Abstract
Efficient homologous recombination permits the directed introduction of specific mutations into the yeast genome. Here we describe a cloning-free, PCR-based allele replacement method that simplifies allele transfer between yeast strains. The desired allele from one strain is amplified by PCR, along with a selectable/counterselectable marker. After transformation, the resident allele in the target strain is replaced by creating a duplication of the new allele. Selection for direct repeat recombinants results in a single copy of the new allele in the target strain. Specifically, the desired allele is amplified by PCR with a pair of adaptamers, which are chimeric oligonucleotides that are used to amplify the allele and differentially tag its 5′ and 3′ ends. These tags allow the directed fusion to two different, but overlapping, regions of an appropriately tagged selectable/counterselectable marker after a second round of PCR amplification. Following cotransformation of the two fusion fragments into yeast, homologous recombination efficiently generates a duplication of the amplified allele flanking the intact selectable marker in the genome. After counterselection, only the desired allele is retained as a result of direct repeat recombination. A simple modification of this method allows the creation of de novo mutations in the genome.
To understand the biological role of a gene, it is often necessary to study different alleles to identify distinct functions. Frequently mutant alleles are isolated in diverse genetic backgrounds. To ensure that a phenotype is attributable to a specific mutation, it is advantageous to analyze alleles in an isogenic strain background. Therefore, it is useful to be able to transfer specific mutations between different strains. In addition, in the case of conserved genes, informative alleles may even exist in different organisms. Consequently, it is desirable to be able to introduce a similar mutation into the homolog of a genetically tractable organism to analyze its phenotype in vivo. Saccharomyces cerevisiae has proven to be an ideal genetic system for these studies. Here, we take advantage of efficient homologous recombination in yeast to develop new approaches to introduce specific mutations into the genome.
Most current allele transfer methods are based on the pioneering work of Scherer and Davis (1979). An allele on a plasmid is integrated at its chromosomal locus, creating a duplication where one copy contains the new allele (“pop-in”). After a subsequent direct repeat recombination event, either the introduced or the resident allele remains in the genome (“pop-out”). To construct the plasmid, the desired allele must be (1) derived from a fragment cloned from its original strain, (2) transferred on the plasmid by gap repair (Orr-Weaver et al. 1983), or (3) cloned onto the plasmid after PCR amplification from the genome. To create a new mutation, the wild-type yeast gene sequence must be specifically altered, for example, using an Escherichia coli-based mutagenesis system (Sambrook et al. 1989). The main disadvantages of classical pop-in, pop-out recombination are that it requires cloning steps tailored for each allele and that the position of the crossover in the direct repeat recombination event determines whether or not the plasmid-borne mutation remains in the genome.
Recently, two methods have been described for transferring specific alleles into the yeast genome (Längle-Rouault and Jacobs 1995; Schneider et al. 1995). In both methods, a single round of PCR is used to generate a linear fragment that contains a short region of the gene sequence as two direct repeats flanking a selectable/counterselectable marker. Both repeats carry the desired mutation, and upon transformation the linear fragment creates a duplication of the short sequence where, ideally, the two copies in the genome contain the mutation. The advantages of these methods are that any mutation can be created easily using PCR primers without any need for cloning. Furthermore, for integrants where both copies of the duplication retain the mutation, the subsequent pop-out event always preserves the desired mutation in the genome. However, these approaches have four disadvantages: (1) Integration occurs at low frequencies, as short stretches of homologous sequences are used to target the mutant allele; (2) the frequency of the subsequent direct repeat recombination event is also very low, as the direct repeat is short; (3) the use of short sequences often results in the failure to incorporate the desired mutation into both repeats; (4) and the integration almost always creates a gene disruption, making this method cumbersome for essential genes.
In this paper we describe an improved PCR-mediated approach that overcomes many limitations of previous allele replacement methods. Specifically, long regions of homology are generated that significantly increase the frequencies of both the integration and subsequent direct repeat recombination events. In addition, the resident chromosomal copy is almost always deleted from the genome. This method can also be applied to transfer alleles into any essential gene, even in haploid cells, as long as the mutation itself does not cause lethality. Finally, an additional advantage is that successful integrants directly exhibit the phenotype of the altered allele, even before selection of pop-out recombinants.
RESULTS
A New Allele Replacement Method
We have developed a new approach for allele replacement that utilizes adaptamers, which are chimeric oligonucleotides complementary to two different DNA sequences. The fusion of two fragments is facilitated by using a pair of matched adaptamers (A and a in Fig. 1), which contain complementary sequences at their 5′ ends. In the example shown in Figure 1, adaptamers A and B are used in a PCR to amplify a fragment and tag its 5′ and 3′ ends. In a separate PCR, the matching adaptamer a is used to tag a second fragment. Subsequently, these two fragments are mixed with a suitable primer and adaptamer. During PCR, annealing at the matched ends results in a fused molecule.
Figure 1.
