40–3.5) |
| Case-only mortality analysis | Cases (n = 1113) | |
| Age at diagnosis, mean ± SD | 69.9 ± 7.5 | |
| PSA at diagnosis (μg/l), median (10th−90th)b | 7.4 (3.8–30.7) | |
| Deaths/metastases due to PCa prostate cancer, n (%) | 179 (16.1) | |
| Diagnosis to PCa prostate cancer death/mets, median years (range) | 5.9 (0.3–21.0) | |
| Follow-up time to censored, median years (range) | 12.7 (0.01–27.2) |
mets, metastases, PCa, prostate cancer.
PSA at baseline was measured in 591 cases and 702 controls who never developed cancer.
PSA at diagnosis is available for 763 cases.
Fig. 1.
Relative positions of SNPs spanning the KLK3 locus and their LD. Asterisks represent SNPs with Hardy–Weinberg Equilibrium < 0.0001. Darker color indicates D′ = 1; numbers in diamonds are the R2 between SNPs.
Six polymorphisms were significantly associated with prostate cancer incidence (P < 0.05; Table II), with odds ratios ranging from 0.73 to 0.88 for each additional copy of the minor allele. However, after adjusting for multiple correlated tests, no SNP remained statistically significantly associated with prostate cancer incidence.
Table II.
Allele frequency of KLK3 SNPs in prostate cancer cases and controls and association with prostate cancer incidence
| SNP | Genotype | Cases, n (%) | Controls, n (%) | OR (95% CI) | Pre-1989 cases, n (%) | OR (95% CI)a | Post-1991 cases, n (%) | OR (95% CI)b | |
| 1 | rs925013 | TT | 591 (59.2) | 801 (62.3) | 1.00 (ref) | 103 (62.8) | 1.00 (ref) | 356 (58.7) | 1.00 (ref) |
| CT | 362 (36.2) | 422 (32.8) | 1.15 (0.97–1.38) | 54 (32.9) | 1.05 (0.70–1.59) | 227 (37.4) | 1.23 (0.98–1.54) | ||
| CC | 46 (4.6) | 62 (4.8) | 1.01 (0.68–1.50) | 7 (4.3) | 0.85 (0.34–2.16) | 24 (4.0) | 0.85 (0.50–1.47) | ||
| P-value | 0.26 | 0.97 | 0.33 | ||||||
| 2 | rs266864 | GG | 644 (65.6) | 819 (63.6) | 1.00 (ref) | 101 (63.1) | 1.00 (ref) | 394 (66.2) | 1.00 (ref) |
| AG | 301 (30.7) | 411 (31.9) | 0.95 (0.79–1.14) | 54 (33.8) | 0.85 (0.56–1.28) | 177 (29.8) | 0.97 (0.76–1.22) | ||
| AA | 37 (3.8) | 58 (4.5) | 0.81 (0.53–1.25) | 5 (3.1) | 0.57 (0.19–1.69) | 24 (4.0) | 0.93 (0.55–1.59) | ||
| P-value | 0.33 | 0.24 | 0.72 | ||||||
| 3 | rs266861 | TT | 577 (58.1) | 744 (57.8) | 1.00 (ref) | 105 (64.8) | 1.00 (ref) | 336 (55.4) | 1.00 (ref) |
| CT | 374 (37.6) | 457 (35.5) | 1.03 (0.87–1.23) | 50 (30.9) | 1.01 (0.66–1.53) | 246 (40.5) | 1.17 (0.94–1.46) | ||
| CC | 43 (4.3) | 86 (6.7) | 0.64 (0.43–0.94) | 7 (4.3) | 0.49 (0.20–1.20) | 25 (4.1) | 0.61 (0.37–1.01) | ||
| P-value | 0.22 | 0.29 | 0.79 | ||||||
| 4 | rs3760721 | CC | 507 (50.6) | 689 (53.4) | 1.00 (ref) | 86 (52.8) | 1.00 (ref) | 311 (50.9) | 1.00 (ref) |
| TC | 423 (42.2) | 505 (39.1) | 1.14 (0.96–1.36) | 65 (39.9) | 0.91 (0.61–1.36) | 260 (42.6) | 1.20 (0.96–1.50) | ||
| TT | 72 (7.2) | 97 (7.5) | 1.01 (0.73–1.40) | 12 (7.4) | 0.89 (0.42–1.87) | 40 (6.6) | 0.88 (0.57–1.36) | ||
| P-value | 0.35 | 0.63 | 0.52 | ||||||
| 5 | rs3760722 | CC | 791 (80.5) | 1066 (82.6) | 1.00 (ref) | 134 (83.2) | 1.00 (ref) | 471 (79.2) | 1.00 (ref) |
| TC | 181 (18.4) | 214 (16.6) | 1.12 (0.90–1.40) | 25 (15.5) | 1.15 (0.68–1.95) | 117 (19.7) | 1.22 (0.93–1.60) | ||
