Abstract
Fibroblast growth factors (FGFs) are a family of structurally related polypeptides that are essential for embryonic development and that function postnatally as homoeostatic factors, in the response to injury, in the regulation of electrical excitability of cells and as hormones that regulate metabolism. In humans, FGF signalling is involved in developmental, neoplastic, metabolic and neurological diseases. Fgfs have been identified in metazoans but not in unicellular organisms. In vertebrates, FGFs can be classified as having intracrine, paracrine and endocrine functions. Paracrine and endocrine FGFs act via cell-surface FGF receptors (FGFRs); while, intracrine FGFs act independent of FGFRs. The evolutionary history of the Fgf family indicates that an intracrine Fgf is the likely ancestor of the Fgf family. During metazoan evolution, the Fgf family expanded in two phases, after the separation of protostomes and deuterostomes and in the evolution of early vertebrates. These expansions enabled FGFs to acquire diverse actions and functions.
Keywords: Development, disease, evolution, FGF, metabolism
Various signalling pathways are activated in a highly coordinated manner to ensure proper development and morphogenesis in vertebrates. Secreted signalling molecules such as fibroblast growth factors (FGFs), bone morphogenetic proteins (BMPs), WNTs and Hedgehogs play crucial roles in development and morphogenesis by acting over variable distances to influence intracellular signalling events in neighbouring cells. FGFs are polypeptide growth factors with diverse biological activities. The mammalian FGF family comprises 22 members. These FGFs can be classified as intracellular FGFs (iFGFs), canonical FGFs and hormone-like FGFs (hFGFs) by their mechanisms of action (1). In this review, we refer to intracellular, canonical and hFGFs as intracrine, paracirne and endocrine FGFs, respectively. Paracrine FGFs mediate biological responses by binding to and activating cell surface tyrosine kinase FGFRs. They act as local paracrine signalling molecules and function in multiple developmental processes including differentiation, cell proliferation and migration (1, 2). Endocrine FGFs are thought to mediate biological responses in an FGFR-dependent manner. However, they function over long distances as endocrine hormones (3, 4). In contrast, intracrine FGFs act as FGFR-independent intracellular molecules that regulate the function of voltage gated sodium channels (5, 6).
The mouse is a widely used mammalian model for studying gene function. Targeted mutagenesis of Fgf genes in mice has elucidated their functions in development and metabolism. In addition, evidence for the involvement of FGF signalling in hereditary, paraneoplastic and metabolic diseases has also accumulated. FGF-signalling disorders contribute to pathological conditions. Several comprehensive reviews on FGFs and FGFRs have been published (1, 2, 7–9). In this article, we provide a succinct review of the FGF family, focusing on its evolutionary history, physiological roles in mice and pathophysiological roles in humans.
The FGF family
‘Invention is often the mother of necessity, rather than vice versa.’ (Jared Diamond, 1937∼).
The prototypic FGFs, FGF1 and FGF2, were originally isolated from the brain and pituitary as mitogens for cultured fibroblasts (10, 11). Several FGFs have since been isolated as growth factors for cultured cells. In addition, several Fgf genes have been identified by homology-based PCR or searches in DNA databases. A few Fgf genes also have been identified as genes responsible for hereditary diseases or cancer (1, 2, 7–9).
The human Fgf gene family comprises 22 members including Fgf1-Fgf23. Fgf15 has not been identified in humans. No other Fgf genes have been identified in the complete human genome sequence. Human FGFs contain ∼150–300 amino acids and have a conserved core of ∼120 amino acids with ∼30–60% identity (12). The mouse Fgf family also comprises 22 members including Fgf1–Fgf23 (1). Fgf19 has not been identified in the mouse and rat. Fgf15 and Fgf19 are likely to be orthologous genes in vertebrates. Except for mouse and rat, the Fgf15/19 orthologues were named Fgf19 in other vertebrates. In this review, we refer to these genes as Fgf15/19.
The FGF subfamilies and their mechanisms of action
Phylogenetic analysis of the human Fgf gene family identify seven subfamilies; Fgf1/2, Fgf4/5/6, Fgf3/7/10/22, Fgf8/17/18, Fgf9/16/20, Fgf11/12/13/14 and Fgf15/19/21/23 (Fig. 1A). Phylogenetic analysis indicates potential evolutionary relationships in the gene family. However, it alone is not sufficient to determine these relationships. Analysis of gene loci on chromosomes indicates more precise evolutionary relationships in a gene family. Their conserved chromosomal locations (synteny) identify seven subfamilies; Fgf/1/2/5, Fgf3/4/6, Fgf7/10/22, Fgf8/17/18, Fgf9/16/20, Fgf11/12/13/14 and Fgf15/19/21/23. Fgf subfamilies indicated by phylogenetic analysis and gene-location analysis are similar to each other, but not identical. For example, gene location analysis indicates that Fgf3 and Fgf5 should be members of an Fgf3/4/6 and Fgf1/2/5 subfamily, respectively (Fig. 1B). The mouse Fgf gene subfamiles are identical to the human Fgf gene subfamilies (1, 12).
