Abstract
Peg3 (paternally expressed gene 3) is the first imprinted gene detected in the proximal region of mouse chromosome 7. Because imprinting is a trait that is generally conserved among mammals, and imprinted domains generally encompass several adjacent genes, expression patterns and chromosomal environment of the human counterpart of Peg3 are of special interest. In this study we have localized human PEG3 ∼2 Mb proximal of the telomere of chromosome 19q, within a region known to carry large numbers of tandemly clustered Krüppel-type zinc finger-containing (ZNF) genes. Peg3 also encodes a Krüppel-type ZNF protein but one that is distinguished from other ZNF gene products by the fact that it carries two novel proline-rich motifs. Comparison between mouse Peg3 and partial human PEG3 gene sequences revealed a high level of conservation between the two species, despite the fact that one of the two proline-rich repeats is absent from the human gene. Our data demonstrate that the human gene is expressed at highest levels in ovary and placenta; mouse Peg3, by contrast, is transcribed at highest levels in the adult brain. These comparative mapping, sequencing, and expression data provide the first clues to the potential activities of PEG3, and generate new tools to aid in the analysis of structure and function of a potentially new imprinted domain located in human chromosome 19q13.4 and mouse chromosome 7.
Peg3 (paternally expressed gene 3) is the first imprinted gene detected in the proximal region of mouse chromosome 7 (Kuroiwa et al. 1996). The Peg3 transcription unit encodes a Krüppel-type (C2H2) zinc finger-containing (ZNF) protein, most of which are thought to function as transcription factors (El-Baradi and Pieler 1991; Pieler and Bellefroid 1994). Genetic studies have indicated that there are three different imprinted regions along the length of mouse chromosome 7 (Mmu7), located in proximal, central, and distal regions of the chromosome, respectively (Searle and Beechey 1990; Cattanach et al. 1992). Studies of several imprinted domains, including those located in central and distal Mmu7, the t-maternal effect (tme) region of Mmu17, and related regions of the human genome, have indicated that imprinted domains are generally large enough to encompass several genes (Efstratiadis 1994; Nicholls 1994; Barlow 1995).
Although exceptions have been noted (Kalscheuer et al. 1993; Riesewijk et al. 1996), these previous studies have also provided evidence that genomic imprinting is generally conserved between orthologous genes in mammalian species (Barlow 1995). Human regions that are syntenically homologous to two of the three Mmu7-imprinted domains are associated with imprinted genetic disorders. Human chromosome 15q11-q13, which contains genes associated with Prader–Willi and Angelman syndromes, represents the human counterpart of the central Mmu7-imprinted region (Nicholls 1993, 1994), whereas Beckwith–Wiedemann syndrome genes are located in a region of 11p15.5 that is closely related to the most distal Mmu7-imprinted domain (Reik 1989; Efstratiadis 1994). However, the human counterpart of Peg3 has not yet been characterized, and gene content and inheritance patterns of the region surrounding the human gene have not been explored. In addition, although the embryonic expression patterns of mouse Peg3 are well-documented, expression of the gene in adult mouse tissues has not been investigated and the transcription pattern of homologous human sequences is unknown. Investigation of these basic characteristics of the human PEG3 gene represent the essential first steps toward understanding the possible role of the gene in imprinted human traits or health-related disorders.
The proximal third of Mmu7 is closely related in gene content, order, and spacing to the long arm of human chromosome 19 (Saunders and Seldin 1990; Stubbs et al. 1996), making it likely that the human counterpart of Peg3 would be found in that human region. To gain more information about the human PEG3 gene and to provide tools for the discovery of other potentially imprinted genes, we have mapped human sequences related to the mouse Peg3 gene within the established physical map of human chromosome 19 (Ashworth et al. 1995). We have also determined the sequence of a human genomic fragment containing Peg3-related sequences and compared it with mouse Peg3. Finally, we have determined the expression patterns of PEG3 in human tissues and have compared these data with transcription patterns of Peg3 in the adult mouse. These studies lay the basic groundwork for investigating whether PEG3 and other human 19q13.4/Mmu7 genes, perhaps including some of the numerous ZNF-containing genes located in both the human and mouse regions, may also be imprinted in one or both species.
RESULTS
Human Gene Mapping
To determine the location of human Peg3-related sequences, we hybridized the mouse Peg3 probe to high density filter arrays of human 19 chromosome-specific cosmids (Olsen et al. 1993). These studies identified 10 cosmids, 7 of which had been fingerprinted and used in the assembly of a physical map of human chromosome 19. Six of the seven cosmids were highly overlapping, clustered at one end of a 170-kb contig located ∼2 Mb proximal of the 19q telomere (Fig. 1). The cosmid numbers of these six clones are as follows: F14378, F16941, F17167, F21492, F21930, and R26864. Previous studies have shown that this telomeric region of human 19q13.4 is exceptionally rich in Krüppel-type ZNF genes (Lichter et al. 1992; Ashworth et al. 1995; Hoffman et al. 1995). Our own previous studies have provided preliminary evidence to suggest that most of these genes are arranged in several independent tandem clusters (Ashworth et al. 1995; J. Kim, M. Shannon, and L. Stubbs, unpubl.). Some of the known human genes localized to this region in previous studies are included in Figure 1 (Ashworth et al. 1995; Stubbs et al. 1996). Peg3-related sequences were found to be positioned between PRKCG and ZNF134, ∼2 and 1 Mb from these two markers, respectively. The overall gene order in this region was determined as follows: Cen–PRKCG–PEG3–ZNF134–ZNF132–ZNF42–Tel.
