Abstract
Y chromosome haplotypes are particularly useful in deciphering human evolutionary history because they accentuate the effects of drift, migration, and range expansion. Significant acceleration of Y biallelic marker discovery and subsequent typing involving heteroduplex detection has been achieved by implementing an innovative and cost-efficient method called denaturing high-performance liquid chromatography (DHPLC). The power of the method resides in its sensitivity and ability to rapidly compare amplified sequences in an automated manner. We have determined the allelic states of 22 Y polymorphisms; 19 of which are unreported, in 718 diverse extant chromosomes; established haplotype frequencies; and deduced a phylogeny. All major geographic regions, including Eurasia, are characterized by mutations reflecting episodes of genetic drift and expansion. Most biallelic markers are localized regionally. However, some show wider dispersal and designate older, core haplotypes. One transversion defines a major haplogroup that distinguishes a previously unknown deep, apparently non-African branch. It provides evidence of an ancient bottleneck event. It is now possible to anticipate the inevitable detailed reconstruction of human Y chromosome genealogy based on several tens to even hundreds of these important polymorphisms.
Despite considerable progress using molecular approaches to analyze mitochondrial and autosomal DNA to decipher evolutionary relationships (Stoneking 1993; von Haeseler et al. 1996; Harding et al. 1997), current attempts to understand prehistoric human migrations have been obscured by subsequent gene flow and admixture events, as well as recurrent mutation and recombination phenomena. Other complicating factors include the influence of natural selection and ascertainment bias (Bowcock et al. 1991). Although allowing the analysis of human history based on populations rather than the history of individual genes (Goldstein et al. 1995; Jorde et al. 1995), autosomal polymorphic tandem repeat motifs, such as microsatellites, are also subject to recombination processes, high mutation rates (Weber and Wong 1993), and the consequent convergent nature of the mutational processes (Shriver et al. 1993). To specifically address the issues of historical migrations, polymorphisms with low mutation rate, which allow the reconstruction of ancestral state and clearly preserve population specific haplotype information over time scales spanning human history, are essential. Principal among these are biallelic polymorphisms that are associated with the nonrecombining portion of the Y chromosome. This haploid system reflects paternal ancestry only. Its potential in exploring issues concerning the evolution of modern humans has long been recognized (Casanova et al. 1985; Ngo et al. 1986).
Recent reviews (Jobling and Tyler-Smith 1995; Hammer and Zegura 1996) about the use of Y chromosome variation in evolutionary studies reveals a scarcity of biallelic polymorphisms. The information obtained from these polymorphisms has provided only limited resolution of Y chromosome phylogenetic relationships even when combined with tandem repeat loci (Ruiz Linares et al. 1996; Santos et al. 1996; Hammer et al. 1997). The reported Y chromosome biallelic polymorphisms include an Alu insertion element (Hammer 1994) and eight base substitutions identified by sequencing (Seielstad et al. 1994; Hammer 1995; Whitfield et al. 1995; Hammer et al. 1997), denaturing high-performance liquid chromatography (DHPLC) (Underhill et al. 1996), or single-strand conformation polymorphism (SSCP) (Zerjal et al. 1997). The reasons for the apparent lack of variability on the Y are uncertain. Besides a possible recent common ancestry (Hammer 1995), the apparent reduction in diversity may be attributed simply to the fact that the effective Y chromosome population size is four times smaller than that of the autosomes. Sex and ethnic differences in migratory behavior (Cavalli-Sforza and Minch 1997) have further impacts. Selection at any locus would generate selective sweeps, also reducing sequence diversity (Dorit et al. 1995; Whitfield et al. 1995). Overall, the shortage of reported Y variation may reflect a combination of low diversity and, not to be underestimated, an inadequate experimental search effort.
Although many techniques exist for the detection of sequence variation (Landegren 1996; Taylor 1997), new economical, automated approaches are required to extensively assess Y chromosome biallelic variation if this unequaled evolutionary record of male genetic history is to be revealed in detail. Accordingly, we have developed an efficient, automated method of assessing Y chromosome biallelic variation, DHPLC (Oefner and Underhill 1995; Ophoff et al. 1996; Underhill et al. 1996; Cotton 1997). DHPLC detects heteroduplexes and is used to both identify new polymorphisms as well as subsequently genotype samples efficiently. We have identified 19 previously unreported Y biallelic markers and constructed haplotypes that display diverse global patterns of frequency and distribution. These results indicate that ample informative variation exists on the Y chromosome and that it is now possible to efficiently extract it. Although this report does not attempt to test hypotheses concerning the origins of modern humans, it does provide a substantive collection of new markers that are of direct value in helping decipher both early and subsequent regional migration patterns.
