Abstract
In this study we describe a new information resource that provides integrated access to information on IMAGE (integrated molecular analysis of genomes and their expression) cDNA library clones and derived expressed sequence tags (ESTs). We have developed an automated procedure that collates data from various public sources into a single ACeDB database. This database is a valuable tool for electronic cloning experiments and gene expression studies. It allows researchers to find information about cDNA libraries, plate addresses, insert sizes, and sequence data for IMAGE clones, the assignment of ESTs to UniGene clusters, and the chromosomal location of those genes in an efficient, graphically oriented manner.
Identification of all human genes and the construction of a genome-wide transcript map are two major goals of the Human Genome Project. Recognizing the potential commercial value of such information, several genome companies have made the characterization of all genes by sequencing cDNA libraries and determining the expression pattern of these genes their business plan (Carey et al. 1995). A large-scale public effort to isolate all human genes began in 1993 when the Integrated Molecular Analysis of Genomes and their Expression (IMAGE) consortium was formed to create, collect, and characterize cDNA libraries from various tissues and different states of normalization (Lennon et al. 1996). This initiative gained significant momentum when Merck & Co. provided funding to the Washington University Genome Sequencing Center to partially sequence clones from the IMAGE cDNA libraries to generate expressed sequence tags (ESTs) (Milner and Sutcliffe 1983; Putney et al. 1983; Adams et al. 1991). As of January 1997, the IMAGE human cDNA collection consisted of >500,000 clones from 50 different cDNA libraries (Hillier et al. 1996). Over 369,000 clones have been sequenced from both 5′ and 3′ ends, and an additional 21,000 clones have been sequenced only from their 5′ ends.
All of this sequence information is available from the dbEST database maintained by the National Center for Biotechnology Information (NCBI) (Boguski et al. 1993). Various groups have reported the clustering of genomic and EST sequence fragments from GenBank (Benson et al. 1996) and dbEST into groups that represent distinct human genes, including UniGene (Boguski and Schuler 1995; Schuler et al. 1996), the Merck Gene Index (Aaronson et al. 1996), and the TIGR Human cDNA Collection (Adams et al. 1995). The latest clustering results from UniGene (based on GenBank release 101) suggest that there are ∼45,918 unique gene clusters. Of those clusters, 97% are represented by at least one EST and 89% are represented by at least one IMAGE cDNA clone.
To move toward the goal of a genome-wide transcript map, a consortium of radiation hybrid (RH) mapping groups (consisting of Cambridge University, Généthon, Oxford University, the Sanger Center, Stanford University, and the MIT/Whitehead Institute) has been coordinating the placement of these unique ESTs onto the RH map (Boguski and Schuler 1995). A recently published transcript map assigns a chromosomal location to >16,000 gene clusters in UniGene (Schuler et al. 1996).
Unfortunately, there is no single resource from which the large amount of information generated by these efforts is accessible in an integrated fashion. Crucial information about cDNA clones and their associated sequences, such as which libraries they came from, clone positions in library microtiter plates, assignment to UniGene clusters, or chromosomal locations, is distributed across a number of databases and flat files available on various Internet servers. For example, information about clone identification numbers, cDNA libraries, and plate addresses of IMAGE clones can be found at Lawrence Livermore National Laboratory’s (LLNL) ftp site, whereas the clone insert size, DNA sequence, GenBank accession number, dbEST identifier, and Genome Database (GDB) (Fasman et al. 1996) identifier are located at the Washington University site. Sequence information, UniGene clustering, and chromosomal assignment are provided by the NCBI.
To address this unsatisfactory situation we have developed an integrated repository of information on IMAGE library clones and derived ESTs. Using this database, researchers can conveniently access all data available on cDNA libraries, clones, and sequences and quickly answer even complex questions such as What is the plate address of the IMAGE clone with the longest cDNA insert that is a member of a UniGene cluster that maps to a given chromosomal location?