Fig. 1.
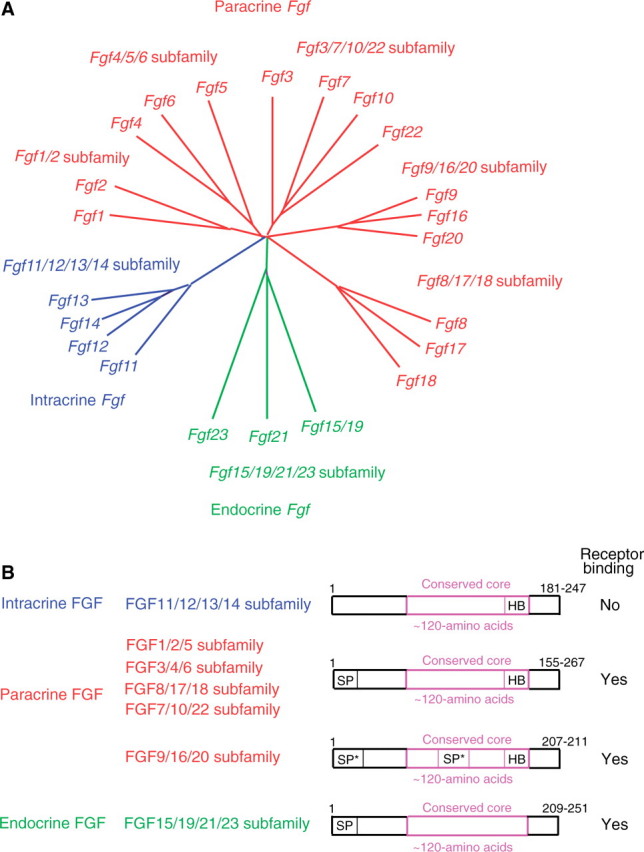
Evolutionary relationships within the human Fgf gene family and schematic representations of FGF structures. (A) Phylogenetic analysis suggests that 22 Fgf genes can be arranged into seven subfamilies containing two to four members each. Branch lengths are proportional to the evolutionary distance between each gene. (B) Gene-location analysis suggests that the Fgf genes can be arranged into seven subfamilies containing three to four members each. FGFs act on target cells in an intracrine, paracrine or endocrine manner. Schematic representations of intracrine, paracrine and endocrine FGF structures are shown. SP, SP* and HB indicate a cleavable secreted signal sequence, an uncleaved bipartite secreted signal sequence and a heparin-binding site, respectively.
FGFs also can be classified as intracrine, paracrine and endocrine FGFs by their mechanisms of action (Fig. 1B). Intracrine FGFs, FGF11-FGF14, are not secreted extracellulary. They act as intracellular molecules in an FGFR-independent manner. They interact with intracellular domains of voltage gated sodium channels and with a neuronal MAPK scaffold protein, islet-brain-2 (5, 13). The only known role for intracrine FGFs is in regulating the electrical excitability of neurons and possibly other cell types (5, 14–16).
Paracrine FGFs comprise members of the FGF/1/2/5, FGF3/4/6, FGF7/10/22, FGF8/17/18 and FGF9/16/20 subfamilies (Fig. 1B). Most are secreted proteins with cleavable N-terminal secreted signal peptides, however FGF9, FGF16 and FGF20 have uncleaved bipartite secreted signal sequences (17). In contrast, FGF1 and FGF2, which have no N-terminal hydrophobic sequences, are not typical secreted proteins. FGF1 and FGF2 might be released from damaged cells or by an exocytotic mechanism that is independent of the endoplasmic reticulum-Golgi pathway (18, 19). All paracrine FGFs mediate biological responses as extracellular proteins by binding to and activating cell surface tyrosine kinase FGFRs with heparin/heparan sulphate as a cofactor. However, it has also been reported that FGF1, FGF2 and FGF3 can directly translocated to the nucleus and act in an intracrine manner (20).
Four Fgfr genes, Fgfr1–Fgfr4, have been identified in humans and mice (2, 9, 12). These genes encode receptor tyrosine kinases (∼800 amino acids) that contain an extracellular ligand-binding domain with three immunoglobulin-like domains (I, II and III), a transmembrane domain and a split intracellular tyrosine kinase domain. Fgfr1–Fgfr3 encode two major versions of immunoglobin-like domain III (IIIb and IIIc) generated by alternative splicing that utilizes one of two unique exons. The immunoglobulin-like domain III is an essential determinant of ligand-binding specificity (21). Thus, seven major FGFR proteins (FGFRs 1b, 1c, 2b, 2c, 3b, 3c and 4) with differing ligand-binding specificity are generated from four Fgfr genes. Paracrine FGFs have a heparin-binding site and interaction with heparin-like molecules is necessary for the stable interaction with FGFRs and local signalling (22). Paracrine FGFs function in development by influencing the intracellular signalling events of neighbouring cells from a distance. The range of FGF signalling is regulated in part by its affinity for extracellular matrix heparan sulphate proteoglycans (23) and in part by dimerization of some FGFs (24, 25). FGF binding to FGFRs induces functional dimerization, receptor transphosphorylation and activation of four key downstream signalling pathways: RAS-RAF-MAPK, PI3K-AKT, STAT and PLCγ (2, 9).