RESULTS
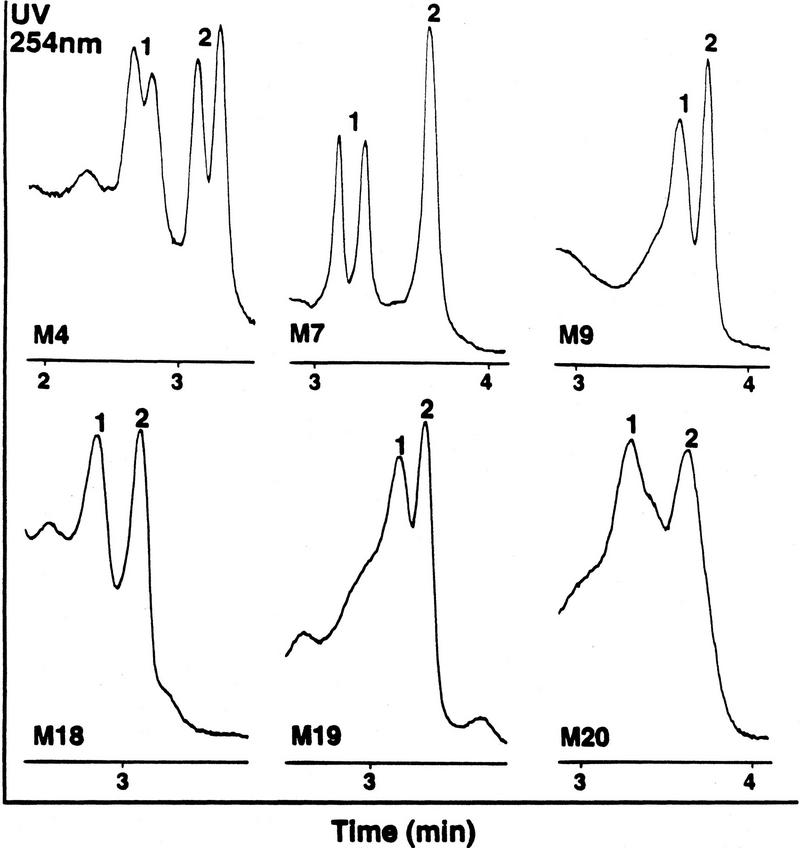
DHPLC has had a pivotal role in acquiring a substantive set of biallelic Y chromosome specific polymorphic markers (Table 1). The new mutations define nucleotide substitution or insertion/deletion events that appear to have occurred only once and therefore describe haplotypes that are common by descent from individual progenitors. Twenty of the 22 mutations listed were identified by DHPLC and 19 of these have not been described previously. DHPLC exploits the differential retention of double-stranded heteroduplex and homoduplex molecules, therefore allowing the automatic comparison of PCR amplicons for variation. DHPLC rapidly detects single-nucleotide mismatches and small insertion–deletion heteroduplexes within an amplified DNA fragment several hundred base pairs in length (Fig. 1). Consequently, the technique provides an efficient and inexpensive approach for identifying new polymorphisms. During the search phase of the process, two screening sets of male individuals, representing five continents, were examined. These two sets differed in the number of individuals and populations surveyed for variation. Initially, 9585 bp were compared in 21 individuals (set I) and nine different single-nucleotide base substitutions were detected. Subsequently, we modified and expanded the screening set to 53 individuals (set II) and surveyed an additional 9316 bp of new sequence in each individual. This effort resulted in the identification of 11 additional biallelic polymorphisms. One variant, M19, was detected unexpectedly while genotyping markers M17 and M18. Conventional sequencing of both human and chimpanzee sequence-tagged sites (STSs) defined the ancestral and derived alleles. In combination with two previously known polymorphisms, DYS287 (M1) and DYS271 (M2), the allelic states of all 22 markers were determined in 718 chromosomes. This considerable amount (22 markers) of biallelic variation on the human Y chromosome has not been obvious previously (Dorit et al. 1995; Whitfield et al. 1995). Estimates of nucleotide diversity, neutrality, and coalescence time were determined independently for set I and set II data. Mutations M1 and M19 were excluded from these estimates because they had not been identified in our screening sets. Nucleotide diversity per site, which was calculated using the UPBLUE estimator (Fu 1994), was estimated to be 3.1 × 10−4 for set I and 2.6 × 10−4 for set II data, respectively. Therefore, a one-nucleotide site difference between two randomly chosen Y chromosomes is anticipated every 3.2 × 103 to 3.8 × 103 nucleotides. A coalescence model (Fu and Li 1997), based on an effective population size of 5000 individuals and a generation time of 20 years, estimated the mean ± 95% confidence interval of age of the most recent common ancestor to be either 162,000 years (69,000–316,00 years for set I data) or 186,000 years (77,000–372,000) for set II data. Our estimate of coalescence time agrees with an estimate reported previously (Hammer 1995). Although the sensitivity of detecting heteroduplexes by DHPLC is quite high, some variation may go undetected. Therefore the estimates of diversity and coalescence time must be considered minimum estimates.
Table 1.
Description of Y Markers Used in Haplotype Construction
References indicate description of polymorphism(
) or sequence tagged site(*). Variation: ancestral → derived allele. The temperature indicates that used to identify and genotype marker by DHPLC. Site is nucleotide position from 5′ end of forward primer. M1 = DYS287, M2 = DYS271, M3 =DYS199, M4 = DYS234, M5 =DYS214, M6 = DYS198, M7 = DYS253, M8 = DYS263, M12 = DYS260, M16 = DYS214, M22 = DYS273.
Figure 1.
Representative chromatograms showing resolution of heteroduplex (peaks 1) from homoduplexes (peaks 2) created on reannealing PCR products of a male reference with males having the polymorphisms M4, M7, M9, M18, M19, and M20, respectively. Column—DNASep, 50 × 4.6-mm ID, Transgenomic, Inc.; linear gradient of 0.45% acetonitrile per minute in 0.1 m triethylammonium acetate, at pH 7.0, at a flow-rate of 0.9 ml/min; mobile phase temperatures: 52°C, 56°C, 54°C, 58°C, 58°C, and 57°C, for M4, M7, M9, M18, M19, and M20, respectively. PCR amplicons were injected directly into the preheated mobile phase.