| TT | 11 (1.1) | 10 (0.8) | 1.45 (0.61–3.44) | 2 (1.2) | 2.13 (0.33–13.67) | 7 (1.2) | 1.50 (0.52–4.32) | ||
| P-value | 0.2 | 0.43 | 0.12 | ||||||
| 6 | rs266882 | GG | 255 (26.4) | 346 (27.1) | 1.00 (ref) | 39 (24.5) | 1.00 (ref) | 152 (26.1) | 1.00 (ref) |
| AG | 483 (50.0) | 622 (48.6) | 1.06 (0.86–1.29) | 73 (45.9) | 0.95 (0.59–1.53) | 306 (52.5) | 1.18 (0.91–1.52) | ||
| AA | 228 (23.6) | 311 (24.3) | 1.01 (0.80–1.28) | 47 (29.6) | 1.39 (0.82–2.37) | 125 (21.4) | 0.92 (0.68–1.26) | ||
| P-value | 0.9 | 0.22 | 0.7 | ||||||
| 7 | rs2292186 | AA | 500 (53.4) | 676 (55.6) | 1.00 (ref) | 86 (56.6) | 1.00 (ref) | 306 (53.4) | 1.00 (ref) |
| AG | 339 (36.2) | 411 (33.8) | 1.11 (0.92–1.34) | 49 (32.2) | 0.86 (0.55–1.33) | 211 (36.8) | 1.13 (0.89–1.43) | ||
| AA | 98 (10.5) | 130 (10.7) | 1.03 (0.78–1.38) | 17 (11.2) | 0.85 (0.45–1.62) | 56 (9.8) | 1.06 (0.73–1.55) | ||
| P-value | 0.48 | 0.48 | 0.45 | ||||||
| 8 | rs266880 | CC | 276 (28.6) | 368 (29.1) | 1.00 (ref) | 42 (26.1) | 1.00 (ref) | 164 (28.4) | 1.00 (ref) |
| GC | 480 (49.7) | 625 (49.5) | 1.02 (0.84–1.25) | 77 (47.8) | 1.02 (0.64–1.63) | 299 (51.7) | 1.10 (0.86–1.42) | ||
| GG | 209 (21.7) | 271 (21.4) | 1.05 (0.82–1.33) | 42 (26.1) | 1.39 (0.81–2.39) | 115 (19.9) | 0.94 (0.69–1.29) | ||
| P-value | 0.71 | 0.25 | 0.8 | ||||||
| 9 | rs1053972 | CC | 482 (49.7) | 665 (53.4) | 1.00 (ref) | 83 (52.5) | 1.00 (ref) | 298 (50.3) | 1.00 (ref) |
| CT | 412 (42.5) | 485 (39.0) | 1.17 (0.98–1.40) | 63 (39.9) | 0.87 (0.58–1.31) | 253 (42.7) | 1.20 (0.96–1.51) | ||
| TT | 76 (7.8) | 95 (7.6) | 1.10 (0.80–1.52) | 12 (7.6) | 0.92 (0.44–1.94) | 42 (7.1) | 0.91 (0.59–1.41) | ||
| P-value | 0.15 | 0.6 | 0.49 | ||||||
| 10 | rs266879 | CC | 401 (40.1) | 525 (40.8) | 1.00 (ref) | 63 (38.7) | 1.00 (ref) | 244 (40.1) | 1.00 (ref) |
| GC | 469 (47.0) | 572 (44.4) | 1.07 (0.90–1.28) | 72 (44.2) | 1.02 (0.67–1.55) | 288 (47.4) | 1.11 (0.88–1.39) | ||
| GG | 129 (12.9) | 191 (14.8) | 0.90 (0.70–1.17) | 28 (17.2) | 1.14 (0.65–2.01) | 76 (12.5) | 0.88 (0.63–1.23) | ||
| P-value | 0.75 | 0.69 | 0.8 | ||||||
| 11 | rs2292185 | GG | 345 (35.7) | 498 (40.1) | 1.00 (ref) | 65 (41.4) | 1.00 (ref) | 206 (35.0) | 1.00 (ref) |
| AG | 471 (48.7) | 569 (45.8) | 1.19 (0.99–1.43) | 69 (44.0) | 0.82 (0.54–1.25) | 293 (49.8) | 1.27 (1.00–1.61) | ||
| AA | 151 (15.6) | 176 (14.2) | 1.22 (0.94–1.58) | 23 (14.7) | 1.10 (0.61–2.00) | 89 (15.1) | 1.17 (0.84–1.63) | ||
| P-value | 0.07 | 0.92 | 0.16 | ||||||
| 12 | rs266878 | CC | 711 (77.3) | 892 (73.7) | 1.00 (ref) | 122 (80.3) | 1.00 (ref) | 422 (76.3) | 1.00 (ref) |
| GC | 183 (19.9) | 266 (22.0) | 0.87 (0.70–1.07) | 29 (19.1) | 0.84 (0.51–1.37) | 113 (20.4) | 0.98 (0.74–1.28) | ||
| GG | 26 (2.8) | 53 (4.4) | 0.60 (0.37–0.98) | 1 (0.7) | 0.11 (0.01–0.86) | 18 (3.3) | 0.67 (0.37–1.21) | ||
| P-value | 0.02 | 0.04 | 0.32 | ||||||
| 13 | rs2271093 | TT | 314 (32.2) | 444 (35.4) | 1.00 (ref) | 56 (35.7) | 1.00 (ref) | 186 (31.2) | 1.00 (ref) |
| CT | 475 (48.7) | 596 (47.5) | 1.13 (0.93–1.36) | 70 (44.6) | 0.70 (0.45–1.09) | 300 (50.3) | 1.26 (0.99–1.60) | ||
| CC | 187 (19.2) | 214 (17.1) | 1.23 (0.96–1.57) | 31 (19.8) | 1.25 (0.72–2.17) | 111 (18.6) | 1.19 (0.87–1.63) | ||
| P-value | 0.08 | 0.81 | 0.16 | ||||||