RESULTS AND DISCUSSION
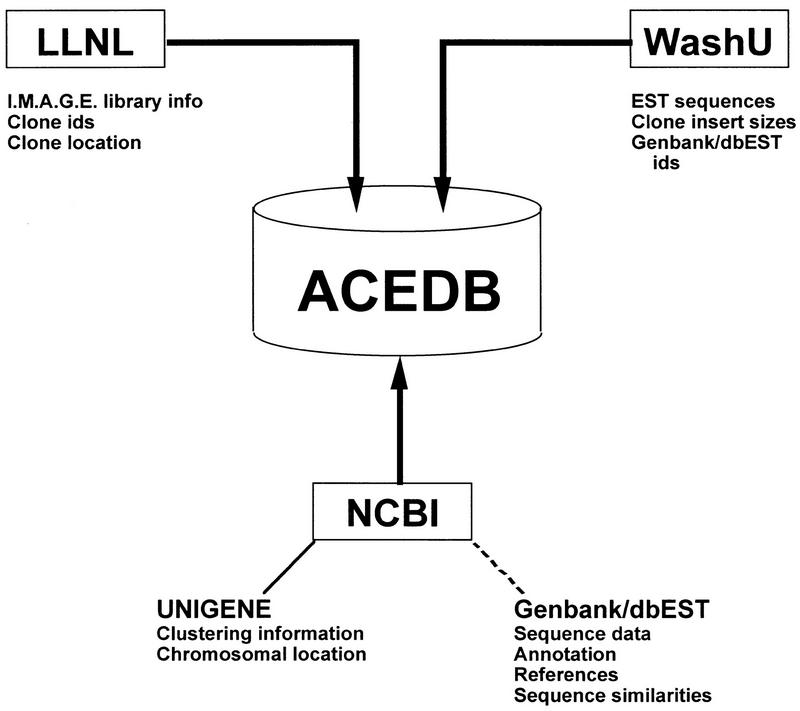
The generation of a single resource containing data on IMAGE cDNA libraries, the ESTs produced from those libraries, and the putative clustering and gene assignments for those ESTs requires the integration of information from a variety of public domain sources. To this end we have implemented a series of UNIX computer programs, each one responsible for extracting and manipulating some of the required data from the primary data sources. Figure 1 illustrates how data from distributed, heterogeneous sources is integrated into a single database. We chose ACeDB (A C. elegans Database) (Eeckman and Durbin 1995; Thierry-Mieg and Durbin 1996) as our database management system. ACeDB is publicly available and widely used in many genomic centers, its basic data model is easy to extend, and it comes with powerful data visualization capabilities. We modified the basic ACeDB data model by adding objects with information specific for IMAGE libraries, clones, EST sequences, and UniGene clusters.
Figure 1.
Data integration overview. Various types of information related to IMAGE. Library clones and derived sequences are collected from LLNL, Washington University Genome Sequencing Center, and NCBI. ACeDB acts as the database management system and visualization tool. The broken line to GenBank/dbEST indicates that information from those databases is stored in ACeDB but imported independently of the integration procedure described in this paper.
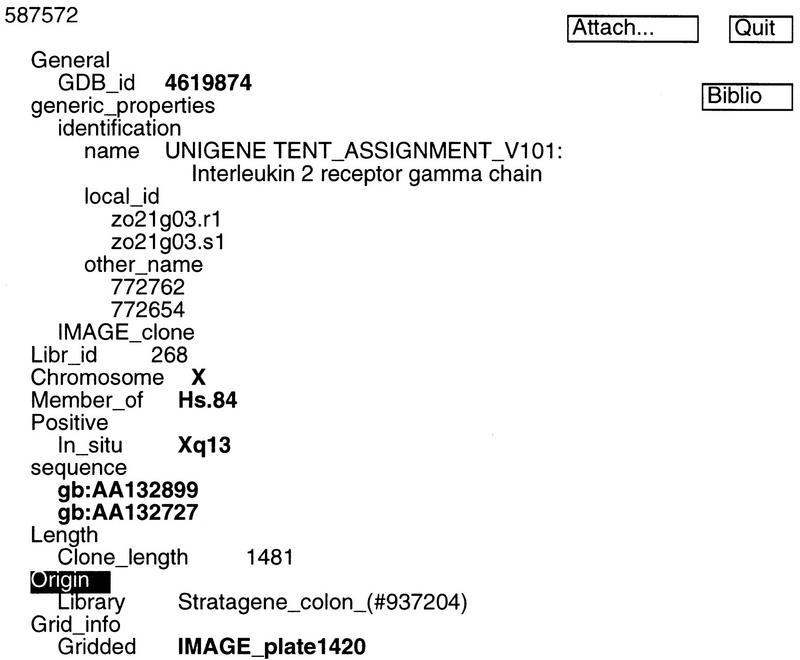
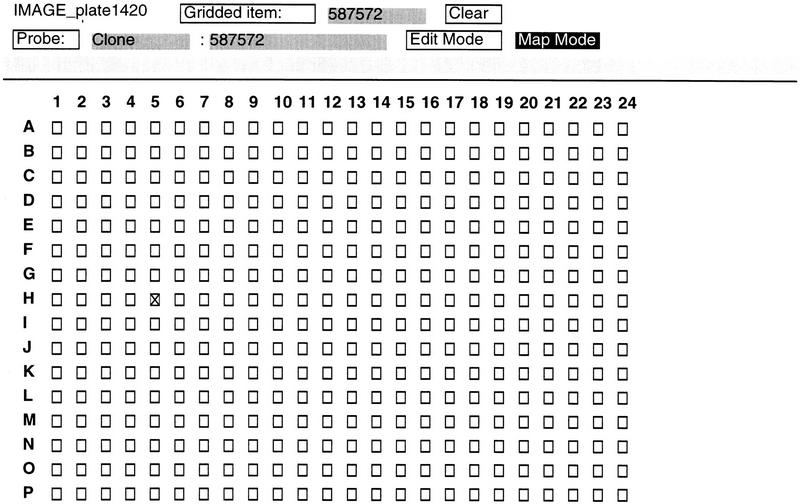
ACeDB provides a convenient framework for browsing and manipulating the integrated results. Users can query the database through one of four ACeDB class objects (chromosome, clone, grid, and sequence). Alternatively, users can query the database using the Global Search function. An example using Interleukin 2 receptor γ chain as key words identified a unigene cluster, Hs.84 (Fig. 2). This display includes information about the number of IMAGE clones in the cluster (10), the number of DNA sequences in the cluster (27), and the IMAGE clone with the largest insert size (587572). Clicking on clone 587572 reveals it has been isolated from a Stratagene colon library, there are two DNA sequences associated with the clone, its chromosomal assignment (Chr. Xq13), its insert size (1481 bp), and its location (plate 1420) (Fig. 3). Clicking on IMAGE_plate1420 brings up a grid display representing plate 1420 in the IMAGE cDNA library (Fig. 4). Each well position in this plate is marked with the LLNL cDNA ID, and the location of clone 587572 is marked as a X in the grid display (H5).
Figure 2.

ACeDB display of information contained in UniGene cluster Hs.84. All members of the cluster and their sequences are displayed.
Figure 3.
ACeDB display of IMAGE clone 587572. This screen is brought up when the user clicks on clone 587572 in Fig. 2.