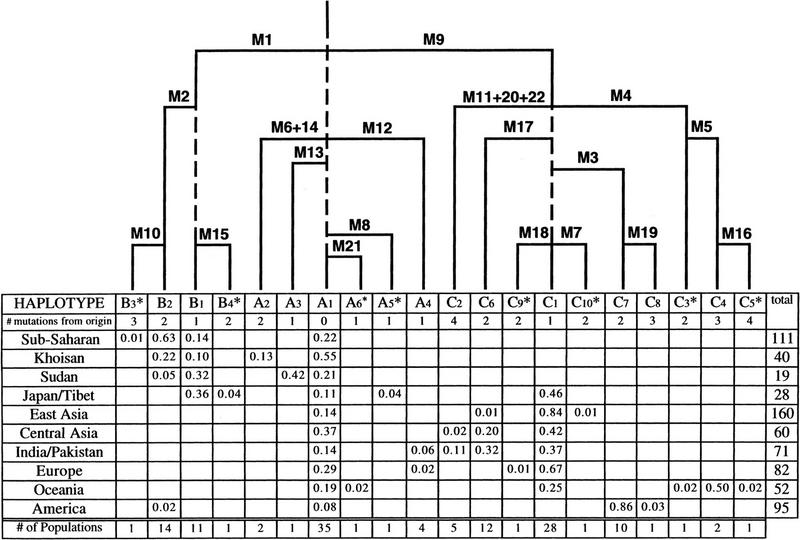
Haplotypes based on 22 biallelic polymorphic markers were constructed and haplotype frequencies determined. There was no evidence of disruption of haplotype integrity by recurrent mutation. Haplotype frequencies are given by geographical affiliation and a simple haplotype-based phylogeny has been inferred (Fig. 2). The tree is reconstructed on standard cladistic considerations together with qualitative insights gleaned from demographic data. The root is determined from the chimpanzee haplotype. Reconstruction of the branches, which is based on the assumption of a minimum number of mutational events, is immediate, and the resulting topology (i.e., the sequential occurrence of fissions) is unambiguous. Such reconstruction is simple enough that it can be done without computer analysis (Jobling and Tyler-Smith 1995). The mutations (M) are numbered, and three haplogroups, A, B, and C, are designated. The 20 haplotypes are identified alphanumerically. Although numerous sequence differences exist between the chimpanzee and human chromosomes within the 19,356 bp of sequence surveyed, 150 human chromosomes share all the chimpanzee alleles at the 22 polymorphic sites used to construct the haplotypes. Therefore, haplotype A1 is shared by some humans and chimpanzees and is used as the root from which all other human haplotypes differentiate. The second group (B) is dictated by M1, an Alu insertion (Hammer and Horai 1995), and the third group (C) by M9, an apparently non-African C → G transversion. Although 150 chromosomes within this data set remain indistinguishable from the putative ancestral haplotype A1, the majority (n = 568) of samples studied are differentiated by one or more of the 22 mutations. Two markers in particular delineate the deeper nodes in the phylogeny. Specifically, mutations M1 and M9 differentiate 120 and 427 chromosomes from the root, respectively. An additional five mutations affiliated with haplogroup A characterize 21 other chromosomes. Also given (Fig. 2) are the frequency of haplotypes by geographic location, number of mutational events from the root for each haplotype, and the number of populations in which a haplotype occurs.
Figure 2.
Relative frequencies of 20 human Y chromosome haplotypes in 10 world regions, and total numbers of males studied per region (last column). The three major haplotypes, A1, B1, and C1, are dashed to indicate the uncertainty in the order of branching points. Haplotypes appearing only once are marked with an asterisk. The principles followed in constructing the tree are described in the text. The mutations leading from one haplotype to another are indicated above each horizontal branch of the tree, and are described in Table 1. In the last row are the numbers of populations (totaling 54, listed in Methods) in which each haplotype has been observed.
We have modified the maximum parsimony tree of Figure 2, to which the root was added from knowledge of the chimpanzee genotype, by giving approximate time positions to the occurrence of mutations. These are indicated by horizontal bars stemming from the parental type that is indicated as a vertical bar. The mutation is identified above the horizontal bar (see also Table 1). When a mutation occurs in individuals already carrying a more frequent mutation (e.g., M16), typical of local differentiation, the succession pattern is definite. The tree topology as inferred by parsimony gives only the sequence of mutations, but not an estimate of their time of origin. We have inferred approximate age on the basis of the overall frequency of mutants. This is similar to results from other methods of tree reconstruction using genetic distances calculated from the frequencies of mutants (Cavalli-Sforza et al. 1994). However, we chose to use the criterion of frequency of mutants in a limited, semiquantitative way. We assume that all markers in which the derived allele is observed on only one individual (i.e., M7, M8, M10, M15, M16, M18, and M21) are the youngest mutations, and we have given the shortest (and approximately equal) length to the vertical segment representing them in the tree. At the other extreme are mutations M1 and M9, which define the major haplogroups B and C, respectively. The topology shows these are the oldest mutations. The chromosomes descended from them differ in frequency, as noted above. In the simplest situation, M9, having greater frequency, should be older than M1. But the frequency difference could be attributable to selection, drift, or even only unbalanced sampling of individuals and populations. We therefore limit ourselves to giving these two mutations, which are obviously the oldest on the basis of the topology rooted by the outgroup method, an equal time depth (their horizontal branches depart at the same height from the common stem). A similar uncertainty exists for M2 versus M15, M6 + M14 versus M12 and M13, as well as several mutations in haplogroup C, and we have given these mutations approximately equal heights, intermediate between the two extremes of the youngest and oldest. This seems a cautious way of adding the frequency criterion to the cladistic approach. We extend this analysis in the Discussion.