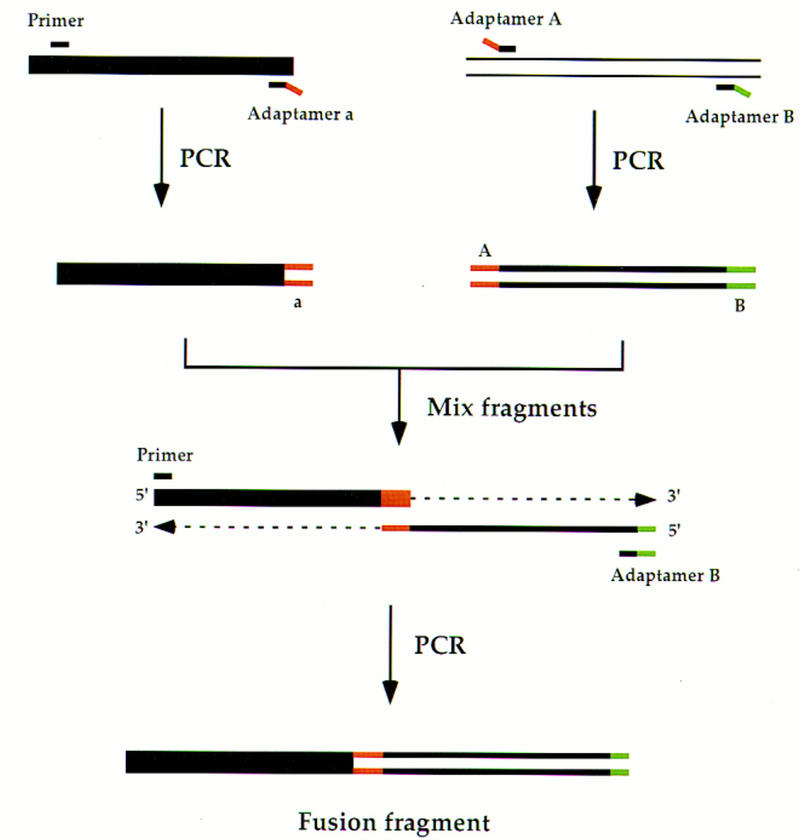
Use of adaptamers to fuse two fragments. The matched adaptamers A and a contain complementary sequence tags at their 5′ ends, as described in the text (indicated as A and a on the PCR products). The 3′ ends of each adaptamer are homologous to two different DNA sequences, respectively. Adaptamer A, in conjunction with adaptamer B, differentially tags one fragment at each end. The primer is designed to permit the PCR amplification of the other fragment, as shown. After amplification, the fragments are mixed and excess primer and adaptamer B are added for an additional PCR step. The complementary sequence tags in adaptamers A and a direct the fusion of the two fragments leading to a chimeric product.
To create the fragments for allele replacement, two sets of matching adaptamers are synthesized (adaptamers A/a and B/b). As shown in Figure 2, a fragment containing the desired allele is amplified using adaptamers A and B (fragment 1). Two truncated, overlapping fragments of a selectable/counterselectable marker (Kluyveromyces lactis URA3) are tagged with the matching adaptamers b and a, respectively (fragments 2 and 3). These two fragments are fused separately with fragment 1 to create fusions L and R after a second round of PCR. Rare, random insertions of each fusion fragment alone into the genome will not generate a functional marker (Schiestl et al. 1993). On the other hand, homologous recombination between the overlapping selectable marker fragments reconstitutes the functional marker (Ma et al. 1987). The K. lactis URA3 gene is used to reduce undesired gene conversion events between the transforming DNA and the endogenous S. cerevisiae ura3 gene (Bailis and Rothstein 1990), as they only display 71% identity (Shuster et al. 1987).
Figure 2.
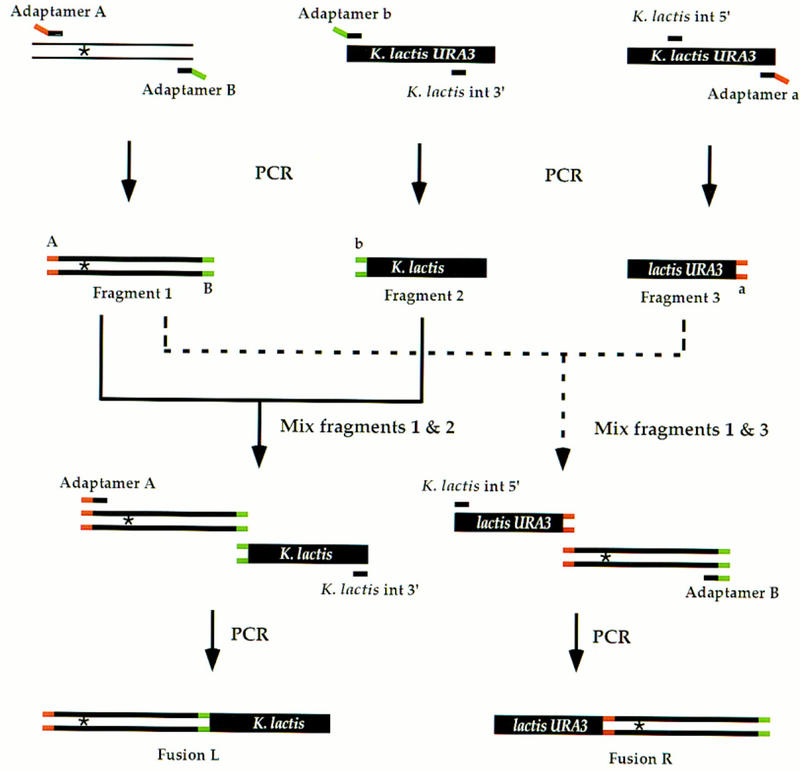
Generating fusion fragments for allele replacement. The gene of interest with an altered site [indicated by an asterisk (*)] is amplified by PCR using adaptamers A and B (fragment 1). Similarly, two overlapping K. lactis URA3 fragments are generated separately by PCR with the K. lactis URA3 adaptamers and two internal K. lactis URA3 primers (fragments 2 and 3). Fragments 2 and 3 do not encode full-length URA3 and thus are represented as K. lactis and lactis URA3, respectively. The ends tagged by adaptamers A, B, a, and b are labeled on the initial PCR products. As described in Fig. 1, fragments 1 and 2 are mixed with the K. lactis int 3′ primer and adaptamer A for an additional PCR step to generate a new fusion product (fusion L). In a separate PCR, fragments 1 and 3 are mixed with the K. lactis int 5′ primer and adaptamer B to generate a second chimeric fragment (fusion R). Both fusions L and R contain the altered site.