| 14 | rs174776 | TT | 742 (75.0) | 915 (71.9) | 1.00 (ref) | 123 (78.3) | 1.00 (ref) | 445 (73.3) | 1.00 (ref) |
| CT | 227 (23.0) | 319 (25.1) | 0.86 (0.71–1.05) | 33 (12.0) | 0.82 (0.51–1.32) | 149 (24.6) | 0.95 (0.74–1.21) | ||
| TT | 20 (2.0) | 38 (3.0) | 0.64 (0.37–1.12) | 1 (0.6) | 0.14 (0.02–1.12) | 13 (2.1) | 0.69 (0.34–1.38) | ||
| P-value | 0.045 | 0.06 | 0.36 | ||||||
| 15 | rs1061477 | TT | 256 (26.7) | 312 (24.7) | 1.00 (ref) | 42 (26.6) | 1.00 (ref) | 151 (26.2) | 1.00 (ref) |
| CT | 457 (47.6) | 608 (48.2) | 0.92 (0.75–1.13) | 70 (44.3) | 0.65 (0.40–1.05) | 284 (49.2) | 1.00 (0.77–1.30) | ||
| CC | 247 (25.7) | 342 (27.1) | 0.89 (0.71–1.13) | 46 (29.1) | 0.84 (0.50–1.42) | 142 (24.6) | 0.90 (0.67–1.22) | ||
| P-value | 0.34 | 0.56 | 0.51 | ||||||
| 16 | rs266877 | GG | 898 (91.5) | 1115 (88.6) | 1.00 (ref) | 147 (92.5) | 1.00 (ref) | 543 (90.5) | 1.00 (ref) |
| AG | 82 (8.4) | 138 (11.0) | 0.73 (0.55–0.98) | 12 (7.6) | 0.50 (0.25–1.02) | 56 (9.3) | 0.88 (0.62–1.27) | ||
| AA | 2 (0.2) | 5 (0.4) | 0.49 (0.09–2.52) | 0 (0.0) | ––– | 1 (0.2) | 0.34 (0.04–3.06) | ||
| P-value | 0.02 | 0.05 | 0.32 | ||||||
| 17 | rs266876 | TT | 540 (57.5) | 668 (55.4) | 1.00 (ref) | 98 (64.9) | 1.00 (ref) | 311 (54.4) | 1.00 (ref) |
| CT | 328 (34.9) | 419 (34.8) | 0.96 (0.80–1.15) | 43 (28.5) | 0.79 (0.50–1.22) | 219 (38.3) | 1.18 (0.93–1.49) | ||
| CC | 71 (7.6) | 118 (9.8) | 0.74 (0.54–1.02) | 10 (6.6) | 0.42 (0.19–0.92) | 42 (7.3) | 0.75 (0.50–1.13) | ||
| P-value | 0.12 | 0.03 | 0.82 | ||||||
| 18 | rs2659122 | AA | 544 (54.5) | 674 (52.4) | 1.00 (ref) | 97 (59.5) | 1.00 (ref) | 317 (52.1) | 1.00 (ref) |
| GA | 387 (38.7) | 504 (39.2) | 0.94 (0.79–1.12) | 55 (33.7) | 0.88 (0.59–1.33) | 249 (41.0) | 1.07 (0.85–1.33) | ||
| GG | 68 (6.8) | 109 (8.5) | 0.77 (0.55–1.06) | 11 (6.8) | 0.58 (0.27–1.25) | 42 (6.9) | 0.80 (0.53–1.21) | ||
| P-value | 0.14 | 0.18 | 0.7 | ||||||
| 19 | rs1058205 | TT | 706 (78.8) | 868 (74.5) | 1.00 (ref) | 111 (77.6) | 1.00 (ref) | 431 (78.2) | 1.00 (ref) |
| CT | 176 (19.6) | 269 (23.1) | 0.81 (0.65–1.01) | 27 (18.9) | 0.73 (0.44–1.21) | 115 (20.9) | 0.96 (0.73–1.25) | ||
| CC | 14 (1.6) | 28 (2.4) | 0.65 (0.34–1.26) | 5 (3.5) | 1.05 (0.33–3.39) | 5 (0.9) | 0.37 (0.14–1.02) | ||
| P-value | 0.03 | 0.38 | 0.2 | ||||||
| 20 | rs1058274 | AA | 473 (48.3) | 603 (47.7) | 1.00 (ref) | 70 (44.6) | 1.00 (ref) | 298 (49.8) | 1.00 (ref) |
| GA | 410 (41.9) | 521 (41.2) | 1.00 (0.84–1.20) | 66 (42.0) | 0.98 (0.65–1.49) | 247 (41.2) | 0.95 (0.76–1.20) | ||
| GG | 96 (9.8) | 141 (11.2) | 0.89 (0.66–1.18) | 21 (13.4) | 1.09 (0.58–2.04) | 54 (9.0) | 0.79 (0.54–1.15) | ||
| P-value | 0.55 | 0.87 | 0.26 | ||||||
| 21 | rs6998 | GG | 430 (43.6) | 587 (46.2) | 1.00 (ref) | 76 (48.4) | 1.00 (ref) | 264 (43.7) | 1.00 (ref) |
| AG | 438 (44.4) | 549 (43.2) | 1.09 (0.91–1.30) | 59 (37.6) | 0.77 (0.50–1.17) | 271 (44.9) | 1.13 (0.90–1.41) | ||
| AA | 118 (12.0) | 136 (10.7) | 1.19 (0.90–1.57) | 22 (14.0) | 1.58 (0.87–2.86) | 69 (11.4) | 1.11 (0.78–1.59) | ||
| P-value | 0.18 | 0.57 | 0.36 | ||||||
| 22 | rs2569735 | GG | 737 (76.0) | 894 (72.7) | 1.00 (ref) | 116 (74.8) | 1.00 (ref) | 452 (75.7) | 1.00 (ref) |
| AG | 211 (21.8) | 295 (24.0) | 0.86 (0.70–1.06) | 36 (23.2) | 0.97 (0.61–1.55) | 132 (22.1) | 0.88 (0.68–1.14) | ||