Endocrine FGFs, FGF15/19, FGF21 and FGF23, are thought to mediate their biological responses in an FGFR-dependent manner. However, they bind to FGFRs and heparin/heparan sulphate with very low affinity. The reduced heparin-binding affinity enables endocrine FGFs to function in an endocrine manner (Fig. 1C) (21, 22). αKlotho is a single-pass transmembrane protein of ∼1,000 amino acids with a short cytoplasmic domain. The phenotypes of αKlotho knockout mice are very similar to those of Fgf23 knockout mice (26). These results indicate that FGF23 and αKlotho may function in a common signal-transduction pathway. αKlotho most efficiently binds to and activates FGFR1c among several isoforms of FGFRs in cultured cells (27), suggesting the FGFR1c can transduce an FGF23/αKlotho signal.
βKlotho is a protein that shares structural similarity and characteristics with αKlotho. The phenotypes of βKlotho knockout mice overlap those of Fgfr4 knockout mice and Fgf15/19 knockout mice (28, 29). FGF15/19 can bind to a βKlotho–FGFR4 complex in cultured cells. FGF15/19 also activates FGF signalling in hepatocytes that primarily express Fgfr4 (30). These results indicate that FGFR4 may be the primary receptor for transduction for an FGF15/19/βKlotho signal.
βKlotho is also essential for FGF21 signalling in cultured cells (31). However, Fgf21 knockout mouse phenotypes are distinct from βKlotho knockout mouse phenotypes (28, 32). In addition, the administration of recombinant human FGF21 to βKlotho knockout mice demonstrated that FGF21 signals can be transduced in the absence of βKlotho (33). These results indicate the existence of a βKlotho-independent FGF21-signalling pathway in which undefined cofactors might be involved.
Evolutionary history of the Fgf family
‘Nothing in biology makes sense except in the light of evolution.’ (Theodosius Dobzhansky, 1900–1975).
The FGF-signalling system has been conserved throughout metazoan evolution. Two Fgf-like genes, egl-17 and let-756, have been identified in the nematode, Caenorhabditis elegans (34). Six Fgf-like genes, Fgf4-like, Fgf5-like, Fgf8-like, Fgf9-like, Fgf10-like and Fgf13-like, which are potential ancestral genes of the human/mouse Fgf subfamilies, have been identified in the ascidian, Ciona intestinalis (35). Ascidians belong to the Subphylum Urochordata, the earliest branch in the Phylum Chordata. These results indicate that most ancestral genes of the human/mouse Fgf subfamilies were generated by gene duplication after the diversion of protostomes and deuterostomes (Fig. 2A).
Fig. 2.
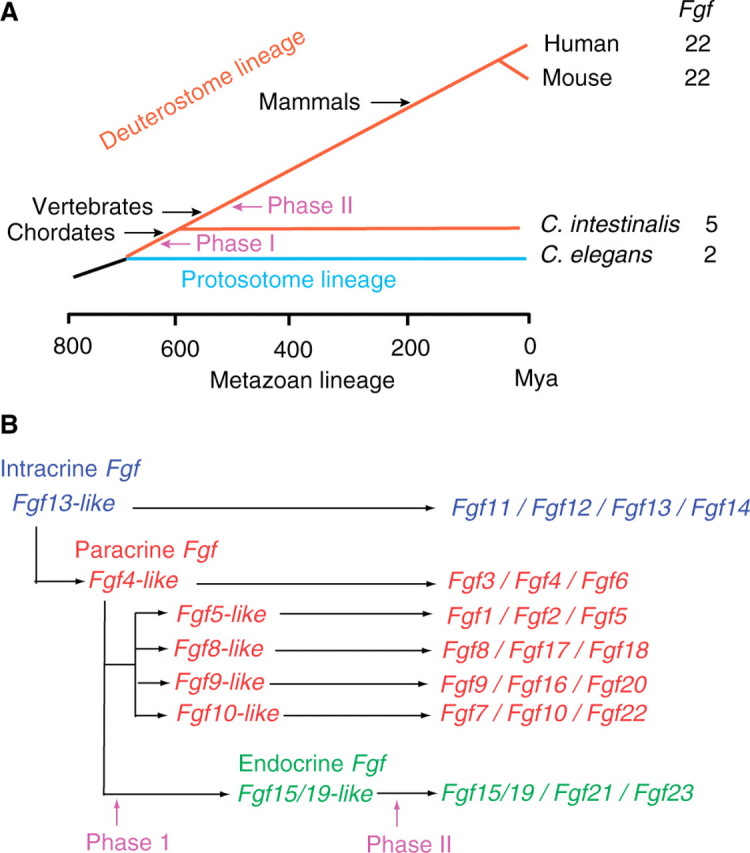
The evolutionary lineage of metazoan organisms and functional evolutionary history of the Fgf gene family. (A) The entire C. elegans, C. intestinalis, mouse and human genomes have been sequenced. The Fgf gene family expanded in two major phases (I and II) during metazoan evolution. Phase I occurred after the separation of protostomes and deutrostomes. Phase II occurred at the early emergence of vertebrates. Mya, million years ago. (B) Fgf13-like is the ancestral gene of the Fgf gene family. Fgf4-like was generated from Fgf13-like by gene duplication during the early stages of metazoan evolution. Fgf5-like, Fgf8-like, Fgf9-like and Fgf10-like were generated from Fgf4-like in phase I by gene duplication. Fgf15/19-like was also generated from Fgf4-like by local gene duplication. Each subfamily further expanded into three or four members via two large-scale genome duplication events in phase II.