To generate a direct repeat of the desired allele flanking the intact selectable marker, ∼100 ng of each of the fusion fragments are cotransformed into yeast (Fig. 3). Generally, 25–100 transformants are obtained, all containing the recombined fragments integrated at the correct chromosomal target locus. The final step requires a pop-out of the direct repeats to leave a single altered copy in the genome. Because the direct repeats are gene length (∼300–3000 bp), the efficiency of recombination is high (10−3–10−4) and recombinants are easily selected on 5-fluoro-orotic acid (5-FOA) medium (Boeke et al. 1987).
Figure 3.
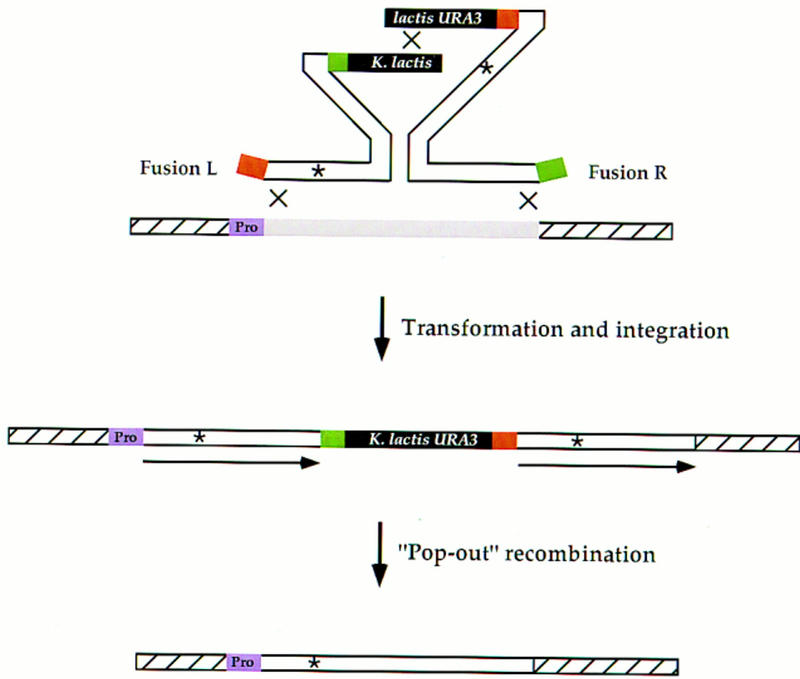
Integration of fusion fragments and subsequent pop-out event for allele replacement. Fusions L and R (Fig. 2) are cotransformed into the appropriate yeast strain. Recombination between the two fusion fragments generates a functional, intact K. lactis URA3 gene. Recombination between each fragment and the homologous chromosomal locus results in a duplication of the gene of interest where both copies contain the altered site (*). During the integration, the tags are deleted from the ends of the fragments. The left copy of the duplication lies adjacent to the endogenous promoter (purple box labeled Pro). After the subsequent pop-out event, the altered site is always preserved in the genome.
This method requires the synthesis of six primers: Two are gene-specific and four may be reused for additional allele transfer experiments. The first pair (adaptamers A and B) are ∼40-mers used to amplify the open reading frame (ORF) of the desired allele. Adaptamer A contains a unique 20-nucleotide (nt) tag at its 5′ end followed by 20 or 21 nts that is identical to the 5′ end of the ORF starting from the ATG start codon. Adaptamer B consists of a unique 20-nt tag followed by 20 nts of the reverse complement of the 3′ end of the ORF, including the termination codon sequence. The remaining four primers for amplifying the two partial K. lactis URA3 fragments are adaptamers (a and b) and two internal primers. The first 20 nts of adaptamer a contains the reverse complement of the unique tag of adaptamer A followed by 20 nts of the reverse complement of the 3′ end of K. lactis URA3 197 bp downstream from the termination codon. The 5′ end of adaptamer b contains the 20-nt reverse complement of the unique tag of adaptamer B followed by 20 nts identical to the sequence starting 283 nts upstream of the ATG start codon of K. lactis URA3. In addition, two internal primers for K. lactis URA3 were designed. One (5′ internal) contains the identical 25 nts starting 105 bp downstream from the K. lactis URA3 start codon. The other (3′ internal) contains the reverse complement of the K. lactis URA3 sequence 552 bp downstream of the ATG. These last four primers are common to every allele replacement and need to be synthesized only once. In addition, pairs of adaptamers (A and B) for every yeast ORF are commercially available (Research Genetics, Huntsville, AL, and Hudson et al., this issue).
Efficiency of the Method
To test the efficiency of the allele replacement method, we applied it to two essential genes: RFA1 and KAR1. Mutations in each gene cause an easily detectable phenotype. rfa1–D228Y mutant strains display increased UV sensitivity and exhibit increased direct repeat recombination (Smith and Rothstein 1995). The rfa1–D228Y mutation is located near the middle of the 1866-bp ORF at position 682 and creates a new AccI restriction site. To transfer this mutation into W303-1A, a wild-type yeast strain, adaptamers A and B for RFA1 were used to amplify the full-length rfa1–D228Y mutant allele. This fragment was fused separately to the two overlapping URA3 fragments described above and in Figure 2. One hundred nanograms each of the chimeric fragments were cotransformed into the wild-type strain, and 25 URA+ transformants were obtained. All 25 transformants displayed increased UV sensitivity. This phenotype indicates that the left repeat contains the full-length copy of the mutated ORF, as it is adjacent to its native promoter. Subsequent PCR analysis followed by the diagnostic restriction enzyme digestion (AccI) showed that both repeats contained the rfa1–D228Y mutation. The absence of wild-type information shows that each transformant integrated at the chromosomal RFA1 locus. Five transformants were chosen for further analysis. Direct repeat recombination events were selected and occurred at a frequency of 10−3. In each case, the event led to loss of the K. lactis URA3 marker and preserved a single copy of the rfa1–D228Y allele in the genome as shown by PCR analysis.