DISCUSSION
The ability to practically develop a significant and almost unlimited assemblage of PCR-compatible, Y chromosome-specific, biallelic polymorphisms was made possible by the development and systematic implementation of DHPLC methodology. The successful detection of 146 out of 150 known autosomal mutations (P.J. Oefner, unpubl.) indicates that DHPLC heteroduplex analysis is highly sensitive, regardless of the mismatch composition. Any inability of DHPLC technology to detect a small fraction of polymorphic loci is of little consequence when the technique is used for polymorphic genetic marker development. Only DNA sequence sufficient for STS primer design is required. Any segment of DNA several hundreds of base pairs in length that is capable of being amplified can now be efficiently and automatically tested for variation. This is achieved for the haploid Y chromosome by mixing nonpurified amplified products with a reference PCR product, denaturing and reannealing the mixture, and then analyzing for the presence or absence of heteroduplexes. The power of the approach is especially realized in those instances when sequence variation is low and consequently only homoduplex patterns are typically observed, a common occurrence for the Y. No conventional sequencing is required as the sequence has already been determined to be monomorphic in all individuals compared. Only those occasional individual samples displaying characteristic heteroduplex signatures need to be sequenced by conventional methods to precisely identify the polymorphisms. DHPLC provides an efficient approach to generate biallelic markers for any haploid or diploid genome. In addition to new biallelic marker development, DHPLC profiles often permit specific allelic determination of samples when used as a genotyping assay. For example, once during genotyping the 718 chromosomes for allele identification involving M17 and M18, a different DHPLC profile was observed, leading to the detection of a new polymorphism (M19).
Our results indicate that a detectable amount of informative genetic variation exists on the human Y chromosome when a sample screening set of diverse population composition is surveyed efficiently over thousands of nucleotides of sequence. This has not been obvious previously (Dorit et al. 1995). Such informative variation permits construction of an increasingly detailed Y chromosome phylogeny. Although still incomplete with only 22 polymorphic biallelic markers, a new, previously unrecognized, phylogenetic structure is emerging. The polymorphisms establish 20 discrete haplotypes. A considerable amount of non-African variation is evident. Excluding haplotypes A1 and B1, which occur both inside and beyond Africa, the 13 observed non-African haplotypes exceed the five haplotypes localized exclusively in Africa. This may simply reflect screening set composition bias or actual varying degrees of regional isolation and gene flow. The Y phylogeny, although bushy with numerous localized haplotypes, consists of a relatively few deep nodes, which permit the recognition of a limited number of haplogroups. Additional biallelic markers are required to improve the resolution of the current phylogeny. About 21% of the human chromosomes sampled remain indistinguishable from the inferred ancestral haplotype, A1. If a common ancestral Y chromosome of modern humans is to be eventually identified, certain populations affiliated with the haplogroup A portion of the phylogeny may be likely candidates. Obviously, the future discovery of more biallelic markers will result in the complete separation of humans from non-human primates that currently share haplotype A1.
The preponderance of genetic evidence supports a recent common African ancestry of modern humans (Batzer et al. 1994; Goldstein et al. 1995; Horai et al. 1995; Tishkoff et al. 1996a). However, the issue remains contentious (Tishkoff et al. 1996b; Wolpoff et al. 1996). Whereas the limited results given here do not resolve the issue, the distribution and diversity of haplotypes provide insights into their possible geographic origins. Two mutations, M1 and M9, distinguish deep nodes in the phylogeny. Mutation M1 is an Alu insertion element (Hammer 1994) that has been studied extensively (Hammer et al. 1997). The geographic origin of M1 is uncertain. Although M1 occurs at highest frequency in Africa, a recent report (Altheide and Hammer 1997) suggests that this mutation may have actually originated in Eurasia. The most compelling new mutation is M9, a C → G transversion, which exquisitely defines a previously unrecognized major patrilineage. Notably, M9 defines haplogroup C, an ancestral lineage that is found in all geographic regions except Africa. Its apparent absence in Africa and extensive distribution and frequency elsewhere suggests that it occurred initially outside of Africa, as a consequence of genetic drift, during an early modern human bottleneck and then dispersed widely. Alternatively, if descendants of the M9 mutant were not lost in Africa by drift or selection, an African origin of M9 may yet be identified. Other scenarios include gene flow from archaic humans and multiregional origins (Hammer and Zegura 1996). Considerable subsequent differentiation defines nine other haplotypes associated with haplogroup C. Although mutant M9 occurs at a higher frequency than mutant M1, it is impossible to determine from the current data which mutation happened first. Many other Y polymorphisms occur within distinctive locations. For example, several important geographic regions are now defined by paternal mutations displaying significant frequency. Examples include M2, which is associated with Africa; M3 with the Americas; M4 and M5 with Oceania; M17 with Central Asia, India, and Pakistan; and M13 with Sudan. Numerous other mutations, although occurring at lower frequency, are affiliated with specific populations and are valuable for forensic use, as well as for high-resolution studies of regional population substructure and gene flow. For example, M6 and M14 are associated with the Khoisan; M11, M20, and M22 with the Pakistani; M12 with Europeans and subcontinent Indians; and M19 with South American populations.