The evolutionary history of the mouse Fgf family has been proposed (Fig. 2B) (1). The ancestral gene of the Fgf family is an ancestral intracrine Fgf gene, Fgf13-like, with a heparin-binding site but no secreted signal sequence (Figs 1B and 2B). An ancestral gene of paracrine Fgfs, Fgf4-like, was generated from Fgf13-like by gene duplication during the early stages of metazoan evolution. During this evolution, Fgf4-like acquired a secreted signal sequence, thus allowing it to function as a paracrine Fgf (Figs 1B and 2B). Ancestral genes, Fgf5-like, Fgf8-like, Fgf9-like and Fgf10-like, of paracrine Fgf subfamilies were also generated from Fgf4-like by gene duplication after the separation of protostomes and deuterostomes. Secreted signal sequences were conserved in Fgf5-like, Fgf8-like and Fgf10-like. A cleavable secreted signal sequence also evolved into an uncleaved bipartite signal sequence in Fgf9-like (36–38). These FGFs with heparin-binding sites function in a paracrine manner. In contrast, no ancestral gene of endocrine Fgfs has been identified in C intestinalis. The ancestral gene of endocrine Fgfs, Fgf15/19-like, appears to have arisen from Fgf4-like by local gene duplication early in vertebrate evolution. During this evolution, Fgf15/19-like lost its high-affinity heparin-binding capacity, thus allowing it to function in an endocrine manner (Figs 1B and 2B). Conserved gene orders are observed among members of each Fgf subfamily, indicating that each subfamily further expanded into three or four members via two large-scale genome duplication events during the evolution of early vertebrates (Fig. 2B).
Physiological roles of FGFs in mice
Most Fgf genes have been disrupted by homologous recombination in mice. Phenotypes range from early embryonic lethality to changes in adult physiology (Table I).
Table I.
Phenotypes in Fgf knockout mice.
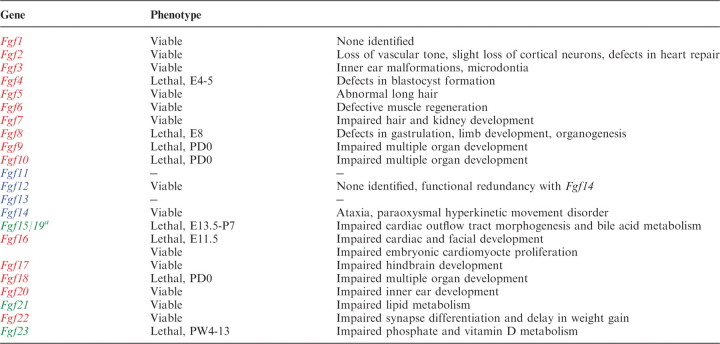 |
Phenotypes of Fgf11, Fgf13 and Fgf20 knockout mice have not been published. aFgf15 is referred to as Fg15/19. E, embryonic day; PD, postnatal day; PW, postnatal week.
Roles of intracrine Fgfs in neuronal functions
Fgf14 knockout mice are viable. However, they develop ataxia and a paraoxysmal hyperkinetic movement disorder (5, 14, 15). In contrast, Fgf12 knockout mice are apparently normal. Fgf12/Fgf14 double knockout mice show severe ataxia and other neurological deficits (5). Phenotypes of Fgf11 and Fgf13 knockout mice have not been reported.
Roles of paracrine Fgfs in development
Paracrine FGFs are expected to act as growth/differentiation factors in developing embryos. Fgf knockout mouse phenotypes mostly indicate roles as growth/differentiation factors.