| AA | 22 (2.3) | 41 (3.3) | 0.64 (0.38–1.09) | 3 (1.9) | 0.56 (0.14–2.20) | 13 (2.2) | 0.58 (0.29–1.13) | ||
| P-value | 0.04 | 0.57 | 0.09 | ||||||
| 23 | rs2569739 | TT | 337 (34.4) | 390 (30.4) | 1.00 (ref) | 51 (31.9) | 1.00 (ref) | 209 (35.0) | 1.00 (ref) |
| CT | 457 (46.7) | 614 (47.9) | 0.86 (0.71–1.04) | 67 (41.9) | 0.69 (0.44–1.08) | 279 (46.7) | 0.86 (0.67–1.09) | ||
| CC | 185 (18.9) | 278 (21.7) | 0.77 (0.61–0.98) | 42 (26.3) | 1.07 (0.63–1.79) | 109 (18.3) | 0.70 (0.52–0.95) | ||
| P-value | 0.03 | 0.97 | 0.02 |
OR, odds ratio; 95% CI, 95% confidence interval.
OR (95% CI) and additive P-value comparing pre-1989 cases to all controls.
OR (95% CI) and additive P-value comparing post-1991 cases to all controls.
There is strong LD in this region; for example, the R2 is 0.96 between two of the SNPs associated with prostate cancer risk, rs266878 and rs174776. After conditioning on rs2569735, which is in strong LD with rs2735839 (R2 = 1.00 in the HapMap CEU panel), the most significant marker in the Eeles et al. (8) GWAS, none of the remaining SNP associations was statistically significant.
None of the KLK3 SNPs genotyped in our study was significantly associated with prostate cancer mortality (supplementary Table I is available at Carcinogenesis Online). SNPs were also not associated with clinical stage at diagnosis or with the incidence of lethal disease (data not shown).
Comparing cases diagnosed before 1989 in the pre-PSA era to all controls, the associations showed similar significance and more extreme effect sizes compared with the results from the overall risk analysis presented above (Table II). Even with decreased power due to a smaller sample size, the minor alleles of rs266878 and rs266876 were statistically significantly associated with a lower risk of prostate cancer in the pre-PSA era. We observed one significant association comparing cases diagnosed in 1992 or later to all controls (rs2569739); however, the magnitude of the odds ratios for all SNPs were qualitatively similar to the overall results (Table II).
Eleven SNPs were significantly associated with PSA levels measured in blood from baseline in control subjects who never developed prostate cancer through follow-up (Table III). The minor allele was associated with higher PSA levels for seven SNPs: rs925013, rs3760721, rs3760722, rs2292186, rs1053972, rs2292185 and rs2271093 (range between homozygote classes: 0.25–0.53), while the minor allele was associated with lower PSA levels for four: rs266878, rs174776, rs1058205 and rs2569735 (range between homozygote classes: −0.35 to −0.61 μg/l). None of the seven alleles associated with higher PSA levels were significantly related to prostate cancer risk, either overall or in analyses limited to the PSA era (odds ratios ranged from 0.85 to 1.50). In contrast, all four alleles associated with lower PSA levels were significantly associated with lower prostate cancer risk overall, with odds ratios of similar magnitude in analyses limited to pre-PSA era cases. After conditioning on rs2569735, the remaining three SNPs were no longer significantly associated with lower PSA levels.
