Abstract
The gene encoding the small nuclear ribonucleoprotein-associated polypeptide N (SNRPN) maps to the Prader–Willi syndrome critical region on chromosome 15 and is expressed preferentially from the paternal allele. A CpG island encompassing the first exon of SNRPN is methylated on the inactive maternal allele. DNA sequence was determined for a cosmid containing the first three exons of SNRPN and extending 20 kb upstream and 15 kb downstream from the CpG island. This region is extremely rich in Alu elements and other repetitive sequences and contains a single CpG island, which includes numerous short direct repeat sequences. Functional analysis of the first exon revealed strong promoter activity for a 260-bp fragment extending 207 bp upstream from the exon. In vitro methylation of this 260-bp fragment abolished promoter activity completely, suggesting that the silencing of the maternal SNRPN allele may be a direct consequence of methylation of the promoter region.
[The sequence data described in this paper have been submitted to the GenBank data library under accession no. U41384.]
The gene encoding the small ribonucleoprotein-associated polypeptide N (SNRPN) is located within chromosome 15q11–q13 in the region associated with Prader–Willi syndrome (PWS) and Angelman syndrome (AS). SNRPN is imprinted with preferential expression from the paternal chromosome (Glenn et al. 1993; Nakao et al. 1994; Reed and Leff 1994). The gene was initially described as having eight exons (Schmauss et al. 1992), but two additional upstream exons were described subsequently and designated as exons α and β (Sutcliffe et al. 1994) or −1 and 0 (Glenn et al. 1996). The imprinted expression of SNRPN is associated with preferential methylation of the maternal allele in the region of exon α, but there is a reciprocal methylation difference between the maternal and paternal alleles in intron 5 (Sutcliffe et al. 1994; Glenn et al. 1993, 1996).
PWS is believed to be associated with deficiency of paternally expressed genes, with most patients having large deletions or maternal uniparental disomy (UPD) for chromosome 15 (Nicholls 1994; Ledbetter and Ballabio 1995). AS is believed to be caused by deficiency of a maternally expressed gene, and most AS patients have large deletions or paternal UPD; a significant fraction have no molecular abnormality identified to date (Nicholls 1994; Ledbetter and Ballabio 1995). Rare PWS and AS patients have “imprinting mutations” that are associated with smaller deletions upstream of SNRPN (Sutcliffe et al. 1994; Buiting et al. 1995). These deletions prevent the appropriate erasure and reestablishment of methylation patterns and are interpreted as evidence for the existence of an imprinting center that acts to establish the methylation and expression pattern appropriate for either a paternal or maternal chromosome. Although SNRPN is the only paternally expressed gene in this region known to encode a protein, other paternally expressed transcripts have been identified including PAR-1 (Prader–Willi Angelman region) (Sutcliffe et al. 1994), PAR-5 (Sutcliffe et al. 1994), and IPW (imprinted in Prader–Willi) (Wevrick et al. 1994). The IPW gene is comprised of three exons and may encode a functional RNA, as there is no substantial open reading frame within the 2.2-kb transcript. The relatively long transcripts of PAR-5 and PAR-1 are only partially characterized with no evidence of open reading frames (Sutcliffe et al. 1994).
Prior to our identification of exon α as the most upstream exon and presumptive 5′ end of SNRPN, exon 1 was believed to be the most upstream exon. Previous analysis of exon 1 revealed detectable promoter activity, and this was interpreted as evidence that exon 1 had a natural promoter and was the first exon of SNRPN (Schmauss et al. 1992). If exon α is the true first exon, it should be associated with promoter activity, and this promoter should display some properties (e.g., response to methylation) related to the imprinted regulation of expression. To analyze the regulation of gene expression for SNRPN, a cosmid containing exons α, β, and 1, and extending 20 kb upstream, was sequenced, and the promoter activity for exons α and 1 was compared with and without in vitro methylation.
RESULTS
Sequence Analysis of Cosmid c102
The 35,882 bp of c102 was sequenced completely (Fig. 1, GenBank accession no. U41384). The repetitive elements were identified with the program CENSOR (Jurka et al. 1995) and included a total of 37 Alu elements of >200 bp, 14 Line 1 repeat fragments, 2 human satellite I DNA (HSTAI) sequences, 2 human K element-interspersed repeats (KER), 5 transposon-like human element long terminal repeat fragments (2 MSTA, 2 MLT2C2, and 1 MLT2D), 13 moderately repetitive DNA elements (MER), and 5 LTR fragments.