Next, we tested the feasibility of this method at the KAR1 locus. kar1-1 is a mutant defective in nuclear fusion (Conde and Fink 1976), resulting from a C → T transition at position 450. Similarly, the kar1-1 mutant allele was fused with the K. lactis URA3 fragments and the resulting PCR products were cotransformed into a wild-type strain selecting for URA+ transformants. The transformation efficiency was similar to that found for rfa1–D228Y and all 25 transformants carried the kar1-1 allele on the left repeat as determined by their defect in diploid formation. Pop-out recombinants were selected on 5-FOA medium for five of the transformants. Twenty recombinants from each exhibited a defect in diploid formation. The failure to recover any recombinants exhibiting wild-type mating behavior suggests that the mutation was incorporated into the second repeat as well.
Finally, we investigated whether the allele transfer method also works for deletion alleles. We used the kar1-Δ15 allele, as it carries a 255-bp deletion in the KAR1 ORF (Spencer et al. 1994). After transformation of the two fused fragments, 100 transformants were obtained on uracil-deficient medium. All of the transformants exhibited a defect in diploid formation showing that they carried the kar1-Δ15 mutation on the left repeat. Ten of these transformants were tested for the presence of the kar1-Δ15 mutation in the second repeat by examining the mating behavior after the pop-out event. Similar to that observed for kar1-1, all of the recombinants exhibited a defect in diploid formation, indicating that the kar1-Δ15 mutation was likely present in the second repeat. These results demonstrate that the allele transfer method is applicable to deletion mutations as well as point mutations.
Efficiency of Transfer of Sequences That Are Close to the Adaptamer
For some genes, the desired site may be located very close to the 5′ or the 3′ end of the gene of interest. Because sequences adjacent to the extreme 5′ and 3′ ends of the two fusion fragments may not be incorporated during integration as a result of the position of the crossover, the integration event may not result in the duplication of the allele. We determined the efficiency of allele transfer by examining the frequency of successful integration of the site as a function of distance from the adaptamer. The rfa1–D228Y allele was used as a test system, as the mutation can easily be detected by colony PCR (Huxley et al. 1990; Ling et al. 1995).
Four new adaptamers, designated A1, A21, A61, and A81, annealing 20, 40, 80, and 100 nucleotides, respectively, from the mutant site were designed for transferring the rfa1–D228Y allele. In combination with adaptamer B, the four fragments were amplified from the rfa1–D228Y mutant strain. Including the nucleotides in the adaptamer, the mutation is 21, 41, 81, and 101 nucleotides from the end of the region of homology on the amplified fragment. After fusion with the K. lactis URA3 fragments, the resulting four pairs of fragments were separately cotransformed into the wild-type strain. All of the URA3+ transformants contain duplications of the amplified RFA1 region. Next, both repeats were analyzed for the presence of the rfa1–D228Y allele and the results are summarized in Table 1. Data are also included for the full-length RFA1 fragment, where there is 681 bp of homology from the 5′ end to the mutation. Nearly 100% of the transformants contained the rfa1–D228Y mutation in both repeats when the mutation was located 81, 101, or 682 bp downstream from the 5′ end of the homologous sequence. Surprisingly, even when the homologous sequence upstream of the mutation was reduced to 20 and 40 bp, 30%–50% of the transformants contained the mutation in both repeats.
Table 1.
Efficiency of Allele Transfer
| Position of the mutation from 5‘ end of homology (bp) | Percent with the mutation in both repeats (no.) |
|---|---|
| 21st | 30 (23) |
| 41st | 53 (32) |
| 81st | 96 (26) |
| 101st | 100 (20) |
| 682nd | 100 (25) |
The numbers in parentheses indicate the total number of transformants analyzed.
A Modification of the Method Allows the Creation of de Novo Mutations
The allele replacement method described here can also be modified to create directed mutations in any gene. For this purpose, two new primers are designed. One is an adaptamer (Amut or Bmut) that contains the desired mutation (shown as a blue dot in adaptamer Amut in Fig. 4) immediately following the unique sequence tag. The other primer is called a mutamer and is used in a second round of PCR to extend the homology to facilitate integration of the mutation. A convenient length for the mutamer is 35–60 nts. The 5′ end, which typically can vary from 20 to 45 nts, are used to extend homology upstream of the mutant site. In the example shown in Figure 4, adaptamers Amut and B are used to amplify and incorporate the desired mutation into a fragment using the wild-type gene of interest as the template. The fusion fragments are generated as described for the allele transfer method except that the the mutamer is substituted for the adaptamer Amut during the amplification of the fusion fragment L. The additional nucleotides added from the mutamer effectively extend the region of homology adjacent to the mutated site to ensure its incorporation into the genome. As before, transformation of the two fusion fragments results in a direct repeat in which, ideally, the mutation is duplicated at its genomic locus.
Figure 4.