Figure 1.
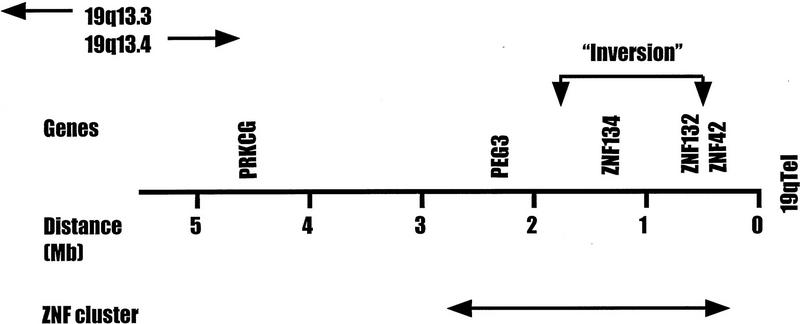
Location of PEG3 on the physical map of human chromosome 19q13.4. The relative position and distances of human PEG3 compared with other gene markers in 19q13.4 are presented. For simplicity, not all genes/markers known in this region are shown. A 2-Mb region of H19q that appears to be filled primarily with Krüppel-type ZNFs is indicated (↔). The small inversion detected in the proximal Mmu7 relative to the human region is indicated by a bracket with arrows. The information about the other gene markers has been presented in the previous study (Stubbs et al. 1996). A current, physical map of human chromosome 19 including this region is available from http://www-bio.llnl.gov/bbrp/genome/genome.html.
Mouse Gene Mapping
Peg3 had been assigned to the proximal portion of Mmu7 in previous studies (Kuroiwa et al. 1996), but the map position within this region was determined only to a relatively low level of resolution. To define the position of the mouse Peg3 gene more precisely, we followed the segregation of sequences related to the pooled Peg3 PCR probes in a 160-member Mus musculus × Mus spretus interspecific backcross already typed for the orthologs of a number of human chromosome 19q13.4 genes (Stubbs et al. 1996). Peg3 cosegregated with Zfp132 in this cross, placing the imprinted mouse gene between Pkcc (representing the mouse ortholog of PRKCG) and a cluster of genetically inseparable genes, Zfp134, Zfp98 (mouse ortholog of ZNF42; Stubbs et al. 1996), and Lig1 (Fig. 2).
Figure 2.
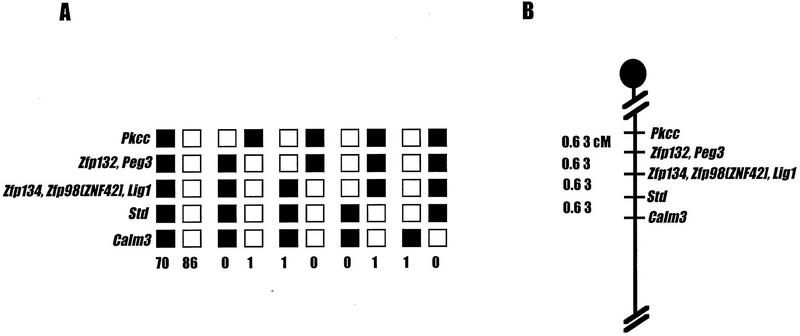
Location of Peg3 on the proximal portion of mouse chromosome 7. (A) The segregation patterns of Peg3 and the neighboring gene markers in 160 interspecific backcross animals. Each column represents a type of parental or recombinant chromosome, with solid boxes representing 129/Sv alleles and open boxes denoting M. spretus alleles for a given gene. The number of animals inheriting a particular type of chromosome is listed at the bottom of each column. Gene markers listed together in the same row showed identical segregation pattern in this interspecific backcross system. Gene markers in parentheses represent human genes. (B) The partial genetic map of the proximal chromosome 7 showing the location of Peg3 relative to other gene markers. The genetic distances (in centimorgans) between markers are listed at left. Peg3 cosegregates with Zfp132.
Previous reports have demonstrated that genes within the Pkcc–Zfp98 interval are inverted in order relative to other genes in the conserved Mmu7/human 19q linkage group. These studies also provided preliminary evidence that genes located within the borders of the Pkcc/PRKCG–Zfp98/ZNF42 region were also organized differently in human and mouse (Stubbs et al. 1996). The relative positions of mouse Peg3 and its human counterpart add further weight to that notion and provide a more detailed picture of the relative organization of genes in this region. In human chromosome 19q13.4, PEG3 and ZNF134 are nearest neighbors, located ∼2.3 and 1.3 Mb from the telomere, respectively. Human ZNF42 and ZNF132 are clustered together closely, located 0.4 and 0.6 Mb from the telomeric end of the chromosome, respectively (Fig. 1). In contrast, Peg3 is linked tightly to Zfp132 in mouse, whereas Znf134 maps nearest to Zfp98 and Lig1 (Fig. 2). These data indicate that an internal inversion, involving the ∼1- to 1.5-Mb segment of DNA that contains Zfp134 and Zfp132, has reversed the relative orders of the two genes in mouse relative to man. These studies would place the proximal breakpoint of this evolutionary inversion in the ∼1-Mb interval located between PEG3 and ZNF134, and the distal “breakpoint” within the 200-kb interval separating ZNF132 from ZNF42 gene sequences (Fig. 1).