Figure 1.
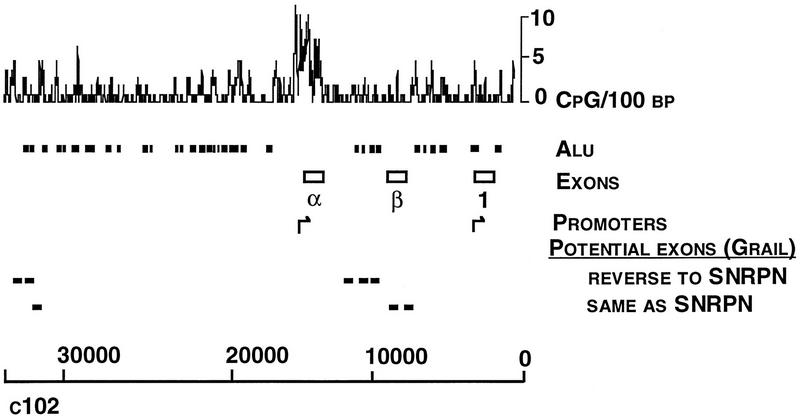
Sequence analysis of cosmid c102. The frequency of CpG dinucleotides and Alu elements are shown with the locations of exons α, β, and 1. Potential additional exons predicted from the GRAIL program are shown as in the same or opposite orientation to the transcription of SNRPN.
Cosmid c102 contains one CpG island associated with exon α of the SNRPN gene. The CpG island extends from base 15,476 to 14,436 in the c102 sequence (GenBank accession no. U41384) and is characterized by 61.6% (G + C) content and lack of CpG suppression (Fig. 1). Sequence analysis using GRAIL (Uberbacher and Mural 1991) and other search tools predicted eight open reading frames >100 bp. However, when these were compared with peptide sequence databases, no homologies to known genes were identified. GRAIL did not predict the presence of exons α and β but did identify exon 1.
The putative promoter region upstream of exon α belongs to the class of TATA-less, (G + C)-rich promoters (Ackerman et al. 1993). No initiator core promoter element (CTCANTCT) (Smale et al. 1990) was identified. In addition, the STEMLOOP program (Wisconsin Package, v. 8, Genetics Computer Group, Madison, WI) predicted the presence of multiple potential stemloop structures, indicating the presence of inverted repeats, which have been proposed to be important in this group of promoters. The COMPARE program of the same software package identified a number of direct repeats ranging from 21 to 43 bp within the SNRPN CpG island (data not shown). The sequence from 14,610 to 14,683 of c102 is similar to Lsau repeats (GenBank accession no. X59423), which are associated with heterochromatic regions of DNA (Hewitt et al. 1994). The same region (14,610–14,683) also has 63% homology to a DNA sequence (GenBank accession no. X06587) associated with hypermethylation in somatic tissue representing all three germ layers and embryonic carcinoma cells but not in sperm (Zhang et al. 1987). Using the PATTERN RECOGNITION program of the same sequence analysis software package, exon α and exon 1 promoter sequences were analyzed for putative transcriptional elements. The exon α promoter contains Sp1, AP-1, USF-II, and E1A consensus binding sites; two JCV repeats; and H4TF1 sequences. Although the region upstream of exon 1 lacked a canonical TATA box, the sequence TCTAAATG at nucleotide −42 to −35 (from 2321 to 2314 in the sequence corresponding to GenBank accession no. U41384) was highly similar to putative TATA box sequence of neuron-specific enolase (Schmauss et al. 1992). Potential trancriptional elements in sequences flanking exon 1 were noted earlier (Schmauss et al. 1992).
Promoter Analysis for Exon α and Exon 1
The promoter activity for the sequences surrounding exons α and 1 was quantitated using an expression vector with a chloramphenicol acetyltransferase (CAT) reporter gene. For each exon, the +1 bp indicates the first nucleotide of published cDNA sequence for that exon (Rokeach et al. 1989; Schmauss et al. 1992). For exon α, construct Exα/260 contained DNA from position −207 to +53 (15,612–15,352 in the sequence corresponding to GenBank accession no. U41384) and demonstrated 59-fold increased activity compared to the promoterless plasmid and 1.7-fold increased activity compared to the SV40 promoter (Fig. 2). A construct containing the same fragment in reverse orientation (Exα/260rev) did not demonstrate promoter activity. Two smaller constructs, Exα/71 containing −18 to +53 and Exα/50 containing +4 to +53, showed intermediate levels of promoter activity of 22-fold and 25-fold, respectively, compared to the promoterless plasmid. These data indicate strong promoter activity for the sequences at the 5′ end of exon α.