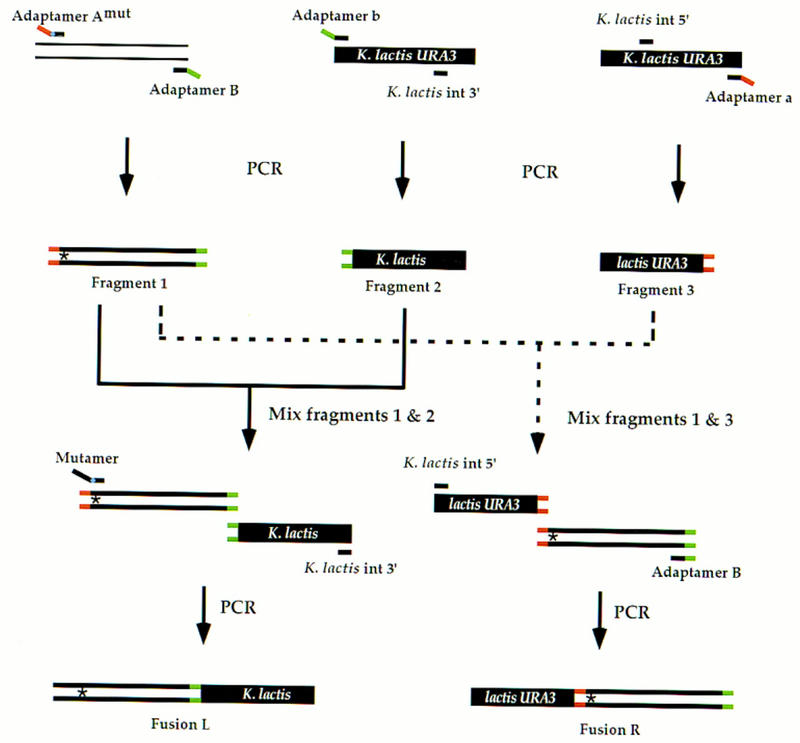
Use of adaptamer Amut and a mutamer to create a de novo mutation. Adaptamer Amut contains the sequence tag described for adaptamer A (see text) followed by an altered nucleotide(s) (indicated by the blue dot on adaptamer Amut) and an additional 20 nts of sequence adjacent to the desired change. In combination with adaptamer B, PCR is used to generate mutated fragment 1. As described in Fig. 2, fragment 1 is fused to fragment 3 generating fusion R. To create fusion L, fragments 1 and 2 are mixed with K. lactis int 3′ and a mutamer. The mutamer consists of an additional 17 nts of sequence upstream of the desired change followed by the desired change itself and 14 nts downstream. The asterisk (*) depicts the introduced mutation. Fusions L and R recombine as described in Fig. 3.
We used this method to create a lysine to arginine missense mutation at amino acid position 706 (K706R) within the conserved Walker box of the Sgs1 helicase (Gangloff et al. 1994; Lu et al. 1996). The mutation was also designed to introduce a new BglII site facilitating its detection. Adaptamer Amut contains the same adaptamer A tag sequence described before and the appropriate nucleotide change for the sgs1 mutation (A → G) along with 19 nts of downstream sequence. The mutamer contains 17 nts upstream of the G followed by 14 nts downstream of the mutant site. Using adaptamer Amut and adaptamer B, the carboxy-terminal half of the SGS1 gene sequence from the K706R mutation to its stop codon was amplified by PCR. As described above, two fusion fragments were generated and after cotransformation, 45 transformants were obtained. Colony PCR analysis followed by BglII digestion indicated that almost 25% of the transformants (11) contained the sgs1–K706R allele in both copies of the duplication. This efficiency, using 17 nucleotides of extended homology, is comparable to that found when 20 nucleotides were placed upstream of the rfa1–D228Y mutation (30% showed duplication of the mutation; Table 1).
DISCUSSION
In this paper we describe a new method that considerably improves existing allele replacement methods and can be modified to permit directed mutagenesis of any site within the yeast genome. One of the major advantages of this method is that no cloning steps are necessary. In addition, many of the required reagents are commercially available. Compared to previously described PCR-based allele-replacement methods (Längle-Rouault and Jacobs 1995; Schneider et al. 1995), the integration and subsequent pop-out recombination frequencies are elevated at least two orders of magnitude because of the increased length of homology. Another important advantage of this method is that after integration, both copies of the duplicated gene carry the alteration. This ensures that the mutation will always be preserved in the genome after the subsequent pop-out event, thereby eliminating further screening of recombinants. Moreover, in most instances, the effects of a specific alteration can be assessed immediately after transformation, as one altered copy recombines adjacent to its endogenous promoter. Finally, with this allele-replacement method, integration of the fusion fragments into the genome does not create a gene disruption (Shortle et al. 1982), which often occurs with the other PCR-based methods. Therefore, the adaptamer method can be used for allele transfer into essential genes in haploid cells provided that the mutation itself does not create a lethal phenotype.
As is the case with other PCR-based methods, undesired second site mutations may be generated by polymerase errors during the PCR amplification. However, this problem can be addressed by using high-fidelity polymerases like Pfu (Stratagene) or Pwo (Boehringer-Mannheim), where the error frequencies of these enzymes are much lower than that of Taq polymerase (Barnes 1992) and/or by keeping the number of PCR cycles at a minimum during amplification. In addition, a new technique has been described that uses the mismatch repair complex from E. coli, the MutHLS proteins to remove the mistakes from PCR products (Smith and Modrich 1997). Finally, it is always advisable to compare the phenotypes of several transformants to ensure that the effect is caused by the allele of interest and not by a secondary mutation.