Table III.
Mean PSA values and P-values (trend) for the association of KLK3 SNPs with baseline PSA in non-cases and PSA at diagnosis in prostate cancer cases
| SNP | Genotype | Mean baseline PSA (controls), μg/l | Mean diagnosis PSA, μg/l | |
| 1 | rs925013 | TT | 1.10 | 8.69 |
| CT | 1.21 | 9.06 | ||
| CC | 1.62 | 13.74 | ||
| Ptrend | 0.02 | 0.09 | ||
| 2 | rs266864 | GG | 1.19 | 9.18 |
| AG | 1.11 | 8.67 | ||
| AA | 0.95 | 7.24 | ||
| Ptrend | 0.14 | 0.24 | ||
| 3 | rs266861 | TT | 1.19 | 9.18 |
| CT | 1.15 | 8.79 | ||
| CC | 0.91 | 7.00 | ||
| Ptrend | 0.14 | 0.23 | ||
| 4 | rs3760721 | CC | 1.09 | 8.91 |
| TC | 1.19 | 8.81 | ||
| TT | 1.45 | 10.72 | ||
| Ptrend | 0.03 | 0.51 | ||
| 5 | rs3760722 | CC | 1.10 | 8.67 |
| TC | 1.40 | 9.89 | ||
| TT | 1.50 | 12.74 | ||
| Ptrend | 0.006 | 0.09 | ||
| 6 | rs266882 | GG | 1.12 | 8.53 |
| AG | 1.22 | 9.23 | ||
| AA | 1.05 | 8.57 | ||
| Ptrend | 0.51 | 0.94 | ||
| 7 | rs2292186 | AA | 1.09 | 8.89 |
| AG | 1.19 | 9.31 | ||
| AA | 1.34 | 9.86 | ||
| Ptrend | 0.05 | 0.38 | ||
| 8 | rs266880 | CC | 1.11 | 8.65 |
| GC | 1.24 | 9.16 | ||
| GG | 1.01 | 8.69 | ||
| Ptrend | 0.48 | 0.90 | ||
| 9 | rs1053972 | CC | 1.09 | 8.83 |
| CT | 1.20 | 8.91 | ||
| TT | 1.49 | 10.59 | ||
| Ptrend | 0.01 | 0.37 | ||
| 10 | rs266879 | CC | 1.17 | 9.18 |
| GC | 1.19 | 8.93 | ||
| GG | 0.98 | 7.80 | ||
| Ptrend | 0.19 | 0.24 | ||
| 11 | rs2292185 | GG | 0.99 | 8.36 |
| AG | 1.21 | 8.93 | ||
| AA | 1.37 | 10.59 | ||
| Ptrend | 0.0004 | 0.05 | ||
| 12 | rs266878 | CC | 1.23 | 9.38 |
| GC | 0.97 | 8.47 | ||
| GG | 0.77 | 6.00 | ||
| Ptrend | 0.0003 | 0.05 | ||
| 13 | rs2271093 | TT | 0.98 | 8.20 |
| CT | 1.24 | 9.18 | ||
| CC | 1.34 | 10.05 | ||
| Ptrend | 0.001 | 0.06 | ||
| 14 | rs174776 | TT | 1.23 | 9.40 |
| CT | 1.01 | 7.83 | ||
| TT | 0.62 | 5.41 | ||
| Ptrend | 0.0001 | 0.01 | ||
| 15 | rs1061477 | TT | 1.17 | 9.44 |
| CT | 1.20 | 8.51 | ||
| CC | 1.03 | 9.12 | ||
| Ptrend | 0.18 | 0.75 | ||
| 16 | rs266877 | GG | 1.16 | 8.87 |
| AG | 0.96 | 8.51 | ||
| AA | 2.04 | 10.00 | ||
| Ptrend | 0.13 | 0.78 | ||
| 17 | rs266876 | TT | 1.15 | 9.29 |
| CT | 1.23 | 8.65 | ||
| CC | 0.96 | 8.71 | ||
| Ptrend | 0.57 | 0.40 | ||
| 18 | rs2659122 | AA | 1.16 | 9.33 |
| GA | 1.17 | 8.61 | ||
| GG | 0.98 | 7.78 | ||
| Ptrend | 0.37 | 0.15 | ||
| 19 | rs1058205 | TT | 1.22 | 9.20 |
| CT | 1.08 | 8.51 | ||
| CC | 0.87 | 8.67 | ||
| Ptrend | 0.05 | 0.40 | ||
| 20 | rs1058274 | AA | 1.15 | 9.20 |
| GA | 1.23 | 8.69 | ||
| GG | 0.93 | 8.09 | ||
| Ptrend | 0.37 | 0.28 | ||
| 21 | rs6998 | GG | 1.10 | 8.36 |
| AG | 1.18 | 9.10 | ||
| AA | 1.27 | 10.26 | ||
| Ptrend | 0.16 | 0.09 | ||
| 22 | rs2569735 | GG | 1.22 | 9.04 |
| AG | 1.00 | 8.26 | ||
| AA | 0.86 | 6.98 | ||
| Ptrend | 0.005 | 0.17 | ||
| 23 | rs2569739 | TT | 1.22 | 9.91 |
| CT | 1.19 | 9.06 | ||
| CC | 0.99 | 7.40 | ||
| Ptrend | 0.07 | 0.01 |
Four SNPs were significantly associated with PSA levels measured in cases at diagnosis; for three, the minor allele was associated with lower levels (rs266878, rs174776, rs2569739) and for one, the minor allele was associated with higher levels (rs2292185). For each, the associations were also at least borderline significant and in the same direction as in the baseline samples among the control subjects. For rs2569739, the association with lower PSA at diagnosis is consistent with lower risk of prostate cancer diagnosis during or after 1992.