The tree of Figure 2 is built with maximum parsimony, made easier by the apparent absence of recurrence or reversion of mutations. The root is added by using the chimpanzee sequence as an outgroup. The times of origin of each mutation were assessed qualitatively by the frequency of their descendants, but we have been careful not to give excessive weight to this quantity. The Y chromosome is a haploid system, and therefore formally similar to a bacterium. In the Luria and Delbrück model (1943) of growth and mutation, the logarithm of the number of mutants (descending from a single mutation) grows linearly with the time since the origin of the mutation. Likewise, with the accumulation of clones from independent similar mutations, the relative frequency of mutants increases linearly with time. These expectations are valid for exponential growth, but human populations can rarely be expected to have regular exponential growth over the long evolutionary periods of interest to us. However, one can expect these frequencies to increase monotonically with time under almost any regime of growth in the absence of drift and selection, and therefore the times of origin of mutants would be ranked in the same order as their frequencies. We have used this criterion cautiously by creating only three major ranks of relative age—the youngest, the intermediate, and the oldest mutants defined by frequency and by cladistic topology. Assignment of relative age was tempered by the realization, already mentioned above, that drift, selection, and an incomplete representation of the world distribution of populations in our sample may affect the frequencies of mutants, subverting the order of frequency that would be expected on the basis of their time of origin. The data suggest, however, that selection and sampling biases are less important than drift. It is true that regional selection may at least in part cause the great differences of mutants observed among continents or subcontinental areas, but our minimal estimate of nucleotide diversity does not support a pronounced global selective sweep effect.
The analysis of the data in Figure 2 allows a first glance at the geographic distribution, a third criterion after tree topology and mutant frequency that can help us understand the evolutionary factors at work. The likely order of settlement of continents was from Africa to Asia, and from Asia to Oceania, Europe, and America (see Cavalli-Sforza et al. 1994). This is reflected in the tree, but the observed geographic distribution of Y chromosome diversity appears to reflect the pronounced impact of genetic drift, which accentuates bottlenecks during range expansion events (Cavalli-Sforza et al. 1993). The G allele associated with M9 is prevalent in Eurasia, the G allele of M4 in Oceania, and the T allele of M3 in America. All other later mutants show some degree of geographic clustering, indicative of the region where they first arose. This behavior is suggestive of two things—first, a greater geographic clustering of Y chromosomes compared with mtDNA and autosomes. This is likely to be attributable to gender based differences in behavior following marriage, with wives usually joining husbands (Cavalli-Sforza and Minch 1997). This difference is reinforced by the prevalence of male over female polygamy. Secondly, the strong differences in mutant frequencies in the various continents may have been due to colonization by very small parties, giving large opportunities to drift. Although regional selection cannot be discounted entirely, no evidence of a pronounced global selective sweep effect exists. Therefore, the observed geographic distribution of Y chromosome diversity appears to reflect the pronounced impact of genetic drift.
In conclusion, DHPLC-based comparative sequencing now makes it feasible to find and characterize sufficient numbers of polymorphisms with which to construct extensive compound haplotypes for phylogenetic analysis. The systematic use of DHPLC to expose the encoded history of Y chromosomes will accelerate our understanding of the global and regional aspects of human evolution as well as reveal ancient human migration and subsequent gene flow episodes. Analysis of multilocus Y chromosome compound haplotypes will provide insights into population structure and affinity that may be useful in interpreting patterns observed in genetic association studies. The continued improved resolution of the Y chromosome phylogeny, resulting from additional biallelic marker development, will provide an independent assessment of issues relating to human origins. Specifically, the prospects of comprehensively examining the proposal that modern humans originated in Africa are propitious.
METHODS
The populations and number of individuals surveyed were as follows. Africa: Biaka Central African Republic Pygmy (23), Mbuti Zaire Pygmy (15), Lisongo (4), Sudanese (19), San (40), Zulu (16), Xhosa (7), Swazi (8), Tswana (18), Soto (18), Pedi (2). Oceania: Australia (9), New Guinea (37), Bougainville Island (6). Asia: Japan (23), Tibet (5). East Asia: Han Chinese (20), Korea (1), Cambodia (18), Yakut (11), Buryat (9), Manchu (18), Blang (4), Hani (3), Taiwan Atayal (38), Ami (38). Central Asia: Khorezmian Uzbek (9), Tajik (8), Kirghiz (9), Turkmen (8), Dungan (8), Uighur (7), Kazak (9), Arab (2). India and Pakistan: Tamil (2), Pathan (10), Hunza (35), Sindhi (12), Balochi (6), Brahui (6). Europe: Basque (41), Italian (18), Sardinian (2), N. Europe (21). America: Karitiana (16), Surui (25), Maya (8), Colombia (8), Quechua (10), Ticuna (13), Guarani (4), Kaingang (3), Moskoke (6), Amerindian (2).
The composition of screening set I consisted of the following. Africa: 2 Zaire Pygmy, 2 Central African Republic Pygmy, 1 Lisongo, 1 San. Asia: 2 Japanese, 1 Chinese, 1 Cambodian, 1 Atayal. Europe: 2 Italian, 1 N. European. America: 1 Surui, 1 Karitiana, 1 Mayan, 1 Colombian Amerindian. Oceania: 1 New Guinean, 1 Australian aborigine, 1 Bougainville Islander.