Comparisons of the Peg3 Sequence with Related Genes
Only small segments of a partial Peg3 amino acid sequence have been published (Kuroiwa et al. 1996). However, a similarity search with the existing amino acid sequence data against public sequence databases permitted us to identify several entries representing closely related mouse and human genes. These database queries identified one mouse gene sequence, called Pw1, with striking similarities to Peg3 (Relaix et al. 1996; GenBank accession no. U48804). The predicted Pw1 amino acid sequence shows an unusually high level of sequence identity with mouse Peg3 throughout all portions of the published sequence. It appears that the two sequences represent a single locus with DNA sequence variation attributable to differences between mouse strains and/or sequencing errors.
Mouse Peg3 sequences also showed significant matches with several human expressed sequence tag (EST; Adams et al. 1991) cDNA sequences. All of these ESTs appear to be derived from one locus based on the high level of nucleotide sequence identity (average 99%) observed between the clones. The cDNA clone from which one of these EST sequences was derived (GenBank accession no. H08927) was analyzed in further detail. Comparison of the complete sequence of this cDNA (1.5 kb in length) with mouse Peg3 indicates that the major portion of this cDNA corresponds to the 3′-untranslated region (UTR) of the human gene, with only a small portion corresponding to the coding region. To determine the map position of the gene corresponding to this cDNA clone, we generated oligonucleotide primers from the 3′-UTR of the cDNA sequence and used these primers to test by PCR for the presence of the gene in the Peg3-positive cosmid clones. These analyses confirmed that the cosmids did contain sequences corresponding to this EST (data not shown).
To obtain more sequences from the upstream coding region of the human gene, we sequenced a 3-kb genomic fragment subcloned from one of the Peg3-cosmids (F 14378) (GenBank accession no. U90336). Sequence analysis indicated that the cosmid subclone contained one contiguous open reading frame (ORF), encoding a predicted protein fragment with 83% overall similarity to mouse Peg3 and Pw1 amino acid sequences (Fig. 3). The ORF translated directly from the DNA sequence of the cosmid subclone indicated that at least the 3′ portion of the human PEG3 is intronless, a feature which has also been observed for mouse Pw1 (Relaix et al. 1996). Peg3 contains two proline-rich repeat regions located between zinc finger-encoding segments of the gene; one is between finger 5 and 6 and the other is between finger 10 and 11 (Kuroiwa et al. 1996). The first proline repeat appears to be absent in human PEG3, whereas a region corresponding in position and sequence to the second mouse proline repeat was present in the human subclone. The amino acid sequence that can be predicted to be encoded by this portion of the human sequence was seen to be nearly identical to the corresponding region of the mouse Pw1 and Peg3 amino acid sequence (Fig. 3). These comparisons also indicated that zinc finger regions of the mouse and human PEG3 genes are highly conserved on the level of both the nucleotide and predicted amino acid sequence; in contrast, interfinger spacer regions are much less similar between the two species. It is also noteworthy that some fingers show more conservation than others. Fingers 7, 8, and 12 of the mouse and human genes are almost identical in amino acid sequence, whereas fingers 5, 6, 9, 10, and 11 are altered considerably in structure in mouse Peg3 relative to the human counterpart of the gene (Fig. 3).
Figure 3.

Sequence alignment of mouse Pw1 (GenBank accession no. U48804) and human PEG3 amino acid sequence (GenBank accession no. 90336). The upper sequence represents the mouse Pw1 and the lower sequence represents the partial human PEG3 amino acid sequence derived from a genomic fragment of one of Peg3 cosmids (F14378). The partial human PEG3 cDNA clone (GenBank accession no. H08927) contains the region corresponding to the last two fingers and 3′ UTR of human PEG3. Zinc finger regions are bold, and finger number and proline repeat regions are indicated at right. Sequence identity (vertical line), conservative substitution (colon), and amino acids with similar properties (dot) are indicated. The two sequences are aligned for maximal match by the BestFit program of the Genetics Computer Group package. We believe that mouse Pw1 and Peg3 represent the same gene, and, for the overall comparison analysis, mouse Pw1 was used instead of Peg3 because its complete sequence is publicly available.
Expression of PEG3 in Human and Mouse Adult Tissues
To provide additional information regarding the possible functions of Peg3 and its human ortholog, we examined expression patterns of the mouse and human genes in a variety of adult tissues. We hybridized Northern blots containing poly(A)+ human RNA samples with a probe derived from the UTR region of the human PEG3 cDNA clone (Fig. 4A). Highest levels of human gene expression, from among 16 different tissues, were seen in the ovary, testis, and placenta. The brain displayed a modest level of PEG3 expression, whereas PEG3 transcript was detected in heart, prostate, pancreas, and intestine at a very low level. Hybridization of a second Northern blot, containing RNA isolated from different portions of the human brain, demonstrated that the gene is expressed uniformly throughout most regions (Fig. 4B). To compare human and mouse gene expression, we hybridized Northern blots carrying poly(A)+ RNA derived from a number of adult mouse tissues with the Peg3 probe (Fig. 4C). Interestingly, the expression pattern of mouse Peg3 differed subtly but significantly from that observed for the human gene, with the highest levels of expression detected in the brain. Another Northern blot analysis with a second probe, the Peg3-2 probe, containing the second proline repeat region also showed the same result (data not shown). Only low levels of Peg3 expression were detected in the mouse ovary, as verified by RNase protection experiments (Fig. 4D). The strikingly different relative levels of Peg3 expression in mouse and human ovary therefore presents a significant contrast between the two species. Three larger mRNA species were also detected in mouse RNA samples with the mouse Peg3 probe, in addition to the predominant 9-kb transcript (Fig. 4C). Although present data do not permit us to identify those larger mouse transcripts or assess their potential relationship to the Peg3 gene, it is worth noting that the same collection of transcripts were detected by each of the three Peg3 PCR probes, which are derived from distinct portions of the mouse gene (Kuroiwa et al. 1996).