We demonstrated that this method works successfully when the length of the amplified fragment is up to 3.1 kb (sgs1–K706R). In principle, even longer fragments should be amplified easily with improved enzymes (e.g., LA Taq polymerase (TaKaRa Biochemicals); rTth DNA polymerase (Perkin Elmer). However, an alternative solution for longer ORFs is readily available by simply fragmenting the gene of interest (see Fig. 5). This requires the synthesis of a new adaptamer. The choice of which of the two adaptamers to replace depends on the position of the desired site within the ORF. When the site lies nearer to the 3′ end, a new adaptamer Aint (defined as an internal position in the ORF) is designed (Fig. 5A). In conjunction with adaptamer B, a shorter 3′ fragment of the gene is amplified. After fusion to the K. lactis URA3 fragments and subsequent transformation, the resulting integration only duplicates the 3′ fragment but does not create a gene disruption (see Fig. 5A). When the desired site is closer to the 5′ end of the ORF, a new adaptamer Bint is used with adaptamer A (Fig. 5B). After integration, a gene disruption will be created, as the resulting duplication of this shortened 5′-amplified fragment gives rise to one 3′-truncated copy and one promoterless copy of the gene. To circumvent gene disruption when the gene of interest is essential, the 5′ end of the duplicated fragment can be expanded to include the promoter. This requires the synthesis of a new adaptamer Apro (defined by sequences upstream of the ORF) that permits amplification of the promoter (Fig. 5B). When adaptamer Apro is used in combination with the adaptamer Bint, the promoter region also will be duplicated resulting in a full-length copy of the gene after integration. Finally, if the desired site lies within the genomic sequences of the adaptamers, it is necessary to synthesize a new adaptamer just outside of the ORF near that site.
Figure 5.
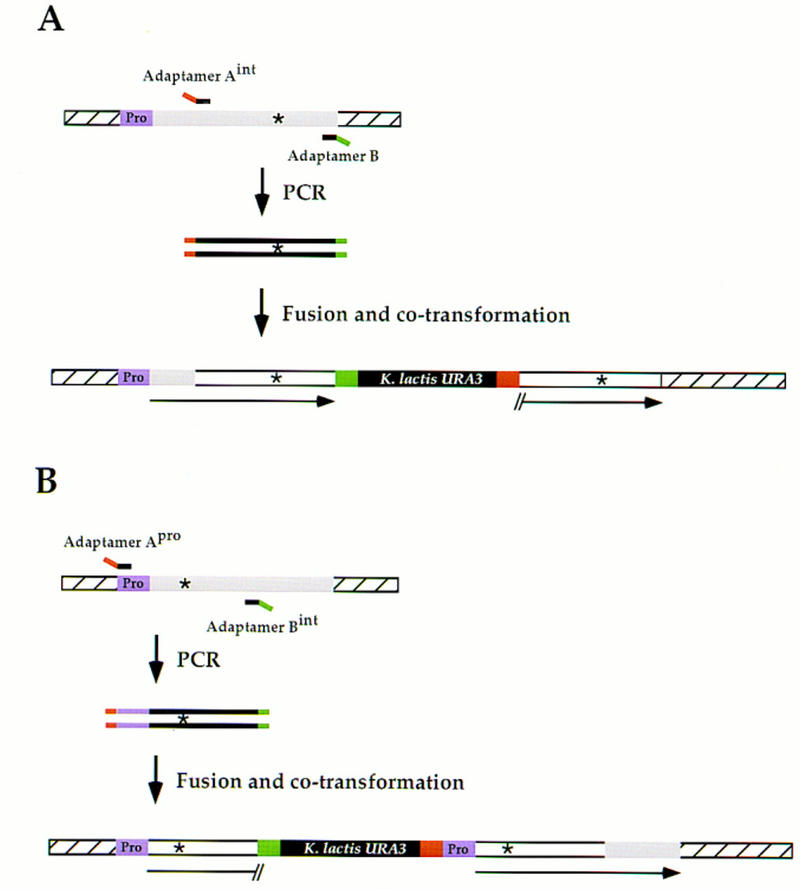
Fragmentation of large sequences for allele transfer. (A) When the mutation is close to the 3′ end of a large gene (>2.5 kb), a new adaptamer Aint is needed to amplify the 3′ portion of the ORF. After amplification using adaptamer Aint and adaptamer B, the fragment is fused to K. lactis URA3 fragments and cotransformed into yeast as described in Figs. 2 and 3. Integration results in a full-length ORF only in the left copy following recombination that fuses the promoter (purple box labeled Pro) and the endogenous, nonamplified region of the ORF (shaded box) with the duplicated 3′-amplified fragment (open box). The fragment on the right is truncated upstream of the sequence homologous to adaptamer Aint. (B) When the mutation is close to the 5′ end in long essential genes, allele transfer requires two new adaptamers. Adaptamer Apro and adaptamer Bint are used to amplify the 5′ portion of the ORF including the promoter. After fusion to K. lactis URA3 and cotransformation (Figs. 2 and 3), integration results in the generation of the full-length ORF with its promoter in the right repeat. The left copy contains a 3′ truncation downstream of the sequence homologous to adaptamer Bint.
The creation of de novo mutations within the ORF using adaptamer Amut or Bmut always leads to the amplification of shortened gene fragments. The choice of which adaptamer to synthesize to create the mutation relies on the same criteria discussed above for fragmentation of large ORFs. Similarly, to avoid disruption of an essential gene when adaptamer Bmut is used, an adaptamer Apro is necessary to create the amplified fragment. In addition, as the phenotype of the new mutation is unknown in most cases, it is advisable to design the alteration so that it can also be easily monitored physically (e.g., creation or destruction of a restriction enzyme site; generation of a deletion or an insertion). Another potential improvement of the de novo mutation method involves the length of the mutamer. Although we showed that only 17 bp of homology upstream of the mutation was sufficient to obtain successful incorporation, the frequency of these events was low (25%). This frequency will likely increase with the length of the mutamer. However, it is important to simultaneously extend the length of the requisite K. lactis internal primer to avoid temperature differences for annealing during generation of the fusion fragment.