Figure 2.
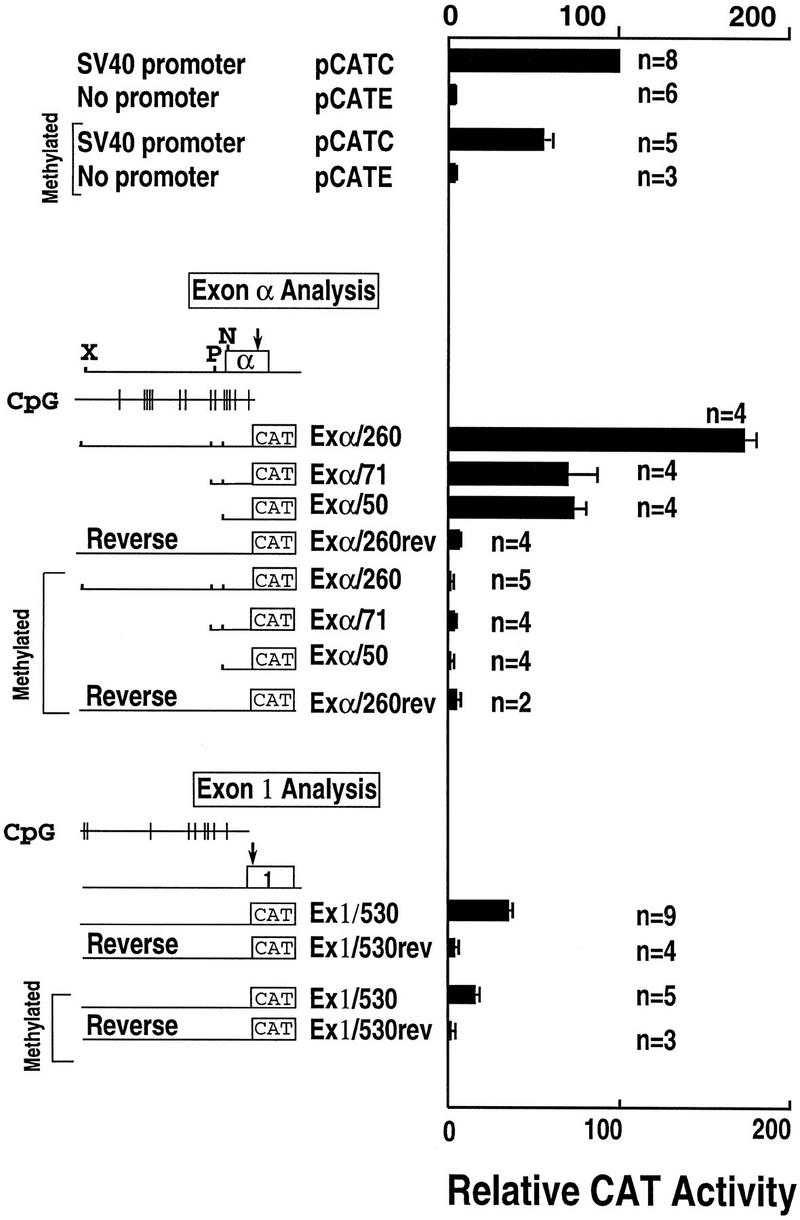
Promoter analysis for exons α and 1. CAT activity is presented relative to β-galactosidase activity for all transfections. The pCAT Control plasmid (pCATC), which includes the SV40 promoter and enhancer, is shown as 100%. The promoterless plasmid containing the SV40 enhancer is designated pCATE. Other constructs are described in the text. Separate transfection experiments (n) were performed with multiple constructs.
A similar construct was prepared for exon 1 (Ex1/530) containing the sequence from -500 to +30 (2770 to 2309 in the sequence corresponding to GenBank accession no. U41384). Consistent with previous reports, this construct yielded modest promoter activity of a 12-fold increase compared to the promoterless plasmid and only 30% as much activity as the SV40 promoter. The same fragment in reverse orientation did not demonstrate promoter activity.