Screening set II was composed of the following. Africa: 2 Zaire Pygmy, 3 Central African Republic Pygmy, 2 Lisongo, 2 San, 2 Sudanese. Asia: 2 Japanese, 2 Han Chinese, 1 Hani, 1 Blang, 2 Cambodian, 1 Atayal, 1 Ami, 2 Tibetan, 1 Korean, 2 Tamil, 2 Hunza, 1 Pathan, 1 Brahui, 1 Balochi, 2 Sindhi, 2 Arab. America: 1 Surui, 1 Karitiana, 1 Mayan, 1 Colombian Amerindian. Europe: 3 Basque, 2 Italian, 1 German, 2 Sardinian. Oceania: 2 New Guinean, 2 Australian aborigine, 2 Bougainville Islander.
The sequence examined in set I was from a subset of Y chromosome STSs mapped to various positions (Vollrath et al. 1992). All other markers were developed in sequences obtained from a Y-specific cosmid that maps to position 5 O (Vollrath et al. 1992). With the exception of DYS287 (M1), DHPLC was used to genotype all markers on 718 chromosomes. The DYS287 locus was assayed as described (Hammer and Horai 1995). Amplification and automated sequencing were performed as described previously (Seielstad et al. 1994; Underhill et al. 1996). Female DNA sample was used as a negative control. Duplexes for DHPLC analysis were created by mixing, denaturing, and reannealing a PCR product from a single arbitrary reference African with an amplicon from one of the 717 other individuals. Reannealing was achieved by reducing temperature at 1°C per minute from 95°C to 65°C, then rapidly ramping to 6°C. Positive heteroduplex controls, prepared from samples with different known alleles, were used during DHPLC genotyping analysis. The chimpanzee allele for each marker was determined in at least one chimpanzee by conventional fluorescent-based sequencing of PCR amplicons. The allelic state of marker M9 was determined in six chimpanzees, four gorillas, and two orangutans by standard sequencing.
DHPLC (Oefner and Underhill 1995; Hayward-Lester et al. 1997) was done using automated HPLC instrumentation to improve analysis productivity. All components were purchased from Rainin Instrument (Woburn, MA). All liquid contact parts were constructed of titanium, fluorocarbon, sapphire, and polyetheretherketone (PEEK), to avoid contamination of the stationary phase with metal cations that cause both a loss of recovery as well as decreased separation efficiency. In detail, the instrumentation consisted of an online degasser (DG-1210), two high-pressure pumps (SD-200), an electronic pressure module (7101-080), a 600-μl dynamic mixer (81-400TIXI), a column oven (CH-1) with air-enforced heating and Peltier cooling, an automated sample injector (AI-1A), a UV-absorbance detector (DYNAMAX, UV-C), and a Macintosh-based system controller and data analysis package. In addition, an 80-cm PEEK tubing of 0.01-inch ID, encased in a tin-alloy block (HEX-440.010, Timberline Instruments, Boulder, CO), was placed immediately before the sample loop in the column oven to thermally precondition the mobile phase. This heat-exchanger is essential for proper resolution of hetero- from homoduplex molecules, with the former eluting generally as one or two additional peaks in front of the homoduplex peak. The stationary phase consisted of 2-μ nonporous alkylated poly(styrene-divinylbenzene) particles (Huber et al. 1993) packed into 50 × 4.6-mm ID columns, which are commercially available (DNASep, Transgenomic, Santa Clara, CA). The mobile phase was 0.1 m triethylammonium acetate buffer at pH 7.0 (TEAA; cat. no. 400613, PE Applied Biosystems, Foster City, CA), containing in addition 0.1 mm tetrasodium ethylenediamine-tetraacetic acid (EDTA; cat. no. ED4SS, Sigma, St. Louis, MO). Crude PCR products, which had been subjected to an additional 3-min 95°C denaturing step followed by gradual reannealing from 95°C–65°C over a period of 30 min in a Perkin Elmer 9600 thermal cycler prior to analysis, were eluted with a linear acetonitrile (cat. no. 9017-03, J.T. Baker, Phillipsburg, NJ) gradient of 1.8% per minute at a flow-rate of 0.9 ml/min. The start- and end-points of the gradient were adjusted according to the size of the PCR products (Huber et al. 1995). Generally, analysis took less than 6 min, including column regeneration and re-equilibration to the starting conditions. The temperature required for successful resolution of heteroduplex molecules was determined empirically by injecting PCR products at increasing mobile phase temperatures until a significant decrease in retention was observed. At this point, at least 95% of all mismatches can be detected, with further improvements in resolution being feasible by substituting dGTP with 7-deaza-2′-dGTP in the PCR reaction mixture when GC-rich domains within an amplicon exceed the average melting temperature >10°C (Hayward-Lester et al. 1997).
Acknowledgments
We thank the 718 men who donated DNA that was provided, in part, by M.E. Ibrahim, R.S. Wells, M. Hsu, J. Chu, E. Mignot, P. Moral, J. Bertranpetit, P. Francalacci, J. Kidd, and P. Parham. Technical aid was provided by P. Shen, R. Hyman, F. Dietrich, B. Sun, V. Doctor, A. Hurlbut, L. Choi, C. Chan, M. Ang, L. Lee, N. Pearson, and M. Blumling. Y.-X. Fu assisted in estimating coalescence time and diversity. This work was supported by National Institute of Health grants HG00205, GM28428, and GM55273.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL under@lotka.stanford.edu; FAX (650) 725-1534.