Figure 4.
ACeDB display of a typical 384-well plate of the IMAGE library collection. Plate 142033 is shown.
The database described in this report has proven to be a useful tool for many research problems. The links between sequence information, UniGene clusters, and IMAGE clone identifier facilitate “cloning in silico” experiments in which researchers can quickly identify cDNA clones from IMAGE libraries that code for genes of interest (Miller and Fuchs 1997). The combination of UniGene cluster information, plate location, and sizing information plus our software’s ability to generate input files for the “Q” Bot picking robot (see Methods) allows the easy identification and reverse picking of cDNA clones representative of most known human genes for gene expression and functional genomics studies. Our long-term plans are to incorporate other functional data, including but not limited to, tissue expression [BODYMAP, Tumor Index of the National Cancer Institute (NCI)] and differential gene expression data.
CONCLUSIONS
A large amount of information on expressed sequences has been generated in the past few years by researchers in the genome community. The IMAGE libraries are a unique resource of cDNA clone material. Partial sequences from these clones have been combined with other information in GenBank and dbEST to form the UniGene collection of >50,000 potentially unique cDNA clusters. Over 30% of these clusters have been chromosomally mapped by RH mapping. The work presented here greatly improves access to all of this information by creating a single database that enables researchers to rapidly and conveniently obtain information about cDNA libraries, plate addresses, insert sizes, and sequence information for IMAGE clones, the assignment of ESTs to UniGene clusters, and the chromosomal location of those genes. Judged by the strong and growing interest in such information, we believe that the system presented should have significant practical impact.
METHODS
Implementation and Availability
ACeDB (Thierry-Mieg and Durbin 1996) version 4.43 for Sun and Silicon Graphics platforms was used as the database management system for this study.
Programs were written either in C or in the UNIX nawk language. Because the relationships between the various data files and the programs that utilize them are complex, we use the standard UNIX utility make to automate the generation of our results.
All programs, ACeDB input files, and a copy of the ACeDB models.wrm file suitable for the .ace files described in this article are publicly available from ftp://ftp.ebi.ac.uk/pub/databases/est_informer and ftp://ncbi.nlm.nih.gov/repository/EST_INFORMER so that researchers can update their databases with future data releases. We expect to make any necessary changes to the programs to handle future modifications of the input files.