Figure 4.
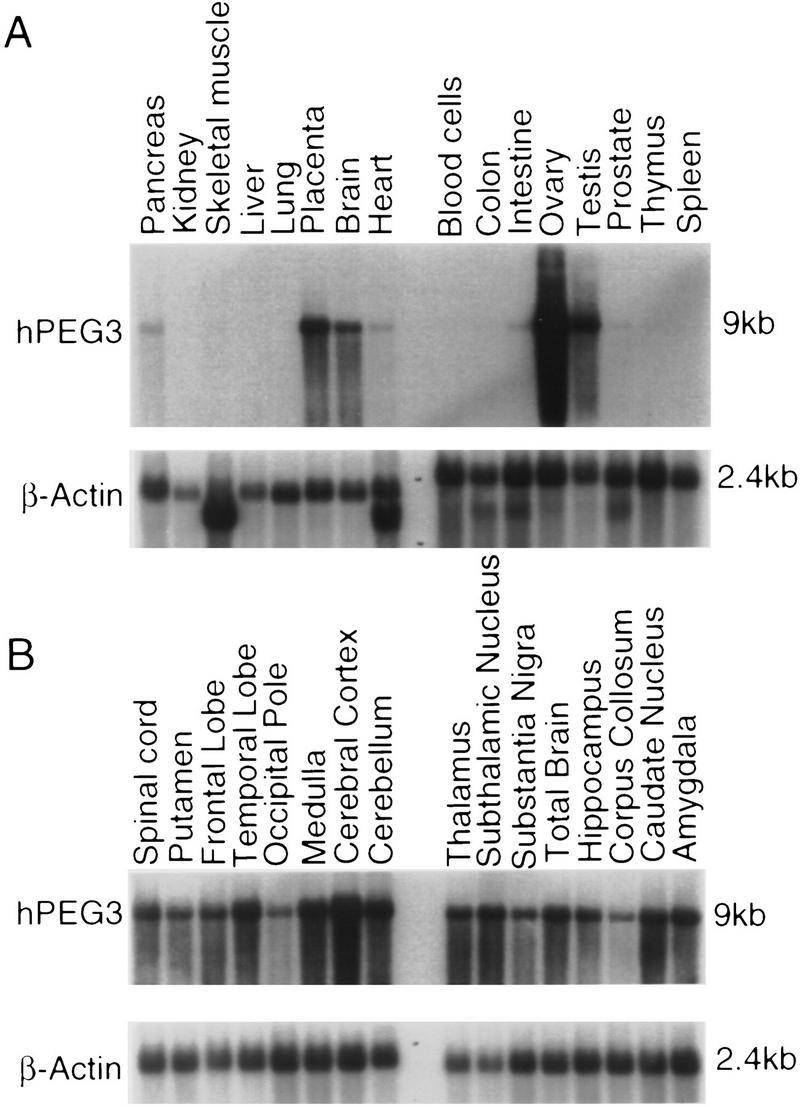
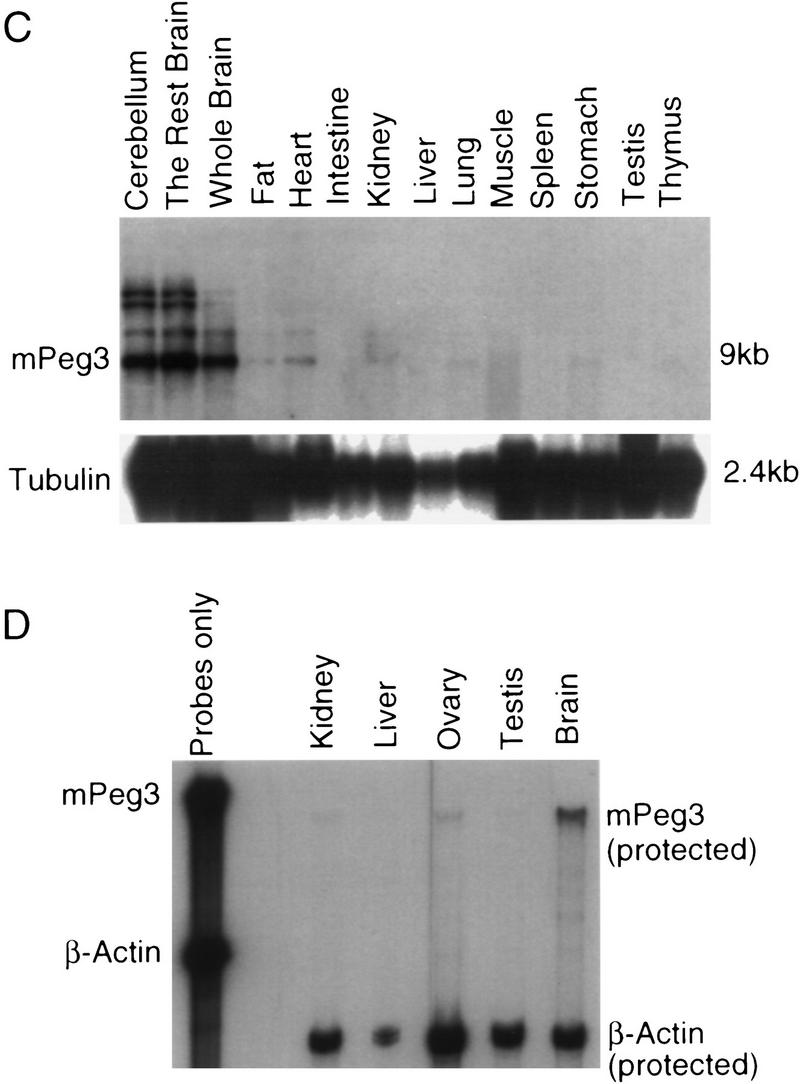
Expression pattern of Peg3 in the different adult tissues of human and mouse. (A) The expression pattern of human PEG3 in 16 different adult tissues. The name of each tissue is listed at the top of each lane, and the size of transcripts are indicated at the right. The size of human and mouse PEG3 are ∼9 kb in length. The same blot was hybridized with a control probe, human β-actin, and the result is shown at the bottom. (B) The expression pattern of human PEG3 in different brain parts. (C) The expression pattern of mouse Peg3 in the adult mouse tissues. The hybridization result of a control probe (tubulin) is shown at the bottom. The hybridization conditions and the probes used for this Northern blot analysis are described in Methods. (D) The relative expression level of mouse Peg3 in kidney, liver, ovary, testes, and brain. The name of each tissue used for this assay is listed at the top, and the probes used for this assay are indicated at right. The β-actin probe was used as a control probe for this assay.
DISCUSSION
In the course of comparative genome mapping, we have identified the human ortholog of Peg3 and have mapped the human gene within a 170-kb cosmid contig located ∼2 Mb from the telomere of human chromosome 19q. Sequence comparison of human and mouse genes has also verified the high level of conservation between them and has suggested that like its murine counterpart, human PEG3 may be intronless. The two proline repeat regions that are present in mouse Peg3 show an interesting contrast in terms of evolutionary conservation. The first proline repeat is absent from the human PEG3 gene, whereas the second repeat is present and highly conserved in both nucleotide and amino acid sequence between the two species. It is interesting to note that the two proline repeat regions appear similar in structure to short tandem repeat sequences and also that these types of tandem repeats in the genome are believed to be susceptible to changes induced by unequal crossing-over and DNA polymerase slippage (Nakamura et al. 1987). Therefore, this differential selection during evolution may indicate that the second proline repeat region has a distinct, and essential, role for PEG3 function.