In this study we found that the frequency of integration into the genome is ∼100-fold greater than that found with the other PCR-based allele replacement methods (Längle-Rouault and Jacobs 1995; Schneider et al. 1995). This is likely attributable to the increased length of the homologous sequences used for targeting. Interestingly, 96% of the integrants exhibit crossovers within the first 80 bp of the homologous sequences (Table 1, line 3). We also found that the presence of the 20-bp nonhomologous tag, which is eliminated by recombination, does not interfere with the integration event. This is supported by the observation that a site 17 bp from the 5′ end without the tag (sgs1–K706R) is incorporated as efficiently (25%) as a tagged site 20 bp from the 5′ end (rfa1–D228Y, 30%). This result agrees with those obtained with another PCR-based allele replacement method, where it was shown that 22% of the transformants carried the alteration in both repeats when it was 24 bp from the ends of the fragment (Längle-Rouault and Jacobs 1995). Finally, although the frequency of integration of longer fragments increases 100-fold, it is still not as high as classical gene disruptions (Rothstein 1983). Perhaps the two fusion fragments used for the integration circularize, as these two fragments contain homologous regions at both ends: the amplified allele and the K. lactis URA3 overlap. Such a circle would integrate into the genome at low efficiency and also decrease the total number of recoverable integration events by removing linear fragments from the “recombination pool.”
In summary, the allele-replacement method described in this paper is convenient for use in yeast, as a set of adaptamers is commercially available for amplifying every ORF in the genome (Research Genetics). In addition, the K. lactis URA3 fragments can be synthesized in batches simplifying future fusions. Moreover, for the new adaptamers and mutamers that need to be synthesized, we showed that only 15 nts are needed for priming most PCR amplifications. In principle, these allele transfer methods can be applied to any organism that undergoes efficient homologous recombination, has genomic DNA sequence information available, and has a suitable selectable/counterselectable marker.
METHODS
Strains and Growth Conditions
Standard yeast genetic methods are employed for the analysis of strains and the preparation of the media (Sherman et al. 1986). The yeast strains used in this study are all derivatives of W303-1A unless otherwise noted. W303-1A is MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 (Thomas and Rothstein 1989). The kar1-1 strain was a gift from G. Fink (Conde and Fink 1976), and the kar1-Δ15 strain was obtained from Phil Hieter (Spencer et al. 1994). The E. coli strains are derivatives of TG1 (Sambrook et al. 1989).
Plasmid Constructions
The K. lactis URA3 gene was amplified from a K. lactis strain by a PCR using adaptamers a and b of K. lactis URA3. The ends of the amplified product were made flush using T4 DNA polymerase (New England Biolabs), and pWJ716 was constructed by cloning the blunt-ended K. lactis URA3 fragment into the SmaI site of pRS414 plasmid (Sikorski and Hieter 1989).
Yeast Transformation
Yeast transformations were performed according to the high-efficiency lithium acetate protocol without any modifications (Schiestl and Gietz 1989). One hundred nanograms each of the two fusion fragments were used to transform yeast cells that routinely yield 105–106 transformants per microgram of uncut circular DNA (pWJ716).
PCR
The primers used in this study are listed in Table 2. Standard conditions were used for PCR amplification (Erlich 1989). To amplify two different but overlapping K. lactis URA3 fragments, 100-μl reactions were composed of 100 pg of pWJ716, 10 mm Tris (pH 8.3), 1.5 mm MgCl2, 50 mm KCl, 0.2 mm each of dNTPs, 1 μm primers, and 5 units of Taq DNA polymerase (Boehringer Mannheim). The rfa1–D228Y, kar1-1, and kar1-Δ15 mutations were amplified by colony PCR in 100-μl reactions from their respective strains. The above described PCR amplifications were performed on a Perkin Elmer 9600 as follows: 3 min at 94°C, then a cycle of 94°C for 30 sec, 54°C for 30 sec, and 72°C for 1 min repeated 35 times. The last cycle was followed by 5 min at 72°C. All PCR products were purified using GeneClean II (Bio101, Inc.).
Table 2.
Primers Used in this Study
| Name | Sequence |
|---|---|
| Adaptamer b for K. lactis URA3 | CATGGCAATTCCCGGGGATCGTGATTCTGGGTAGAAGATCG |
| Adaptamer a for K. lactis URA3 | CATGGTGGTCAGCTGGAATTCGATGATGTAGTTTCTGGTT |
| K. lactis internal 5‘ primer | CTTGACGTTCGTTCGACTGATGAGC |
| K. lactis internal 3‘ primer | GAGCAATGAACCCAATAACGAAATC |
| Adaptamer ARfa1 for RFA1 | AATTCCAGCTGACCACCATGATGAGCAGTGTTCAACTTTC |
| Adaptamer BRfa1 for RFA1 | GATCCCCGGGAATTGCCATGTTAAGCTAACAAAGCCTTGG |
| Adaptamer A1 for RFA1 | AATTCCAGCTGACCACCATGTATTCAATGTCAACTTCTTG |
| Adaptamer A21 for RFA1 | AATTCCAGCTGACCACCATGCAATCAAAGAGGTGATGG |
| Adaptamer A61 for RFA1 | AATTCCAGCTGACCACCATGAAGAGTTTCCTACAAGGGAG |
| Adaptamer A81 for RFA1 | AATTCCAGCTGACCACCATGAACGTTTGGACTATCAAAGC |
| Adaptamer AKar1 for KAR1 | AATTCCAGCTGACCACCATGATGAATGTAACTTCTCCAAA |
| Adaptamer BKar1 for KAR1 | GATCCCCGGGAATTGCCATGTTAAAACCTATAATACACAT |
| Adaptamer Amut for SGS1 | AATTCCAGCTGACCACCATGGATCTCTTTGCTATCAACTTC |
| Adaptamer BSgs1 for SGS1 | GATCCCCGGGAATTGCCATGCTTTCTTCCTCTGTAGTGACC |
| Mutamer for sgs1 | GCCAACAGGGGGTGGTAGATCTCTTTGCTATC |
| RFA1–2A | CAGAGCATCCAAATGAAACC |
| RFA1–3B | TTTGGATAATACCGAGGACG |
The fusion fragments were generated by a modification of the PCR protocol described above. Specifically, 100-μl reactions were prepared from 10–25 ng each of the mutant allele and the appropriate K. lactis URA3 fragment, 10 mm Tris (pH 8.3), 1.5 mm MgCl2, 50 mm KCl, 0.2 mm each of the dNTPs, 1 μm primers, and 5 units of Taq polymerase (Boehringer Mannheim). The PCR amplifications were performed on a Perkin Elmer 9600 using the following conditions: 3 min at 94°C, then 10 cycles of 94°C for 30 sec, 54°C for 15 sec, and 72°C for 4 min, followed by another 20 cycles of 94°C for 30 sec, 60°C for 15 sec, and 72°C for 4 min with addition of 30 sec at every cycle for the elongation step. The last cycle was followed by 5 min at 72°C. Again the PCR products were gel-purified from agarose gels using GeneClean II (Bio101, Inc.).