A summary of the associations of KLK3 SNPs with overall risk of prostate cancer, risk of prostate cancer in the pre-PSA and screening eras, PSA levels in controls at study baseline and PSA at diagnosis is presented in Table IV.
Table IV.
Summary of associations of KLK3 SNPs with risk of PCa, risk of PCa in the pre-PSA and screening eras, PSA levels in controls at study baseline and PSA at diagnosis
| SNP | Overall risk | Pre-1989 risk | Post-1991 risk | Baseline PSA (controls) | PSA at diagnosis | |
| 1 | rs925013 | — | — | — | ↑ | (↑) |
| 2 | rs266864 | — | — | — | — | — |
| 3 | rs266861 | — | — | — | — | — |
| 4 | rs3760721 | — | — | — | ↑ | — |
| 5 | rs3760722 | — | — | — | ↑ | (↑) |
| 6 | rs266882 | — | — | — | — | — |
| 7 | rs2292186 | — | — | — | ↑ | — |
| 8 | rs266880 | — | — | — | — | — |
| 9 | rs1053972 | — | — | — | ↑ | — |
| 10 | rs266879 | — | — | — | — | — |
| 11 | rs2292185 | (↑) | – | — | ↑ | ↑ |
| 12 | rs266878 | ↓ | ↓ | — | ↓ | ↓ |
| 13 | rs2271093 | (↑) | — | — | ↑ | (↑) |
| 14 | rs174776 | ↓ | (↓) | — | ↓ | ↓ |
| 15 | rs1061477 | — | — | — | — | — |
| 16 | rs266877 | ↓ | (↓) | — | — | — |
| 17 | rs266876 | — | ↓ | — | — | — |
| 18 | rs2659122 | — | — | — | — | — |
| 19 | rs1058205 | ↓ | — | — | ↓ | — |
| 20 | rs1058274 | — | — | — | — | — |
| 21 | rs6998 | — | — | — | — | (↑) |
| 22 | rs2569735 | ↓ | — | (↓) | ↓ | — |
| 23 | rs2569739 | ↓ | — | ↓ | (↓) | ↓ |
Direction of the arrow indicates the direction of the association. Arrows in parentheses signify P < 0.10, arrows without parentheses signify P < 0.05, dashes represent no significant association.
Three SNPs associated with lower PSA levels or prostate cancer risk (or both) were also associated with higher baseline total IGF-1 levels in non-cases (rs266878, rs174776 and rs266876; supplementary Table II is available at Carcinogenesis Online).
Discussion
Several polymorphisms in the KLK3 gene are associated with risk of prostate cancer in this large prospective nested case–control study. The minor alleles for six SNPs were significantly associated with lower risk; for four of these SNPs, the minor alleles were also associated with significantly lower PSA levels measured in blood provided at the study baseline by controls. The associations for two SNPs were significant in analyses restricted to cases diagnosed before the PSA era.
The association of SNPs in the KLK3 locus with prostate cancer risk and PSA levels has been previously reported in both candidate gene studies and GWAS. The minor allele for rs2735839 was associated with reduced risk in the initial GWAS (8) and several additional reports (16–20). Although we did not genotype rs2735839, it is in complete LD (R2 = 1) with rs2569735. In this study, the minor allele for rs2569735 was significantly associated with reduced overall prostate cancer risk.