Databases and Input Files
IMAGE Consortium
The IMAGE data files (id_to_names files) are retrieved from LLNL (URL: ftp://humpty.llnl.gov/pub/image/outgoing). Each of these files is a tab-delimited text file that specifies the plate numbers and well positions of individual cDNAs within a subset of the complete IMAGE cDNA library (Lennon et al. 1996).
EST Data
The EST names file is obtained from Washington University Genome Sequencing Center at URL ftp://genome.wustl.edu/pub/gsc1/est. This database is a tab-delimited text file describing each EST sequence produced by the Genome Sequencing Center and is used to establish the identities of those ESTs within the GenBank nucleotide sequence database and dbEST that are derived from IMAGE consortium cDNAs (Hillier et al. 1996).
UniGene
The UniGene database is available from the NCBI at URL ftp://ncbi.nlm.nih.gov/pub/schuler/unigene. Each release of UniGene consists of four primary data files. Only one is used for this project. The Hs.data file contains a summary of all the information that has been computed for each UniGene cluster (Boguski and Schuler 1995). This file documents the gene sequences that belong to each cluster, the chromosomal locations that have been determined for each cluster as a part of the Human Transcript Map project (Schuler et al. 1996), a gene assignment for the cluster, and a summary of previously computed similarity search results.
Construction of an Integrated ACeDB Database
The process of data integration begins with the retrieval of the raw data files from their respective repositories (Fig. 1). Once all of the original data files have been retrieved, a simple C program, make_plate_file, is used to combine all of the IMAGE id_to_name files into a single file for future processing. We refer to this file as the plate file.
The next step involves producing a version of UniGene that consists entirely of ESTs derived from IMAGE consortium cDNA collection. A C program, unigene2image, is used to filter the Hs.data file using the names file from Washington University. Because the names file contains the accession number of every human EST sequence produced at the Genome Sequencing Center, and those ESTs are, by definition, from the IMAGE consortium, we have found that we can utilize this database quite efficiently to throw out sequences within UniGene that are derived from sources other than IMAGE. Some additional useful information can be extracted from the names file at this time. The LLNL clone ID number is recorded for each accession number in the file, as well as the insert size for that clone. Finally, the orientation of each EST is recorded in the Hs.data entry for each EST sequence. All of this information is abstracted into an intermediate results file, with the following format:
| ⧣ID | Hs.xxx |
| ⧣TITLE | unigene cluster title |
| ⧣SCOUNT | nn |
| <ACCNO> | clone: <LLNL Clone> ⧣> <end> insert_length: <n> |
| … |
Each line beginning with the # character is considered a comment and starts the definition of a cluster. These lines are extracted directly from the Hs.data file. They are followed by an entry for each member of that cluster derived from an IMAGE cDNA, containing the EST sequence accession number and orientation, source cDNA, and insert size information.
These intermediate results are processed futher by a second C program, processimage. Using the insert size information in combination with the implicit clustering of the IMAGE cDNAs in UniGene, it is possible to select a representative cDNA for each cluster. Because we know from the definition of UniGene that the clusters are generated solely on the basis of 3′ identity (Boguski and Schuler 1995), by selecting the cDNA clone with the largest insert size from each cluster, we are able to represent the greatest amount of coding sequence with a minimum of redundancy.
Several command line options can be used to modify the behavior of the processimage program. In its default mode of operation, processimage outputs a file in a format suitable for the Q Bot reverse-picking robot (Genetics Ltd., Christchurch, UK). In this format, each line represents a single (plate,row,column) triple defining the position of a clone to be selected from the original IMAGE cDNA library. The identity of the clone to be selected is determined using the procedure described above. The location of that clone within the library is deduced from the plate file. A second output format can be specified using the −ace option. This format generates a text file in ACeDB format that lists, for each UniGene cluster, the identity of the longest cDNA clone representing that cluster. Finally, a −maxlen option can be used to establish an upper bound on the insert sizes of clones in the new library, which is useful when this material is to be the target of later PCR experimentation.