Effect of Methylation on Promoter Activity
The expression of SNRPN is known to be imprinted with the maternal allele being repressed and methylated. For this reason, the effect of methylation on the CpG sequences within the SNRPN promoter region was analyzed using in vitro methylation with SssI methylase. Methylation of all of the promoter constructs from exon α caused complete loss of promoter activity (Fig. 2). Treatment of constructs with SssI methylase in the absence of the methyl donor S-adenosyl methionine did not have any effect on reporter gene activity (data not shown). The complete abolition of promoter activity for these constructs was in contrast to the SV40 promoter, which demonstrated a 46% reduction in promoter activity upon methylation. Promoter activity for the exon 1 construct was moderately reduced by methylation to 40% of the unmethylated value.
DISCUSSION
DNA sequence was determined for a cosmid that extends 20 kb upstream and 15 kb downstream from the presumptive first exon for SNRPN. Additionally, functional analysis for promoter activity was performed for the first and the third exon. The sequence indicates that the region is extremely rich in Alu elements and other repetitive sequences, and a single CpG island is located at the site of exon α. The presence of direct repeat sequences is reported to be a feature of imprinted regions (Neumann et al. 1995). This observation has led to speculation that secondary DNA structure could be a feature of imprinting control at some level, possibly influencing chromatin structure or accessibility to transcriptional machinery in a methylation-dependent manner (Neumann et al. 1995). A number of direct repeats ranging from 21 to 43 bp were identified within the SNRPN CpG island at the 5′ end of the gene. Additionally, there is a sequence 650 bp downstream of exon α that has 63% homology to Lsau repeats; these repeats are associated with heterochromatin and are hypermethylated in somatic tissues and embryonic carcinoma cells but not in sperm (Zhang et al. 1987; Hewitt et al. 1994). These observations are intriguing and may lend support to the notion that such sequences could be involved in the control of methylation and imprinted transcription.
Functional analysis was performed on the first and third exons of SNRPN, as the latter had been previously considered to be the most upstream exon and was reported to have promoter activity (Schmauss et al. 1992). Exon α was found to have strong promoter activity that was completely abolished by in vitro methylation. Interestingly, a construct containing only exonic sequences (+3 to +53) retained substantial promoter activity, although reduced to less than half of that seen with the construct with the strongest activity. This has been observed with other genes utilizing (G + C)-rich, TATAless promoters, and may represent a general feature of this class of gene (Ackerman et al. 1993). These results, in combination with the location within a CpG island, are strongly suggestive that exon α represents the normal promoter region for SNRPN. Although some promoter activity was found in the region of the third exon as reported previously, this promoter activity is associated with an Alu repeat sequence, is weaker than that for exon α, and is only partially repressed by methylation. The promoter activity associated with the third exon may not be of biological significance. Recently, Dittrich et al. (1996) reported that exons mapping centromeric to the 5′ end of SNRPN, well beyond the centromeric end of c102, form alternative, comparatively low-abundance transcripts involving internal exons of SNRPN but not exon α. These transcripts are proposed to be involved in the switching of the imprint during gametogenesis but not in the direct regulation of gene expression from the primary SNRPN transcript. The ATG that initiates translation of the SNRPN peptide sequence is located within the fourth exon (Schmauss et al. 1992). There are two additional ATG codons located within exon α (Rokeach et al. 1989), the latter of which starts a 71-amino-acid open reading frame terminating upstream of the exon 4 ATG. The significance of this open reading frame is currently unknown. The complete inhibition of promoter activity by in vitro methylation of this CpG island is consistent with the observation that the maternal copy of the SNRPN locus is normally methylated and repressed. Normal in vivo methylation of SNRPN on the maternal chromosome affects virtually every CpG dinucleotide in the exon α region (Zeschnigk et al. 1997); in vitro methylation with SssI methylase would similarly methylate every CpG dinucleotide within the construct. The data suggest that methylation is the cause rather than the consequence of transcriptional silencing of the repressed allele in the case of SNRPN.Although the sensitivity of the exon α promoter to methylation is striking, other CpG-rich promoters, including human α-globin, mouse phosphoglycerate kinase (PGK-γ), herpes simplex virus thymidine kinase (HSV–TK), and an LTR from the murine myeloproliferative sarcoma virus (PCMV) have demonstrated a similar response to in vitro methylation (Boyes and Bird 1991).