Fgf1 knockout mice are viable and normal (39). Fgf2 knockout mice are also viable, but have decreased vascular tone and reduced numbers of neurons in deep cortical layers (40–42). Fgf2 knockout mice show impaired recovery from ischaemic injury to the heart (43, 44). Fgf3 knockout mice are viable, but have phenotypes that include inner ear malformation and microdontia (45–48). Fgf4 and Fgf8 knockout mice die at early embryonic stages. Fgf4 and Fgf8 have essential roles in blastocyst formation and gastrulation, respectively (49, 50). Conditional inactivation of Fgf8 has identified additional roles in limb bud development and organogenesis. Fgf5, Fgf6 and Fgf7 knockout mice are viable. Abnormal long hair is observed in Fgf5 knockout mice (51). Fgf6 knockout mice have defects in muscle regeneration (52). Fgf7 knockout mice have impaired hair and kidney development (53, 54). Fgf9, Fgf10 and Fgf18 knockout mice die shortly after birth. Fgf10 is critical for epithelial–mesenchymal interactions necessary for the development of epithelial components of multiple organs (55–58). Fgf9 and Fgf18 have essential roles in the development of mesenchymal components of multiple organs (59–64). Fgf16 knockout mice on a C57BL/6 genetic background are viable, but have impaired embryonic cardiomyocyte proliferation (65). Fgf16 knockout phenotypes may be more severe on a Black Swiss genetic background where they die at embryonic day (E) 10.5 with severely impaired cardiac and facial development (66, 67). Fgf17 and Fgf22 knockout mice are viable, but show impaired hindbrain development and impaired synaptic differentiation, respectively (68, 69). In addition, Fgf22 knockout mice also show a clear delay in weight gain upon sexual maturity (R. Grose et al., unpublished data). Fgf20 knockout mice are viable but have profound hearing loss (D.M. Ornitz et al., ARO abstract 423, 2009).
Roles of endocrine Fgfs in development and metabolism
Hormones are usually responsible for communication between tissues in an endocrine manner. However, several hormones are produced in developing tissues that are unrelated to the endocrine gland of origin in adults. These hormones are synthesized locally, and serve as differentiation factors in embryos (70). Endocrine FGFs also act as differentiation factors in embryos and as hormones in adults (4).
Fgf15/19 knockout mice develop normally until E10.5, but then gradually die. The phenotype indicates that FGF15/19 is required for proper morphogenesis of the cardiac outflow tract at embryonic stages (71). Although most Fgf15/19 knockout mice die by post-natal day (P) 7, a few survive and appear phenotypically normal. However, fecal bile acid excretion was found to be increased in surviving Fgf15/19 knockout mice, indicating that intestinal FGF15/19 plays a crucial role in regulating hepatic bile acid synthesis (29). Fgf21 knockout mice are seemingly normal, but show hypertrophy and decreased lipolysis in adipocytes. In contrast, Fgf21 knockout mice fasted for 24 h show increased lipolysis in adipocytes and increased serum non-esterified fatty acid levels. Their phenotypes indicate that Fgf21 is important for the metabolic regulation of lipolysis in white adipose tissue (32). Fgf21 knockout mice fed a ketogenic diet show partial impairments in ketogenesis (72). However, we have observed that ketogenesis is not impaired in Fgf21 knockout mice fed a ketogenic diet (N. Itoh et al., unpublished data). Fgf23 knockout mice survive until birth, but then gradually die, usually by 12 weeks of age (26). The mice show hyperphosphataemia and increased active vitamin D levels. Fgf23, which is expressed in osteocytes, signals to the kidney where it regulates serum phosphate and active vitamin D levels. FGF23 may have other target organs including parathyroid gland and osteoblasts (73, 74).
Although roles of FGFs in embryogenesis have been revealed from Fgf knockout mouse phenotypes, their contributions to adult physiology remain relatively unexplored. The widespread expression of Fgf genes in the adult tissues suggests multiple roles in tissue homeostasis and repair (75). In addition to endocrine FGFs, which have well defined endocrine roles in the adult (4), emerging reports indicate homoeostatic and regenerative roles for canonical paracrine FGF signalling (76–78).
FGF-signalling disorders in human diseases
As described earlier, FGF-signalling is crucial to development and metabolism. In addition, FGF-signalling disorders also result in human hereditary, paraneoplastic and metabolic diseases (Tables II and III).
Table II.
Human hereditary diseases caused by Fgf mutations.
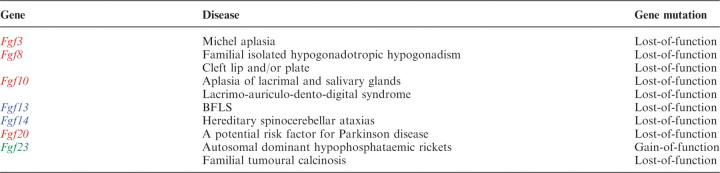 |
Table III.
Human paraneoplastic or metabolic diseases caused by endocrine FGF-signalling disorders.
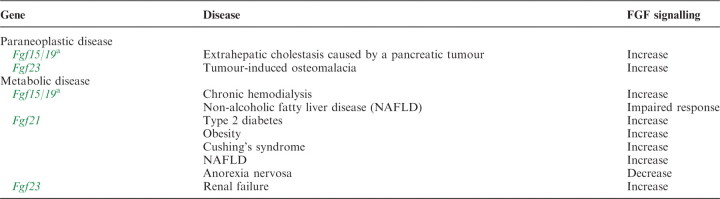 |
aFgf19 is referred to as Fg15/19.