What remained unclear after previous studies is whether these SNPs represent causal associations with risk or if the associations observed are due solely to an effect on likelihood of diagnosis by virtue of an association with PSA levels. By limiting controls to men with low PSA levels, the original study probably inadvertently enriched the control group with the variant allele, leading to an exaggerated lower odds ratio. We addressed this question by determining if the likelihood of being diagnosed with prostate cancer in the era before the widespread adoption of PSA screening in 1989 or after 1991, differed based on genotype. We reasoned that with non-selected controls, the likelihood of initial diagnosis would be unrelated to PSA level before PSA screening was adopted. In contrast, in the PSA screening era, one would expect that SNPs associated with lower PSA may appear to be associated with lower disease risk even in the absence of any true causal relationship. Despite slight differences in statistical significance, perhaps attributable to sample size, the magnitude of the associations restricting to pre-PSA cases was strikingly similar and even somewhat stronger compared with the overall analysis. Moreover, we observed only one significant association with prostate cancer risk in the PSA era, despite the larger number of cases and greater statistical power. The association of the minor allele of rs2569739 with lower PSA at diagnosis is consistent with the association with lower risk of prostate cancer diagnosis after 1991 that was observed. Further, none of the SNPs associated with higher PSA levels were significantly related to risk overall, in the pre-PSA or PSA eras. While the association of this one SNP with PSA levels and prostate cancer risk in the PSA screening era points toward a potential detection bias, we believe that overall our results suggest that the association of SNPs with PSA levels and detection bias cannot completely explain the association with prostate cancer risk.
An alternative analysis to address this issue would be to examine the association between SNPs and risk, adjusting for baseline PSA level, as was performed in Ahn et al. (12). If the SNPs associated with risk are no longer statistically significant when PSA level is introduced to the model, this would suggest that the entire association with risk is through PSA levels and screening. However, we were unable to perform this analysis as only approximately half of the cases and controls have baseline PSA data available. Also, such an analysis is predicated on the assumption that PSA itself bears no causal relation to prostate cancer incidence.
PSA can biologically interact with IGF-binding factors, which may result in higher levels of free IGF-1. Since IGF-1 has also been associated with prostate cancer risk, we explored the association of KLK3 SNPs with IGF-1 levels. Three SNPs associated with lower PSA levels or prostate cancer risk were also associated with higher total IGF-1 levels. The IGF-1 assay we employed measures both free and bound IGF-1 (the majority is bound), and we were unable to measure free IGF-1 independently. One may speculate that the lower PSA levels in men with these variants may lead to reduced degradation of IGFBP5 and less cleavage of IGFBP3, leading to higher total, though less free, IGF-1.
After conditioning on rs2569735, which defines the 3′ boundary of the KLK3 locus, the other SNPs that were marginally associated with prostate cancer risk or PSA levels were no longer significantly associated with those end points. This is consistent with the region containing a single causal variant that is in complete LD with the most significant marker in the KLK3 region, rs2735839, reported in the original GWAS (8). However, we caution that our sample size limits the power of these conditional analyses. Larger collaborative efforts will probably be needed to better understand the covariance between genetic variation at this locus, prostate cancer risk and PSA levels.
Determining the mechanism for how KLK3 polymorphisms affect prostate cancer risk, whether through their effect on PSA levels or other pathways, has important implications for further study. The finding that several highly prevalent genetic variants affect PSA levels could change the interpretation of levels in screening by suggesting a different baseline PSA level for an individual determined by genotype; however, the data are insufficient to support genetic screening tests based on these findings. If the KLK3 SNPs were only affecting risk by influencing PSA levels and therefore creating a detection bias through screening, less follow-up of these genetic markers would be needed and more effort could be focused on other known risk SNPs. Although our data are insufficient to completely rule out a detection bias, our findings suggest that the association of these SNPs with prostate cancer risk cannot entirely be explained by such bias and support further mechanistic studies of their biological function.
Supplementary material
Supplementary Tables I and II can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute (CA-42182, CA-34944, CA-40360, CA141298, CA-097193); National Heart, Lung, and Blood Institute, Bethesda, MD, USA (HL-26490, HL-34595); National Research Service Award (T32 CA009001-32 to K.L.P.). S.I.H. was supported by a National Kidney Foundation Clinician Scientist Award, grant (CA-101075) from the National Cancer Institute, a genotyping subsidy award from The Broad Institute Center for Genotyping and Analysis funded by grant (U54 RR020278) from the National Center for Research Resources Center, and the R. Glenn Davis (DCI) Endowment (University of Florida).