REFERENCES
- Altheide TK, Hammer MF. Evidence for a possible Asian origin of YAP+ chromosomes. Am J Hum Genet. 1997;61:462–466. doi: 10.1016/S0002-9297(07)64077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Stoneking M, Alegria-Hartman M, Bazan H, Hass DH, Shaikh TH, Novick GE, Ioannou PA, Scheer WD, Herrera RJ, Deininger PL. African origin of human-specific polymorphic Alu insertions. Proc Natl Acad Sci. 1994;91:12288–12292. doi: 10.1073/pnas.91.25.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowcock AM, Kidd JR, Mountain JL, Hebert JM, Carotenuto L, Kidd KK, Cavalli-Sforza LL. Drift, admixture, and selection in human evolution: A study with DNA polymorphisms. Proc Natl Acad Sci. 1991;88:839–843. doi: 10.1073/pnas.88.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova M, Leroy P, Boucekkine C, Weissenbach J, Bishop C, Fellous M, Purrello M, Fiori G, Siniscalco M. A human Y linked DNA polymorphism and its potential for estimating genetic and evolutionary distance. Science. 1985;230:1403–1406. doi: 10.1126/science.2999986. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Minch E. Paleolithic and Neolithic lineages in the Europoean mitochondrial gene pool. Am J Hum Genet. 1997;61:251–254. doi: 10.1016/S0002-9297(07)64303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. Demic expansion and human evolution. Science. 1993;259:639–646. doi: 10.1126/science.8430313. [DOI] [PubMed] [Google Scholar]
- ————— . The history and geography of human genes. Princeton, NJ: Princeton University Press; 1994. [Google Scholar]
- Cotton RGH. Slowly but surely towards better scanning for mutations. Trends Genet. 1997;13:43–46. doi: 10.1016/s0168-9525(97)01011-1. [DOI] [PubMed] [Google Scholar]
- Dorit RL, Akashi H, Gilbert W. Absence of polymorphism at the YFZ locus on the human Y chromosome. Science. 1995;268:1183–1185. doi: 10.1126/science.7761836. [DOI] [PubMed] [Google Scholar]
- Fu Y-X. A phylogenetic estimator of effective population size or mutation rate. Genetics. 1994;136:685–692. doi: 10.1093/genetics/136.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y-X, Li W-H. Estimating the age of the common ancestor of a sample of DNA sequences. Mol Biol Evol. 1997;14:195–199. doi: 10.1093/oxfordjournals.molbev.a025753. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Ruiz Linares A, Cavalli-Sforza LL, Feldman MW. Genetic absolute dating based on microsatellites and the origin of modern humans. Proc Natl Acad Sci. 1995;92:6723–6727. doi: 10.1073/pnas.92.15.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF. A recent insertion of an Alu element on the Y chromosome is a useful marker for human population studies. Mol Biol Evol. 1994;11:749–761. doi: 10.1093/oxfordjournals.molbev.a040155. [DOI] [PubMed] [Google Scholar]
- ————— A recent common ancestry for human Y chromosomes. Nature. 1995;378:376–378. doi: 10.1038/378376a0. [DOI] [PubMed] [Google Scholar]
- Hammer MF, Horai S. Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet. 1995;56:951–962. [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Zegura SL. The role of the Y chromosome in human evolutionary studies. Evol Anthropol. 1996;5:116–134. [Google Scholar]
- Hammer MF, Spurdle AB, Karafet T, Bonner MR, Wood ET, Novelletto A, Malaspina P, Mitchell RJ, Horai S, Jenkins T, Zegura SL. The geographic distribution of human Y chromosome variation. Genetics. 1997;145:787–805. doi: 10.1093/genetics/145.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding RM, Fullerton SM, Griffiths RC, Bond J, Cox MJ, Schneider JA, Moulin DS, Clegg JB. Archaic African and Asian lineages in the genetic ancestry of modern humans. Am J Hum Genet. 1997;60:772–789. [PMC free article] [PubMed] [Google Scholar]
- Hayward-Lester A, Chilton BS, Underhill PA, Oefner PJ, Doris PA. Quantification of specific nucleic acids, regulated RNA processing and genomic polymorphisms using reversed-phase HPLC. In: Ferré F, editor. Gene Quantification. Basel, Switzerland: Birkhäuser Verlag; 1997. pp. 44–77. [Google Scholar]
- Horai S, Hayasaka K, Kondo R, Tsugane K, Takahata N. Recent African origin of modern humans revealed by complete sequences of hominoid mitochondrial DNAs. Proc Natl Acad Sci. 1995;92:532–536. doi: 10.1073/pnas.92.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber CG, Oefner PJ, Bonn GK. High-resolution liquid chromatography of oligonucleotides on nonporous alkylated styrene-divinylbenzene copolymers. Anal Biochem. 1993;212:351–358. doi: 10.1006/abio.1993.1340. [DOI] [PubMed] [Google Scholar]
- Huber CG, Oefner PJ, Bonn GK. Rapid and accurate sizing of DNA fragments by ion-pair chromatography on alkylated nonporous poly(styrene-divinylbenzene) particles. Anal Chem. 1995;67:578–585. [Google Scholar]