Expression patterns of PEG3/Peg3 in adult tissues of human and mouse are similar but also differ in several significant respects. The prominent levels of PEG3/Peg3 expression in the brains of both species suggest that PEG3 may have an important and conserved role in neuronal cells. The detection of human PEG3 in placenta is consistent with the notion that most imprinted genes are involved in embryogenesis (Barlow 1995). At least one current theory centers the “battle” between maternal and paternal gene expression on questions of embryonic growth and development (Moore and Haig 1991; Haig 1993). However, Peg3 has been shown to be expressed in a differential fashion not only in embryonic tissues, but also in adult mouse brain (Kuroiwa et al. 1996). The notion that adult behavior might also be influenced by expression of a gene transcribed only from the paternal chromosome may add a new twist to the proposed “parental tug-of-war” (Moore and Haig 1991). Despite other similarities in tissue-specific patterns of expression, the high level of PEG3 transcription detected in human ovary is not observed in the mouse (Fig. 4). An understanding of the biological significance of these observations must await further study.
The close proximity of human and mouse PEG3/Peg3 to other, clustered Krüppel-type zinc finger genes is intriguing, because the imprinted domains that have been studied are large—spanning distances ranging from several hundred kilobases to two megabases—and include a number of neighboring genes (Efstratiadis 1994; Nicholls 1994; Barlow 1995; Saitoh et al. 1996). Mouse genetic studies have indicated that in addition to Peg3, proximal Mmu7 may also contain maternally expressed imprinted genes, because mutant mice with paternal duplication of the proximal Mmu7 region displayed reduced postnatal viability and rates of growth (Cattanach et al. 1992). These data suggest that additional imprinted genes may be found near Peg3 in the proximal region of Mmu7, and the ZNF genes that surround Peg3 in Mmu7 represent excellent candidates as additional members of this newly mapped imprinted region. Because Peg3 and other ZNFs are putative transcription factors (El-Baradi and Pieler 1991), it will be interesting to see whether many downstream genes controlled by these transcription factors are also affected indirectly by the imprinting.