Supplementary Material
Acknowledgments
We thank Daniel Mirel and The Broad Institute Center for Genotyping and Analysis for expert design and execution of the SNP genotyping reported herein.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- GWAS
genome-wide association study
- IGF
insulin-like growth factor
- IGFBP
insulin-like growth factor-binding protein
- LD
linkage disequilibrium
- PHS
Physicians’ Health Study
- PSA
prostate-specific antigen
- SNP
single-nucleotide polymorphism
References
- 1.Stamey TA, et al. Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N. Engl. J. Med. 1987;317:909–916. doi: 10.1056/NEJM198710083171501. [DOI] [PubMed] [Google Scholar]
- 2.Amundadottir LT, et al. A common variant associated with prostate cancer in European and African populations. Nat. Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 3.Freedman ML, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc. Natl Acad. Sci. USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gudmundsson J, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat. Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 5.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 6.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 7.Gudmundsson J, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat. Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eeles RA, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 9.Eeles RA, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudmundsson J, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeager M, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahn J, et al. Variation in KLK genes, prostate-specific antigen and risk of prostate cancer. Nat. Genet. 2008;40 doi: 10.1038/ng0908-1032. 1032–4; author reply, 1035–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, et al. Association of prostate cancer risk variants with clinicopathologic characteristics of the disease. Clin. Cancer Res. 2008;14:5819–5824. doi: 10.1158/1078-0432.CCR-08-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kader AK, et al. Individual and cumulative effect of prostate cancer risk-associated variants on clinicopathologic variables in 5,895 prostate cancer patients. Prostate. 2009;69:1195–1205. doi: 10.1002/pros.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eeles R, et al. Reply to “Variation in KLK genes, prostate-specific antigen and risk of prostate cancer”. Nat. Genet. 2008;40:1035–1036. doi: 10.1038/ng0908-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooker S, et al. Replication of prostate cancer risk loci on 8q24, 11q13, 17q12, 19q33, and Xp11 in African Americans. Prostate. 2010;70:270–275. doi: 10.1002/pros.21061. [DOI] [PubMed] [Google Scholar]
- 17.Pal P, et al. Tagging SNPs in the kallikrein genes 3 and 2 on 19q13 and their associations with prostate cancer in men of European origin. Hum. Genet. 2007;122:251–259. doi: 10.1007/s00439-007-0394-3. [DOI] [PubMed] [Google Scholar]
- 18.Lai J, et al. PSA/KLK3 AREI promoter polymorphism alters androgen receptor binding and is associated with prostate cancer susceptibility. Carcinogenesis. 2007;28:1032–1039. doi: 10.1093/carcin/bgl236. [DOI] [PubMed] [Google Scholar]
- 19.Severi G, et al. Variants in the prostate-specific antigen (PSA) gene and prostate cancer risk, survival, and circulating PSA. Cancer Epidemiol. Biomarkers Prev. 2006;15:1142–1147. doi: 10.1158/1055-9965.EPI-05-0984. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald LM, et al. Analysis of recently identified prostate cancer susceptibility loci in a population-based study: associations with family history and clinical features. Clin. Cancer Res. 2009;15:3231–3237. doi: 10.1158/1078-0432.CCR-08-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallagher DJ, et al. Susceptibility loci associated with prostate cancer progression and mortality. Clin. Cancer Res. 2010;16:2819–2832. doi: 10.1158/1078-0432.CCR-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiklund F, et al. Association of reported prostate cancer risk alleles with PSA levels among men without a diagnosis of prostate cancer. Prostate. 2009;69:419–427. doi: 10.1002/pros.20908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cramer SD, et al. Association between genetic polymorphisms in the prostate-specific antigen gene promoter and serum prostate-specific antigen levels. J. Natl Cancer Inst. 2003;95:1044–1053. doi: 10.1093/jnci/95.14.1044. [DOI] [PubMed] [Google Scholar]
- 24.Maeda H, et al. Prostate-specific antigen enhances bioavailability of insulin-like growth factor by degrading insulin-like growth factor binding protein 5. Biochem. Biophys. Res. Commun. 2009;381:311–316. doi: 10.1016/j.bbrc.2009.01.096. [DOI] [PubMed] [Google Scholar]
- 25.Cohen P, et al. Biological effects of prostate specific antigen as an insulin-like growth factor binding protein-3 protease. J. Endocrinol. 1994;142:407–415. doi: 10.1677/joe.0.1420407. [DOI] [PubMed] [Google Scholar]
- 26.Rowlands MA, et al. Circulating insulin-like growth factor peptides and prostate cancer risk: a systematic review and meta-analysis. Int. J. Cancer. 2009;124:2416–2429. doi: 10.1002/ijc.24202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan JM, et al. Insulin-like growth factor-I (IGF-I) and IGF binding protein-3 as predictors of advanced-stage prostate cancer. J. Natl Cancer Inst. 2002;94:1099–1106. doi: 10.1093/jnci/94.14.1099. [DOI] [PubMed] [Google Scholar]
- 28.Final report on the aspirin component of the ongoing Physicians' Health Study. Steering Committee of the Physicians' Health Study Research Group. N. Engl. J. Med. 1989;321:129–135. doi: 10.1056/NEJM198907203210301. [DOI] [PubMed] [Google Scholar]
- 29.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 30.David A, et al. Unusual alternative splicing within the human kallikrein genes KLK2 and KLK3 gives rise to novel prostate-specific proteins. J. Biol. Chem. 2002;277:18084–18090. doi: 10.1074/jbc.M102285200. [DOI] [PubMed] [Google Scholar]
- 31.Michael IP, et al. Intron retention: a common splicing event within the human kallikrein gene family. Clin. Chem. 2005;51:506–515. doi: 10.1373/clinchem.2004.042341. [DOI] [PubMed] [Google Scholar]
- 32.Gann PH, et al. Strategies combining total and percent free prostate specific antigen for detecting prostate cancer: a prospective evaluation. J. Urol. 2002;167:2427–2434. [PubMed] [Google Scholar]
- 33.Conneely KN, et al. So many correlated tests, so little time! Rapid adjustment of P values for multiple correlated tests. Am. J. Hum. Genet. 2007;81 doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.