- Jobling MA, Tyler-Smith C. Fathers and sons: The Y chromosome and human evolution. Trends Genet. 1995;11:449–456. doi: 10.1016/s0168-9525(00)89144-1. [DOI] [PubMed] [Google Scholar]
- Jorde LB, Bamshad MJ, Watkins WS, Zenger R, Fraley AE, Krakowiak PA, Carpenter KD, Soodyall H, Jenkins T, Rogers AR. Origins and affinities of modern humans: A comparison of mitochondrial and nuclear genetic data. Am J Hum Genet. 1995;57:523–538. doi: 10.1002/ajmg.1320570340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landegren U, editor. Laboratory protocols for mutation detection. Oxford, UK: Oxford University Press; 1996. p. 192. [Google Scholar]
- Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo KY, Vergnaud H, Johnsson C, Lucotte G, Weissenbach J. A DNA probe detecting multiple haplotypes of the human Y chromosome. Am J Hum Genet. 1986;38:407–418. [PMC free article] [PubMed] [Google Scholar]
- Oefner PJ, Underhill PA. Comparative DNA sequencing by denaturing high-performance liquid chromatography (DHPLC) Am J Hum Genet. 1995;57:A266. doi: 10.1002/0471142905.hg0710s48. [DOI] [PubMed] [Google Scholar]
- Ophoff RA, Terwindt GM, Vergouwe MN, van Eijk R, Oefner PJ, Hoffman SMG, Lamerdin JE, Mohrenweiser HW, Bulman DE, Ferrari M, Haan J, Lindhout D, van Ommen G-JB, Hofker MH, Ferrari MD, Frants RR. Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell. 1996;87:543–552. doi: 10.1016/s0092-8674(00)81373-2. [DOI] [PubMed] [Google Scholar]
- Ruiz Linares A, Nayar K, Goldstein DB, Hebert JM, Seielstad MT, Underhill PA, Lin AA, Feldman MW, Cavalli-Sforza LL. Geographic clustering of human Y-chromosome haplotypes. Ann Hum Genet. 1996;60:401–408. doi: 10.1111/j.1469-1809.1996.tb00438.x. [DOI] [PubMed] [Google Scholar]
- Santos FR, Bianchi NO, Pena SDJ. Worldwide distribution of human Y-chromosome haplotypes. Genome Res. 1996;6:601–611. doi: 10.1101/gr.6.7.601. [DOI] [PubMed] [Google Scholar]
- Seielstad MT, Hebert JM, Lin AA, Underhill PA, Ibrahim M, Vollrath D, Cavalli-Sforza LL. Construction of human Y-chromosomal haplotypes using a new polymorphic A to G transition. Hum Mol Genet. 1994;3:2159–2161. doi: 10.1093/hmg/3.12.2159. [DOI] [PubMed] [Google Scholar]
- Shriver MD, Jin L, Chakraborty R, Boerwinkle E. VNTR allele frequency distributions under the stepwise mutation model: A computer simulation approach. Genetics. 1993;134:983–993. doi: 10.1093/genetics/134.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneking M. DNA and recent human evolution. Evol Anthropol. 1993;2:60–73. [Google Scholar]
- Taylor GR, editor. Laboratory methods for the detection of mutations and polymorphisms in DNA. Boca Raton, FL: CRC Press Inc.; 1997. p. 333. [Google Scholar]
- Tishkoff SA, Dietzsch E, Speed W, Pakstis AJ, Kidd JR, Cheung K, Bonné-Tamir B, Santachiara-Benerecetti AS, Moral P, Krings M, Pääbo S, Watson E, Risch N, Jenkins T, Kidd KK. Global patterns of linkage disequilibrium and modern human origins. Science. 1996a;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Kidd KK, Risch N. Interpretations of multiregional evolution. Science. 1996b;274:706–707. doi: 10.1126/science.274.5288.706. [DOI] [PubMed] [Google Scholar]
- Underhill PA, Jin L, Zemans R, Oefner PJ, Cavalli-Sforza LL. A pre-Columbian human Y chromosome-specific transition and its implications for human evolution. Proc Natl Acad Sci. 1996;93:196–200. doi: 10.1073/pnas.93.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollrath D, Foote S, Hilton A, Brown LG, Beer-Romero P, Bogan JS, Page D. The human Y chromosome: A 43-interval map based on naturally occurring deletions. Science. 1992;258:52–59. doi: 10.1126/science.1439769. [DOI] [PubMed] [Google Scholar]
- von Haeseler A, Sajantila A, Pääbo S. The genetical archaeology of the human genome. Nature Genet. 1996;14:135–140. doi: 10.1038/ng1096-135. [DOI] [PubMed] [Google Scholar]
- Weber JL, Wong C. Mutation of human short tandem repeats. Hum Mol Genet. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- Whitfield LS, Sulston JE, Goodfellow PN. Sequence variation of the human Y chromosome. Nature. 1995;378:379–380. doi: 10.1038/378379a0. [DOI] [PubMed] [Google Scholar]
- Wolpoff MH. Interpretations of multiregional evolution. Science. 1996;274:704–706. doi: 10.1126/science.274.5288.704d. [DOI] [PubMed] [Google Scholar]
- Zerjal T, Dashnyam B, Pandya A, Kayser M, Roewer L, Santos FR, Schiefenhövel W, Fretwell N, Jobling MA, Harihara S, Shimizu K, Semjidmaa D, Sajantila A, Salo P, Crawford MH, Ginter EK, Evgrafov OV, Tyler-Smith C. Genetic relationships of Asian and Northern Europeans, revealed by Y-chromosomal DNA analysis. Am J Hum Genet. 1997;60:1174–1183. [PMC free article] [PubMed] [Google Scholar]