Further studies will be required to determine whether the highly conserved PEG3 gene is also imprinted in humans. However, if human PEG3 is also found to be imprinted, the observation of an evolutionary inversion in this region may be helpful in determining the limits of the Mmu7 and potential 19q13.4 imprinted domains. The evolutionary rearrangement of genes in this region may also prove useful in understanding the mode and action-range of DNA sequences that may serve to regulate imprinting in this region. Because sequences controlling the imprinting process, termed IC (Imprinting Center; Dittrich et al. 1996), are generally thought to operate on genes within relatively limited, contiguous chromosomal domains, an inversion in the interval between PEG3 and ZNF42 could potentially result in altered patterns of imprinting for human PEG3 and/or its neighbors in 19q13.4 relative to their counterparts in the mouse genome. Conversely, if selective pressure favors the maintenance of a fixed pattern of imprinting in this region, separation of an IC from imprinted genes in one species would be unlikely; breakpoints of the inverted region could therefore prove useful in defining the physical extent of a putative PEG3-containing conserved imprinted domain. Examination of the imprinting status of PEG3 and neighboring genes in both human and mouse, now in progress, will soon provide the first clues regarding the structure, function, and evolutionary history of this newly discovered, potential imprinted domain.
METHODS
Human PEG3 Mapping
To localize human Peg3-related sequences, we hybridized PCR products corresponding to mouse gene sequences to filter arrays of human chromosome 19-specific cosmids (Olsen et al. 1993). The three mouse genomic DNA fragments used, corresponding to the transcribed region of mouse Peg3, were amplified by PCR using oligonucleotide primer sequences and PCR conditions taken from a report published previously (Kuroiwa et al. 1996). These three PCR products, Peg3-1, Peg3-2, and Peg3-3 probe, were subcloned into the TA cloning vector (Invitrogen, Inc.) and sequenced from both directions. The three fragments were pooled, radiolabeled, and hybridized to the human cosmids as described previously (Stubbs et al. 1990), except that the hybridization buffer contained formamide at a final concentration of 45%. Results of Peg3 probe hybridizations were integrated into the metric physical map of human chromosome 19q13.4, established from data published previously (Ashworth et al. 1995; Stubbs et al. 1996). Contigs in this map were generated using a restriction digest fingerprinting strategy (Carrano et al. 1989; Branscomb et al. 1990). Position of contigs in the map were determined by high-resolution fluorescence in situ hybridization (FISH) mapping methods (Gordon et al. 1995).
Mouse Peg3 Mapping
To map mouse Peg3 more precisely in proximal Mmu7, we followed the segregation of sequences detected by pooled mouse PCR-generated probes (described above) in the progeny of a M. musculus × M. spretus interspecific backcross typed previously for a large number of 19q3.4 gene orthologs (Stubbs et al. 1996). The Peg3 probe detected a fragment of 2.6 kb and 1.8 kb in HincII-digested M. spretus and M. musculus DNA samples, respectively. This variant fragment was used to trace the segregation of Peg3 alleles in the backcross progeny. Radiolabeled probes were hybridized to blots carrying restriction digests of DNA samples prepared from each of the 160 backcross progeny, as described (Stubbs et al. 1990), and washed in a solution containing 0.2× SSC, 0.2% SDS, at 65°C. Mapping data were stored and calculated, with standard errors, using standard statistical methods (Silver 1985) with the aid of the Map Manager data analysis program (Manly 1993).
Sequencing and Sequence Analysis
A human Peg3-related cDNA clone (GenBank accession no. H08927) was obtained from the Research Genetics/IMAGE Consortium collection. The 1.5-kb human cDNA insert was digested with HindIII and subcloned into predigested pBluescript vector. The subcloned fragments were sequenced from both directions using a fluorescence-based cycle-sequencing DNA sequencing kit (Perkin Elmer Cetus) and reactions analyzed on an ABI 373 automated sequencer. For human PEG3 genomic DNA sequencing, one of the Peg3-positive human cosmids (F 14378) was digested independently with PstI and HindIII, and each digest subcloned into the pBluescript vector. Subclones containing the coding region of human PEG3 were selected by hybridization with the mouse Peg3 probe and sequenced, as described above. Database search and sequence alignments were analyzed using GCG software, version 7 (Genetics Computer Group, Inc).
Northern Blot Analysis
For human gene expression studies, we hybridized commercial poly(A)+ RNA blots (Multiple Northern Blot, Clonetech) with a 600-bp HindIII fragment from the 3′ UTR of the human Peg3-related cDNA clone. The human β-actin probe, provided with the human Northern blots, was used as a loading control. Total RNA was isolated from different tissues dissected from eight-week-old mice, using a total RNA isolation kit (Rapid total RNA isolation kit, 5prime-3prime, Inc), and poly(A)+ RNAs were purified using oligo(dT) columns (Rapid poly(A)+ mRNA preparation kit, 5prime-3prime, Inc). Two micrograms of poly(A)+ RNA was separated on a 1.0% formaldehyde–agarose gel, transferred to a nylon membrane (Hybond, Amersham), and cross-linked to the blot by UV irradiation. For mouse Northern blot analysis, we used the Peg3-3 probe, containing the last two zinc fingers plus the 3′ UTR of the mouse Peg3 gene. The mouse tubulin cDNA was used as a control probe for the mouse poly(A)+ RNA blot. Hybridization and washing conditions were the same as described for human PEG3 Northern analysis.