One feature common to virtually all imprinted genes is parental-specific methylation, which has been proposed for some genes to act as the mark that discriminates alleles allowing for the establishment and maintenance of imprinted expression. Although parental-specific methylation is characteristic of imprinted gene expression, there is variation with regard to the occurrence of the methylation on the active or inactive allele. For genes such as H19 and SNRPN, there is substantial methylation at the 5′ end of the gene on the repressed allele, and methylation may inhibit transcription directly (Razin and Cedar 1991; Bird 1992; Bartolomei et al. 1993; Glenn et al. 1993; Sutcliffe et al. 1994; Tremblay et al. 1995). The results of in vitro methylation studies reported here are consistent with this interpretation for SNRPN. For other genes such as Igf2r, there are multiple regions of parental-specific methylation with a reciprocal pattern such that some regions are methylated on the active allele and other regions on the inactive allele (Stöger et al. 1993). Although the most prominent methylation for SNRPN occurs on the repressed allele at the CpG island, there is a site of reciprocal methylation on the expressed paternal allele within intron 5 (Glenn et al. 1993). Not all methylation differences seen in the adult organism can act as the so-called mark or epigenetic imprint, because they are not retained during the period of global demethylation in the developing embryo (Stöger et al. 1993; Leighton et al. 1995; Tremblay et al. 1995). Overall, the data for SNRPN are quite analogous to results for H19 and for genes subject to X inactivation in that hypermethylation of the 5′ end of the gene is associated with tight repression.
METHODS
Sequencing and Analysis of Cosmid c102
Sequencing of cosmid 102 was performed according to previously published protocols (Andersson et al. 1994; Muzny et al. 1994). Intact cosmid DNA was sheared by sonication to produce random strand breakage. The ends of the sonicated fragments were repaired enzymatically, and adaptors were added using DNA ligase. The M13 vector was prepared by uracil DNA glycosylase cloning of PCR products, using PCR with uracil-containing primers, followed by uracil DNA glycosylase treatment to produce overhangs. The DNA fragments in the desired size range (1–2.5 kb) were purified and annealed to the M13 vector. Sequencing reactions were performed according to a cycle sequencing protocol using fluorescent primers and Applied Biosystem 373A automated DNA sequencers. The sequence reads were edited and assembled using a combination of software, including the Staden XDAP package and SEQPREP software developed at the Molecular Biology Computing Resource Center at Baylor College of Medicine. Gaps between contigs were closed by PCR-based strategies (Muzny et al. 1994). Various computer software packages including GRAIL (Uberbacher and Mural 1991), GCG (Sequence Analysis Software Package, v. 8, Genetics Computer Group, Madison, WI), and CENSOR (Jurka et al. 1995) were used for the analysis of sequence.
Plasmid Construction and Transfections
The exon α construct Exα/260 and the exon 1 construct Ex1/530 were prepared by inserting PCR-amplified genomic fragments from the 5′-flanking sequence of exons α and 1 into the pCAT Enhancer plasmid (Promega). The exon α fragment was amplified from the pX4.2 plasmid (Sutcliffe et al. 1994) using an exon α primer (5′-ACCGCTCCTCAGACAGATGC-3′) and a primer in the vector. The PCR fragment was cloned into a pCR II vector (Invitrogen, San Diego, CA) and then subcloned into the pCAT Enhancer plasmid in both orientations. The exon 1 fragment was amplified from plasmid pH2.7 (a 2.7-kb HindIII fragment containing exon 1) using a primer within exon 1 (5′-CTGCTGTCTAGAACGCCTCGG-3′) and a primer upstream of exon 1 (5′-ACATCTACTTGTCTAGAGGAT-3′). An XbaI site was introduced into the exon primer. The PCR fragment was digested with XbaI and subcloned into pCAT Enhancer plasmid in both orientations. The reporter plasmid pCAT Enhancer contains the CAT-coding region plus the SV40 enhancer but no promoter. The Exα/71 construct was prepared by digesting the Exα/260 construct with PstI and religating. Similarly, the Exα/50 construct was prepared by digesting the Exα/260 construct with NotI and religating. To asses the bidirectionality of these promoter regions, both the exon α and exon 1 promoter regions were subcloned in both orientations. Prior to transfection into HeLa cells, the promoter constructs were digested with restriction enzymes (EcoRV for Exα/260, PstI for Exα/71, NotI for Exα/50 and HindIII for exon 1 constructs) immediately 5′ to the inserted genomic sequences.