Intracrine FGF-signalling disorders
Börjeson–Forssman–Lehmann syndrome (BFLS) is a syndromic X-linked mental retardation disease. Fgf13 is a candidate causative gene for BFLS (79). Hereditary spinocerebellar ataxias (SCAs) are a clinically and genetically heterogeneous group of neurodegenerative disorders. One SCA with early onset tremour, dyskinesia and slowly progressive cerebellar ataxia is caused by Fgf14 mutations (80–82).
Paracrine FGF-signalling disorders
Michel aplasia is a unique autosomal recessive syndrome characterized by type I microtia, microdontia and profound congenital deafness associated with a complete absence of inner ear structures. Michel aplasia is caused by mutations in Fgf3 (47).
Nonsense mutations in Fgf8 are found in familial-isolated hypogonadotropic hypogonadism with variable degrees of gonadotropin-releasing hormone deficiency and olfactory phenotypes. These findings confirm that loss-of-function mutations in Fgf8 cause human gonadotropin-releasing hormone deficiency (83). Cleft lip and/or palate (CLP) appear when the two halves of the palatal shelves fail to fuse completely. A missense mutation in Fgf8 was found in a patient with CLP. This mutation is predicted to cause loss-of-function by destabilizing the N-terminal conformation, which is important for FGFR-binding affinity and specificity (84).
Aplasia of lacrimal and salivary glands (ALSG) is an autosomal dominant congenital anomaly characterized by aplasia, atresia or hypoplasia of the lacrimal and salivary systems. Lacrimo-auriculo-dento-digital syndrome (LADD) is an autosomal-dominant multiple congenital anomaly disorder characterized by aplasia, atresia or hypoplasia of the lacrimal and salivary systems, cup-shaped ears, hearing loss and dental and digital anomalies. Both ALSG and LADD are caused by Fgf10 mutations (85, 86).
Fgf20 was originally identified as a neurotrophic factor preferentially expressed in dopaminergic neurons within the substantia nigra pars compacta of rat brain (87). Parkinson disease (PD) is caused by a pathogenic process responsible for the loss of dopaminergic neurons within the substantia nigra pars compacta. A pedigree disequilibrium test and a case–control association study indicated that Fgf20 is potentially a risk factor for PD (88).
Endocrine FGF signalling disorders
Serum FGF15/19 levels are markedly increased in patients with extrahepatic cholestasis caused by a pancreatic tumour. FGF15/19 is abundantly expressed in the liver of cholestatic patients, but not in the normal liver. FGF15/19 signalling may be involved in some of the adaptations that protect the liver against bile salt toxicity (89). Serum FGF15/19 levels are also significantly increased in patients on chronic hemodialysis (90). Hepatic lipid metabolism is disturbed in patients with NAFLD. The hepatic response to FGF15/19 is impaired in NAFLD patients with insulin resistance. This impaired response may contribute to the disturbance of lipid homeostasis in NAFLD (91).
Serum FGF21 levels are increased in patients with type 2 diabetes and obesity, Cushing’s syndrome or NAFLD (92–94). In contrast, serum FGF21 levels are decreased in patients with anorexia nervosa (95).
FGF23-signalling disorders also result in diseases (96). Autosomal dominant hypophosphatemic rickets (AHDR) is caused by gain-of-function mutations of Fgf23 (97). FGF23 is partially cleaved by intracellular proteolysis. The cleaved FGF23 forms lose their biological activity. Fgf23 mutations in ADHR result in impaired proteolysis of FGF23 and increased serum levels of active FGF23 (98). Reduced FGF23 signalling also causes human hereditary diseases. Familial tumoural calcinosis (FTC) is characterized by ectopic calcification and hyperphosphataemia. Loss-of-function mutations of Fgf23 result in FTC. These mutations destabilize the tertiary structure of FGF23 and increase its susceptibility to degradation (99). Tumours that over produce FGF23 also cause tumour-induced osteomalacia, which is a paraneoplasitc disease characterized by hypophosphataemia caused by renal phosphate wasting (100). In addition, serum FGF23 levels are also greatly increased in patients with renal failure, partly owing to decreased renal clearance. These results suggest that FGF23 has a compensatory role in the disease (101).