Next, a number of additional C programs are used to reformat each of the data files described up to this point into a format suitable for use with ACeDB. name2ace reformats the information contained within the name file into a series of annotated ACE clone objects. plate2ace provides additional information for these clone objects and produces a number of grid objects representing the 384-well plates that make up the IMAGE consortium libraries. unigene2ace extracts the name, summary, and membership information from the Hs.data file into a series of cluster objects. unigene_clones takes the intermediate file produced by the unigene2image program and provides additional information that defines which cDNA clones belong in each cluster. Finally, a nawk script called unigene-hs2ace reformats the chromosomal location information that is contained in the Hs.data file into additional annotation for each cluster. These ace files are then read into ACeDB using a modified models.wrm database definition file. Any inconsistencies in the primary data files (e.g., clones with DNA sequences but no plate addresses) are reported back to their curators for correction.
Automated Data Generation and Database Maintenance
All of these primary data sources are modified on a semiregular basis. To ensure that our results remain relevant, we have taken some care to automate the process described above. We use the unix utility expect (Libes 1994), in combination with the cron utility, to check the various raw data files for changes at their remote sites. If changes are detected, then the database curator is notified and may retrieve the new data files using a second set of automated retrieval routines.
Given modifications to one or more of the prerequisite data files, some or all of the results may need to be regenerated. We have written a makefile used by the standard unix utility make to automate the regeneration of only those results affected by the change in the source data. These new results are then loaded into ACeDB. Although we have not found this to be necessary, the existing ACeDB utility acediff could be used to track changes in the results from release to release.
To minimize the impact of changes on the results, we use the same cluster identifiers as the UNIGENE database, which remain as constant as possible.
Acknowledgments
We thank LLNL, Washington University Genome Sequencing Center, and NCBI for providing the various input files for our programs. In particular, we thank LaDeana Hillier at the Washington University Genome Sequencing Center for her help in resolving a number of discrepancies in those files. We thank Don Holt for suggestions and discussion, and Chaunbo Xu for the expect scripts which automate the download of the source data files.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ehl21107@glaxowellcome.com; FAX (919) 483-0315.
REFERENCES
- Aaronson JS, Eckman B, Blevins RA, Borkowski JA, Myerson J, Imran S, Elliston KO. Toward a development of a gene index to the human genome: An assessment of the nature of high-throughput EST sequence data. Genome Research. 1996;6:829–845. doi: 10.1101/gr.6.9.829. [DOI] [PubMed] [Google Scholar]
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polyeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O, et al. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995;377 (Suppl.):3–174. [PubMed] [Google Scholar]
- Benson DA, Bogusky M, Lipman DJ, Ostell J. GenBank. Nucleic Acids Res. 1996;24:1–5. doi: 10.1093/nar/24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, Schuler GD. ESTablishing a human transcript map. Nature Genet. 1995;10:369–371. doi: 10.1038/ng0895-369. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Lowe MJ, Tolstoshev CM. dbEST–Database for “expressed sequence tags.”. Nature Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Carey, J., J.O.C. Hamilton, J. Flynn, and G. Smith. 1995. The gene kings. Business Week May 18, pp. 72–78.
- Eeckman FH, Durbin R. ACeDB and macace. Methods Cell Biol. 1995;48:583–605. [PubMed] [Google Scholar]
- Fasman KH, Letovsky SI, Cottingham RW, Kingsbury DT. Improvements to the GDB Human Genome Data Base. Nucleic Acid Res. 1996;24:57–63. doi: 10.1093/nar/24.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L, Lennon G, Backer M, Bonaldo MF, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, et al. Generation and analysis of 280,000 human expressed sequence tags. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- Lennon G, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. Consortium: An integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- Libes D. Exploring Expect: ATCL-based toolkit for automating interactive programs. Sebastopol, CA: O’Reilly and Associates; 1994. [Google Scholar]
- Milner RJ, Sutcliffe JG. Gene expression in rat brain. Nucleic Acids Research. 1983;11:5497–5520. doi: 10.1093/nar/11.16.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GS, Fuchs R. Post-processing of BLAST results using databases of clustered sequences. Comput Appl Biosci. 1997;13:81–87. doi: 10.1093/bioinformatics/13.1.81. [DOI] [PubMed] [Google Scholar]
- Putney SD, Herlihy WC, Schimmel P. A new troponin T and cDNA clones for 13 different muscle proteins, found by shotgun sequencing. Nature. 1983;302:718–721. doi: 10.1038/302718a0. [DOI] [PubMed] [Google Scholar]
- Schuler GS, Boguski MS, Stewart EA, Stein LD, Gyapay G, Rice K, White RE, Rodriguez-Tomé P, Aggarwal A, Bajorek E, et al. A gene map of the human genome. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- Thierry-Mieg, J. and R. Durbin. 1996. ACeDB: A C. elegans database. http://www.sanger/ac/uk/acedb.