RNase Protection Assay
To compare the expression level of mouse Peg3 ovary and other tissues, a RNase protection assay was performed. The TA clone containing the Peg3-2 probe corresponding to the second proline repeat region of mouse Peg3 was used as a template for generating an antisense RNA probe with T7 RNA polymerase. The antisense RNA probe for the mouse β-actin gene was also generated and used as a control probe. Fifteen micrograms of each tissue total RNA was mixed with the Peg3 and the β-actin antisense RNA probes, hybridized overnight at 42°C, and digested with an RNAse mixture (RPA II, Ambion). The remaining RNA probes were precipitated and separated on a 6% polyacrylamide gel containing 8 m urea.
Acknowledgments
We thank Bill Dunn, Xiaojia Ren, and Loren Houser for assistance and advice with mapping, expression, and DNA sequence analysis, respectively, and Mark Shannon, Cymbeline Culiat, and Rob Nicholls for helpful discussions and critical reading of the manuscript. We especially thank Rob Nicholls for initially drawing our attention to Peg3 as the ortholog of a potential 19q imprinted gene. This work was supported by the U.S. Department of Energy (under contract DEAC05-96OR22464 with Lockheed-Martin Energy Systems, Inc., and under contract W-7405-Eng-48 with Lawrence Livermore National Laboratory).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL stubbsl@bioax1.bio.ornl.gov; FAX (423) 574-1283.
REFERENCES
- Adams MD, Kelly JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, Kerlavage AR, McCombi WR, Venter JC. Complementary DNA sequencing: Expressed sequence tags and Human Genome Project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Ashworth LK, Batzer MA, Brandriff B, Branscomb E, deJong P, Garcia E, Garnes J, Gordon L, Lamerdin JE, Lennon G, Mohrenweiser H, Olsen A, Slezak T, Carrano AV. A metric physical map of human chromosome 19. Nature Genet. 1995;11:422–427. doi: 10.1038/ng1295-422. [DOI] [PubMed] [Google Scholar]
- Barlow DP. Gametic imprinting in mammals. Science. 1995;270:1610–1613. doi: 10.1126/science.270.5242.1610. [DOI] [PubMed] [Google Scholar]
- Brandriff BF, Gordon LA, Fertitta A, Olsen AS, Christensen M, Ashworth LK, Nelson DO, Carrano AV, Mohrenweiser HW. Human chromosome 19p: A fluorescence in situ hybridization map with genomic distance estimates for 79 intervals spanning 20 Mb. Genomics. 1994;23:582–591. doi: 10.1006/geno.1994.1546. [DOI] [PubMed] [Google Scholar]
- Branscomb E, Slezak T, Pae R, Galas D, Carrano AV, Waterman M. Optimizing restriction fragment fingerprinting methods for ordering large genomic libraries. Genomics. 1990;8:351–366. doi: 10.1016/0888-7543(90)90293-4. [DOI] [PubMed] [Google Scholar]
- Carrano AV, Lamerdin J, Ashworth LK, Watkins B, Branscomb E, Slezak T, Raff M, de Jong PJ, Keith D, McBride L, Meister KS, Kronick M. A high-resolution, fluorescence-based, semiautomated method for DNA fingerprinting. Genomics. 1989;4:129–136. doi: 10.1016/0888-7543(89)90291-7. [DOI] [PubMed] [Google Scholar]
- Cattanach BM, Barr JA, Evans EP, Burtenshaw M, Beechey CV, Leff SE, Brannan CI, Copeland NG, Jenkins NA, Jones J. A candidate mouse model for Prader-Willi syndrome which shows an absence of Snrpn expression. Nature Genet. 1992;2:270–274. doi: 10.1038/ng1292-270. [DOI] [PubMed] [Google Scholar]
- Dittrich B, Buiting K, Korn B, Rickard S, Buxton J, Saitoh S, Nicholls RD, Poustka A, Winterpacht A, Zabel B, Horsthemke B. Imprint switching on human chromosome 15 may involve alternative transcripts of the SNRPN gene. Nature Genet. 1996;14:163–170. doi: 10.1038/ng1096-163. [DOI] [PubMed] [Google Scholar]
- El-Baradi T, Pieler T. Zinc finger proteins: What we know and what we would like to know. Mech Dev. 1991;35:155–169. doi: 10.1016/0925-4773(91)90015-x. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A. Parental imprinting of autosomal mammalian genes. Curr Opin Genet Dev. 1994;4:265–280. doi: 10.1016/s0959-437x(05)80054-1. [DOI] [PubMed] [Google Scholar]
- Gordon LA, Bergmann A, Christensen M, Ganganan L, Lee DA, Ashworth LK, Nelson DO, Olsen AS, Mohrenweiser HW, Carrano AV, Brandriff BF. A 30-Mb metric fluorescence in situ hybridization map of human chromosome 19q. Genomics. 1995;30:187–194. doi: 10.1006/geno.1995.9886. [DOI] [PubMed] [Google Scholar]
- Haig D. Genetic conflicts in human pregnancy. Quart Rev Biol. 1993;68:495–532. doi: 10.1086/418300. [DOI] [PubMed] [Google Scholar]
- Hoffman, S.M.G., C. Amemiya, and H.W. Mohrenweiser. 1995. “Distribution and organization of zinc finger genes on chromosome 19.” Report of the D.O.E. Human Genome Program Contractor-Grantee Workshop IV, p. 49. Santa Fe, NM.