Colony PCR for detection of the rfa1–D228Y allele was performed using RFA1–2A and RFA1–3B primers (Smith and Rothstein 1995) under standard PCR conditions described above for amplifying the K. lactis URA3 fragments. The total volume of the PCR was 10 μl, and cells that are barely visible on a toothpick were used as the template.
Acknowledgments
We thank Fred Chang, Stan Fields, Jim Hopper, Dan Lockshon, Steve Sturley, Marcel Wehrli, Justin Weinstein, Xiaolan Zhao, and Hui Zou for constructive comments on the manuscript. We also thank Jane Love for help in conceiving the initial concept. This work was supported by the Carlsberg Foundation (U.H.M.), the Danish Natural Science Research Council (U.H.M.), and National Institutes of Heath grants GM50237 (R.R.) and HG01620 (R.R.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rothstein@cuccfa.ccc.columbia.edu; FAX (212) 923-2090.
REFERENCES
- Bailis AM, Rothstein R. A defect in mismatch repair in Saccharomyces cerevisiae stimulates ectopic recombination between homeologous genes by an excision repair dependent process. Genetics. 1990;126:535–547. doi: 10.1093/genetics/126.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes WM. The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion. Gene. 1992;112:29–35. doi: 10.1016/0378-1119(92)90299-5. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Conde J, Fink GR. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich HA. PCR technology: Principles and applications for DNA amplification. New York, NY: Stockton Press; 1989. [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, J.R., E.P. Dawson, K.L. Rushing, C.H. Jackson, D. Lockshon, D. Conover, C. Lanciault, J.R. Harris, S.J. Simmons, R. Rothstein, and S. Fields. 1997. The complete set of predicted genes from Saccharomyces cerevisiae in a readily usuable form. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Huxley C, Green ED, Dunham I. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 1990;6:236. doi: 10.1016/0168-9525(90)90190-h. [DOI] [PubMed] [Google Scholar]
- Längle-Rouault F, Jacobs E. A method for performing precise alterations in the yeast genome using a recycable selectable marker. Nucleic Acids Res. 1995;23:3079–3081. doi: 10.1093/nar/23.15.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling M, Merante F, Robinson BF. A rapid and reliable DNA preparation method for screening a large number of yeast clines by polymerase chain reaction. Nucleic Acids Res. 1995;23:4924–4925. doi: 10.1093/nar/23.23.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Mullen JR, Brill SJ, Kleff S, Romeo AM, Sternglanz R. Human homologues of yeast helicase. Nature. 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Rothstein RJ. One-step gene disruption in yeast. In: Wu R, Grossman L, Moldave K, editors. Methods in enzymology. New York, NY: Academic Press; 1983. pp. 202–211. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scherer S, Davis RW. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Dominska M, Petes TD. Transformation of Saccharomyces cerevisiae with nonhomologous DNA: Illegitimate integration of transforming DNA into yeast chromosomes and in vivo ligation of transforming DNA to mitochondrial DNA sequences. Mol Cell Biol. 1993;13:2697–2705. doi: 10.1128/mcb.13.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Shortle D, Haber JE, Botstein D. Lethal disruption of the yeast actin gene by integrative DNA transformation. Science. 1982;217:371–373. doi: 10.1126/science.7046050. [DOI] [PubMed] [Google Scholar]
- Shuster JR, Moyer D, Irvine B. Sequence of the Kluyveromyces lactis URA3 gene. Nucleic Acids Res. 1987;15:8573. doi: 10.1093/nar/15.20.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Modrich P. Removal of polymerase-produced mutant sequences from PCR products. Proc Natl Acad Sci. 1997;94:6847–6850. doi: 10.1073/pnas.94.13.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Rothstein R. A mutation in the gene encoding the Saccharomyces cerevisiae single-stranded DNA-binding protein Rfa1 stimulates a RAD52-independent pathway for direct-repeat recombination. Mol Cell Biol. 1995;15:1632–1641. doi: 10.1128/mcb.15.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F, Hugerat Y, Simchen G, Hurko O, Connelly C, Hieter P. Yeast kar1 mutants provide an effective method for YAC transfer to new hosts. Genomics. 1994;22:118–126. doi: 10.1006/geno.1994.1352. [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. The genetic control of direct-repeat recombination in Saccharomyces: The effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics. 1989;123:725–738. doi: 10.1093/genetics/123.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]