Conclusion
The prototypic FGFs, FGF1 and FGF2, were originally isolated as mitogens for fibroblasts from the brain and pituitary >20 years ago. Many FGF proteins or Fgf genes have since been isolated as growth factors for cultured cells or identified by homology-based PCR and/or homology-based searches in DNA databases, respectively. The human/mouse Fgf family comprises 22 members. FGFs are now recognized as polypeptide growth factors with diverse biological activities and act as intracellular or extracellular signalling molecules in an intracrine, paracrine or endocrine manner. Fgf knockout mice indicate that FGFs play crucial roles in development and metabolism. In addition, the roles of FGFs in human diseases indicate that FGF-signalling disorders contribute to pathological conditions. Although Fgf1 and Fgf2 are genes for prototypic FGFs, they are not ancestral genes of the Fgf family in evolution. The ancestral gene of the Fgf family is an ancestral gene of the intracrine Fgf subfamily, Fgf13-like. The evolutional history of the Fgf gene family indicates that Fgf genes acquired a diversity of roles and functions with the expansion of the Fgf gene family by gene duplication after the diversion of protostomes and deuterostomes and by two genome-duplication events during the evolution of early vertebrates. Secreted signalling molecules such as BMPs, WNTs and Hedgehogs also play crucial roles in development by influencing the intracellular signalling events of their neighbours from a distance. FGFs, along with these signalling molecules, have roles in diverse biological processes of multicellular organisms. However, the interaction/cooperation of FGFs with BMPs, WNTs and Hedgehogs mostly remain unclear. Further understanding of the roles of FGFs will provide clues to their mechanisms of interaction/cooperation.
Funding
A Grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan; Takeda Science Foundation, Japan (NI); National Institutes of Health and the March of Dimes Foundation (DMO).
Conflict of interest
None declared.
Acknowledgements
We would like to thank members of our laboratories and the many collaborators in our FGF projects.
Glossary
Abbreviations
- BMP
bone morphogenetic protein
- FGF
fibroblast growth factor
- FGFR
FGF receptor
- MAPK
mitogen-activated protein kinase
- PCR
polymerase chain reaction
- PI3K
phosphatidylinositol 3-kinase
- PLCγ
phospholipase Cγ
- STAT
signal transducer and activator of transcription
References
- 1.Itoh N., Ornitz D.M. Functional evolutionary history of the mouse Fgf gene family. Dev. Dyn. 2008;237:18–27. doi: 10.1002/dvdy.21388. [DOI] [PubMed] [Google Scholar]
- 2.Beenken A., Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kharitonenkov A. FGFs and metabolism. Curr. Opin. Pharmacol. 2009;9:805–810. doi: 10.1016/j.coph.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Itoh N. Hormone-like (endocrine) Fgfs: Their evolutionary history and roles in development, metabolism, and disease. Cell Tissue Res. 2010;342:1–11. doi: 10.1007/s00441-010-1024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldfarb M., Schoorlemmer J., Williams A., Diwakar S., Wang Q., Huang X., Giza J., Tchetchik D., Kelley K., Vega A., Matthews G., Rossi P., Ornitz D.M., D’Angelo E. Fibroblast growth factor homologous factors control neuronal excitability through modulation of voltage-gated sodium channels. Neuron. 2007;55:449–463. doi: 10.1016/j.neuron.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laezza F., Lampert A., Kozel M.A., Gerber B.R., Rush A.M., Nerbonne J.M., Waxman S.G., Dib-Hajj S.D., Ornitz D.M. FGF14 N-terminal splice variants differentially modulate Nav1.2 and Nav1.6-encoded sodium channels. Mol. Cell. Neurosci. 2009;42:90–101. doi: 10.1016/j.mcn.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itoh N. The Fgf families in humans, mice, and zebrafish: their evolutional processes and roles in development, metabolism, and disease. Biol. Pharm. Bull. 2007;30:1819–1825. doi: 10.1248/bpb.30.1819. [DOI] [PubMed] [Google Scholar]
- 8.Krejci P., Prochazkova J., Bryja V., Kozubik A., Wilcox W.R. Molecular pathology of the fibroblast growth factor family. Hum. Mutat. 2009;30:1245–1255. doi: 10.1002/humu.21067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner N., Grose R. Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 10.Gospodarowicz D. Purification of a fibroblast growth factor from bovine pituitary. J. Biol. Chem. 1975;250:2515–2520. [PubMed] [Google Scholar]
- 11.Gospodarowicz D., Bialecki H., Greenberg G. Purification of fibroblast growth factor activity from bovine brain. J. Biol. Chem. 1978;253:3736–3743. [PubMed] [Google Scholar]