- Kalscheuer VM, Mariman EC, Schepens MT, Rehder H, Ropers H-H. The insulin-like growth factor type-2 receptor gene is imprinted in the mouse but not in humans. Nature Genet. 1993;5:74–78. doi: 10.1038/ng0993-74. [DOI] [PubMed] [Google Scholar]
- Kuroiwa Y, Kaneko-Ishino T, Kagitani F, Kohda T, Li L-L, Tada M, Suzuki R, Yokoyama M, Shiroishi T, Wakana S, Barton SC, Ishino F, Surani MA. Peg3 imprinted gene on proximal chromosome 7 encodes for a zinc finger protein. Nature Genet. 1996;12:186–190. doi: 10.1038/ng0296-186. [DOI] [PubMed] [Google Scholar]
- Lichter P, Bray P, Ried T, Dawid IB, Ward DC. Clustering of C2-H2 zinc-finger motif sequences within telomeric and fragile site regions of human chromosomes. Genomics. 1992;13:999–1007. doi: 10.1016/0888-7543(92)90013-i. [DOI] [PubMed] [Google Scholar]
- Manly KF. A MacIntosh program for the storage and analysis of experimental genetic mapping data. Mamm Genome. 1993;4:303–313. doi: 10.1007/BF00357089. [DOI] [PubMed] [Google Scholar]
- Moore T, Haig D. Genomic imprinting in mammalian development: A parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Leppert M, O’Connel P, Wolf R, Holm T, Culver M, Martin C, Fujimoto E, Hoff M, Kumlin E, White R. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science. 1987;235:1616–1622. doi: 10.1126/science.3029872. [DOI] [PubMed] [Google Scholar]
- Nicholls RD. Genomic imprinting and candidate genes in the Prader-Willi and Angelman syndromes. Curr Opin Genet Dev. 1993;3:445–456. doi: 10.1016/0959-437x(93)90119-a. [DOI] [PubMed] [Google Scholar]
- ————— New insights reveal complex mechanisms involved in genomic imprinting. Am J Hum Genet. 1994;54:733–740. [PMC free article] [PubMed] [Google Scholar]
- Olsen A, Conbs J, Garcia E, Elliot J, Amemiya C, deJong P, Treadgill G. Automated production of high density cosmid and YAC colony filters using a robotic workstation. BioTechniques. 1993;14:116–123. [PubMed] [Google Scholar]
- Pieler T, Bellefroid E. Perspectives on zinc finger protein function and evolution—An update. Mol Biol Rep. 1994;20:1–8. doi: 10.1007/BF00999848. [DOI] [PubMed] [Google Scholar]
- Reik W. Genomic imprinting and genetic disorders in man. Trends Genet. 1989;5:331–336. doi: 10.1016/0168-9525(89)90138-8. [DOI] [PubMed] [Google Scholar]
- Relaix F, Weng X, Marazzi G, Yang E, Copeland N, Jenkins N, Spence SE, Sassoon D. Pw1, a novel zinc finger gene implicated in the myogenic and neuronal lineages. Dev Biol. 1996;177:383–396. doi: 10.1006/dbio.1996.0172. [DOI] [PubMed] [Google Scholar]
- Riesewijk AM, Schepens MT, Mariman EM, Ropers H-H, Kalscheuer VM. The MAS proto-oncogene is not imprinted in humans. Genomics. 1996;35:380–382. doi: 10.1006/geno.1996.0372. [DOI] [PubMed] [Google Scholar]
- Saitoh S, Buiting K, Rogan PK, Buxton JL, Driscoll DJ, Arnemann J, Konig R, Malcom S, Horsthemke B, Nicholls RD. Minimal definition of the imprinting center and fixation of a chromosome 15q11-q13 epigenotype by imprinting mutations. Proc Natl Acad Sci. 1996;93:7811–7815. doi: 10.1073/pnas.93.15.7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Seldin MF. A molecular genetic linkage map of mouse chromosome 7. Genomics. 1990;8:525–535. doi: 10.1016/0888-7543(90)90040-2. [DOI] [PubMed] [Google Scholar]
- Searle AG, Beechey CV. Genome imprinting phenomena on mouse chromosome 7. Genet Res. 1990;56:237–244. doi: 10.1017/s0016672300035333. [DOI] [PubMed] [Google Scholar]
- Silver J. Confidence limits for estimates of gene linkage analysis of recombinant inbred strains. J Heredity. 1985;76:436–440. doi: 10.1093/oxfordjournals.jhered.a110140. [DOI] [PubMed] [Google Scholar]
- Stubbs L, Huxley C, Hogan B, Evans T, Fried M, Duboule D, Lehrach H. The hox-5 and surfeit gene clusters are linked in the proximal portion of mouse chromosome 2. Genomics. 1990;6:645–650. doi: 10.1016/0888-7543(90)90499-k. [DOI] [PubMed] [Google Scholar]
- Stubbs LJ, Carver EA, Shannon ME, Kim J, Geisler J, Generoso EE, Stanford BG, Dunn WC, Mohrenweiser H, Zimmermann W, Watt SM, Ashworth LK. Detailed comparative map of human chromosome 19q and related regions of the mouse genome. Genomics. 1996;35:499–508. doi: 10.1006/geno.1996.0390. [DOI] [PubMed] [Google Scholar]