HeLa cells were subcultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum with a plating density of 1 × 105 cells/60-mm plate 24 hr prior to transfection. Plasmids were transfected with 1 μg of CAT construct and 8 μl lipofectamine (GIBCO BRL, Gaithesburg, MD) in serum-free medium (Optimem, GIBCO BRL, Gaithesburg, MD), and 0.1 μg of β-galactosidase expression vector pCMV β (Promega) by a cationic liposome-mediated transfection procedure under the conditions recommended by the manufacturer (GIBCO BRL) (Felgner et al. 1987). Cells were harvested 48 hr after transfection, and cellular extracts were prepared by freeze–thawing three times. Transfected cell extracts were assessed for β-galactosidase activity (Latchman 1994), heated at 65°C to inactivate deacetylase, and analyzed for CAT activity.
CAT Assays
Cell extracts were assayed for CAT activities as described elsewhere (Sambrook et al. 1989; Latchman 1994). Assays for β-galactosidase activity were performed, and activity ratios (CAT/β-galactosidase) were determined. CAT assay results were quantified using a Betascope Betascanner (Intelligenetics). Statistical values and significance tests were performed with the software INSTAT.
In Vitro Methylation of the Promoter Construct
The CpG dinucleotide sequences in these constructs were methylated in vitro by using SssI methylases and S-adenosyl methionine under conditions recommended by the manufacturer (New England Biolabs, Beverly, MA). Complete methylation was ascertained by digesting the methylated DNA with an excess (20 U/mg) of restriction enzyme pair HpaII or MspI. Only completely methylated DNA preparations were used.
Acknowledgments
This work is supported by National Institutes of Health grant R01 HG01459 (to R.A.G.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL abeaudet@bcm.tmc.edu; FAX (713) 798-8515.
REFERENCES
- Ackerman SL, Minden AG, Yeung C. The minimal self-sufficient element in a murine G-C-rich promoter is a large element with imperfect dyad symmetry. Proc Natl Acad Sci. 1993;90:11865–11869. doi: 10.1073/pnas.90.24.11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson B, Povinelli CM, Wentland MA, Shen Y, Muzny DM, Gibbs RA. Adaptor-based uracil DNA glycosylase cloning simplifies shotgun library construction for large-scale sequencing. Anal Biochem. 1994;218:300–308. doi: 10.1006/abio.1994.1182. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Webber AL, Brunkow ME, Tilghman SM. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes & Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Buiting K, Saitoh S, Gross S, Bittrich B, Schwartz S, Nicholls RD, Horsthemke B. Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nature Genet. 1995;9:395–400. doi: 10.1038/ng0495-395. [DOI] [PubMed] [Google Scholar]
- Dittrich B, Buiting K, Korn B, Rickard S, Buxton J, Saitoh S, Nicholls RD, Poustka A, Winterpacht A, Zabel B, Horsthemke B. Imprint switching on human chromosome 15 may involve alternative transcripts of the SNRPN gene. Nature Genet. 1996;14:163–170. doi: 10.1038/ng1096-163. [DOI] [PubMed] [Google Scholar]
- Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, Northrop JP, Ringold GM, Danielsen M. Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn CC, Porter KA, Jong MTC, Nicholls RD, Driscoll DJ. Functional imprinting and epigenetic modification of the human SNRPN gene. Hum Mol Genet. 1993;2:2001–2005. doi: 10.1093/hmg/2.12.2001. [DOI] [PubMed] [Google Scholar]
- Glenn CC, Saitoh S, Jong MTC, Filbrandt MM, Surti U, Driscoll DJ, Nicholls RD. Gene structure, DNA methylation, and imprinted expression of the human SNRPN gene. Am J Hum Genet. 1996;58:335–346. [PMC free article] [PubMed] [Google Scholar]
- Hewitt JE, Lyle R, Clark LN, Valleley EM, Wright TJ, Wijmenga C, van Deutekom JCT, Francis F, Sharpe PT, Hofker M, Frants RR, Williamson R. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet. 1994;3:1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- Jurka J, Knonowski P, Dagman V, Pelton P. CENSOR-a program for identification and elimination of repetitive elements from DNA sequences. Comput Chem. 1995;20:119–122. doi: 10.1016/s0097-8485(96)80013-1. [DOI] [PubMed] [Google Scholar]
- Latchman DS. Transcriptional regulation of gene expression and characterization of gene promoters and transcription factors. Methods Mol Genet. 1994;5:34–52. [Google Scholar]
- Ledbetter DH, Ballabio A. Molecular cytogenetics of contiguous gene syndromes: Mechanisms and consequences of gene dosage imbalance. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic and molecular bases of inherited disease. New York, NY: McGraw-Hill; 1995. pp. 811–39. [Google Scholar]
- Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- Muzny DM, Richards S, Shen Y, Gibbs RA. PCR based strategies for gap closure in large-scale sequencing projects. In: Ventor JC, editor. Automated DNA sequencing and analysis techniques. New York, NY: Academic Press; 1994. p. 182. [Google Scholar]
- Nakao M, Sutcliffe JS, Durtschi B, Mutirangura A, Ledbetter DH, Beaudet AL. Imprinting analysis of three genes in the Prader-Willi/Angelman region: SNRPN, E6-associated protein, and PAR-2 (D15S225E) Hum Mol Genet. 1994;3:309–315. doi: 10.1093/hmg/3.2.309. [DOI] [PubMed] [Google Scholar]
- Neumann B, Kubicka P, Barlow DP. Characteristics of imprinted genes. Nature Genet. 1995;9:12–13. doi: 10.1038/ng0195-12. [DOI] [PubMed] [Google Scholar]
- Nicholls RD. New insights reveal complex mechanisms involved in genomic imprinting. Am J Hum Genet. 1994;54:733–740. [PMC free article] [PubMed] [Google Scholar]
- Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55:451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed ML, Leff S E. Maternal imprinting of human SNRPN, a gene deleted in Prader-Willi syndrome. Nature Genet. 1994;6:163–167. doi: 10.1038/ng0294-163. [DOI] [PubMed] [Google Scholar]
- Rokeach LA, Jannatipour M, Haselby JA, Hoch SO. Primary structure of a human small nuclear ribonucleoprotein polypeptide as deduced by cDNA analysis. J Biol Chem. 1989;264:5024–5030. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmauss C, Brines ML, Lerner MR. The gene encoding the small nuclear ribonucleoprotein-associated protein N is expressed at high levels in neurons. J Biol Chem. 1992;267:8521–8529. [PubMed] [Google Scholar]
- Smale ST, Schmidt MC, Berk AJ, Baltimore D. Transcriptional activation by Sp1 as directed through TATA or initiator: Specific requirement for mammalian transcription factor IID. Proc Natl Acad Sci. 1990;87:4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöger R, Kubicka P, Liu C-G, Kafri T, Razin A, Cedar H, Barlow DP. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Nakao M, Christian S, Orstavik KH, Tommerup N, Ledbetter DH, Beaudet AL. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nature Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nature Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- Uberbacher EC, Mural RJ. Locating protein-coding regions in human DNA sequences by a multiple sensor-neural network approach. Proc Natl Acad Sci. 1991;88:11261–11265. doi: 10.1073/pnas.88.24.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevrick R, Kerns JA, Francke U. Identification of a novel paternally expressed gene in the Prader-Willi syndrome region. Hum Mol Genet. 1994;3:1877–1882. doi: 10.1093/hmg/3.10.1877. [DOI] [PubMed] [Google Scholar]
- Zeschnigk M, Schmitz B, Dittrich B, Buiting K, Horsthemke B, Doerfler W. Imprinted segments in the human genome—Different DNA methylation patterns in the Prader-Willi/Angelman syndrome region as determined by the genomic sequencing method. Hum Mol Genet. 1997;6:387–395. doi: 10.1093/hmg/6.3.387. [DOI] [PubMed] [Google Scholar]
- Zhang X, Loflin PT, Gehrke CW, Andrews PA, Ehrlich M. Hypermethylation of human DNA sequences in embryonal carcinoma cells and somatic tissues but not in sperm. Nucleic Acids Res. 1987;15:9429–9449. doi: 10.1093/nar/15.22.9429. [DOI] [PMC free article] [PubMed] [Google Scholar]