- 12.Itoh N., Ornitz D.M. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Schoorlemmer J., Goldfarb M. Fibroblast growth factor homologous factors and the islet brain-2 scaffold protein regulate activation of a stress-activated protein kinase. J. Biol. Chem. 2002;277:49111–49119. doi: 10.1074/jbc.M205520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao M., Xu L., Laezza F., Yamada K., Feng S., Ornitz D.M. Impaired hippocampal synaptic transmission and plasticity in mice lacking fibroblast growth factor 14. Mol. Cell. Neurosci. 2007;34:366–377. doi: 10.1016/j.mcn.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 15.Shakkottai V.G., Xiao M., Xu L., Wong M., Nerbonne J.M., Ornitz D.M., Yamada K.A. FGF14 regulates the intrinsic excitability of cerebellar Purkinje neurons. Neurobiol. Dis. 2009;33:81–88. doi: 10.1016/j.nbd.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dover K., Solinas S., D'Angelo E., Goldfarb M. Long-term inactivation particle for voltage-gated sodium channels. J. Physiol. 2010;588:3695–3711. doi: 10.1113/jphysiol.2010.192559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Revest J.M., DeMoerlooze L., Dickson C. Fibroblast growth factor 9 secretion is mediated by a non-cleaved amino-terminal signal sequence. J. Biol. Chem. 2000;275:8083–8090. doi: 10.1074/jbc.275.11.8083. [DOI] [PubMed] [Google Scholar]
- 18.Mohan S.K., Rani S.G., Yu C. The heterohexameric complex structure, a component in the non-classical pathway for fibroblast growth factor 1 (FGF1) secretion. J. Biol. Chem. 2010;285:15464–15475. doi: 10.1074/jbc.M109.066357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nickel W. Pathways of unconventional protein secretion. Curr. Opin. Biotechnol. 2010;21:621–626. doi: 10.1016/j.copbio.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Antoine M., Reimers K., Dickson C., Kiefer P. Fibroblast growth factor 3, a protein with dual subcellular localization, is targeted to the nucleus and nucleolus by the concerted action of two nuclear localization signals and a nucleolar retention signal. J. Biol. Chem. 1997;272:29475–2981. doi: 10.1074/jbc.272.47.29475. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X., Ibrahimi O.A., Olsen S.K., Umemori H., Mohammadi M., Ornitz D.M. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goetz R., Beenken A., Ibrahimi O.A., Kalinina J., Olsen S.K., Eliseenkova A.V., Xu C., Neubert T.A., Zhang F., Linhardt R.J., Yu X., White K.E., Inagaki T., Kliewer S.A., Yamamoto M., Kurosu H., Ogawa Y., Kuro-o M., Lanske B., Razzaque M.S., Mohammadi M. Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol. Cell. Biol. 2007;27:3417–3428. doi: 10.1128/MCB.02249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalinina J., Byron S.A., Makarenkova H.P., Olsen S.K., Eliseenkova A.V., Larochelle W.J., Dhanabal M., Blais S., Ornitz D.M., Day L.A., Neubert T.A., Pollock P.M., Mohammadi M. Homodimerization controls the fibroblast growth factor 9 subfamily’s receptor binding and heparan sulfate-dependent diffusion in the extracellular matrix. Mol. Cell. Biol. 2009;29:4663–4678. doi: 10.1128/MCB.01780-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalinina J., Byron S.A., Makarenkova H.P., Olsen S.K., Eliseenkova A.V., Larochelle W.J., Dhanabal M., Blais S., Ornitz D.M., Day L.A., Neubert T.A., Pollock P.M., Mohammadi M. Homodimerization controls the fibroblast growth factor 9 subfamily's receptor binding and heparan sulfate-dependent diffusion in the extracellular matrix. Mol. Cell Biol. 2009;29:4663–4678. doi: 10.1128/MCB.01780-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harada M., Murakami H., Okawa A., Okimoto N., Hiraoka S., Nakahara T., Akasaka R., Shiraishi Y., Futatsugi N., Mizutani-Koseki Y., Kuroiwa A., Shirouzu M., Yokoyama S., Taiji M., Iseki S., Ornitz D.M., Koseki H. FGF9 monomer/dimer equilibrium regulates extracellular matrix affinity and tissue diffusion. Nat. Genet. 2009;41:289–298. doi: 10.1038/ng.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimada T., Kakitani M., Yamazaki Y., Hasegawa H., Takeuchi Y., Fujita T., Fukumoto S., Tomizuka K., Yamashita T. Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. J. Clin. Invest. 2004;113:561–568. doi: 10.1172/JCI19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urakawa I., Yamazaki Y., Shimada T., Iijima K., Hasegawa H., Okawa K., Fujita T., Fukumoto S., Yamashita T. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444:770–774. doi: 10.1038/nature05315. [DOI] [PubMed] [Google Scholar]
- 28.Ito S., Fujimori T., Furuya A., Satoh J., Nabeshima Y., Nabeshima Y. Impaired negative feedback suppression of bile acid synthesis in mice lacking betaKlotho. J. Clin. Invest. 2005;115:2202–2208. doi: 10.1172/JCI23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C.L., McDonald J.G., Luo G., Jones S.A., Goodwin B., Richardson J.A., Gerard R.D., Repa J.J., Mangelsdorf D.J., Kliewer S.A. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Kurosu H., Choi M., Ogawa Y., Dickson A.S., Goetz R., Eliseenkova A.V., Mohammadi M., Rosenblatt K.P., Kliewer S.A., Kuro-o M. Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J. Biol. Chem. 2007;282:26687–2695. doi: 10.1074/jbc.M704165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kharitonenkov A., Dunbar J.D., Bina H.A., Bright S., Moyers J.S., Zhang C., Ding L., Micanovic R., Mehrbod S.F., Knierman M.D., Hale J.E., Coskun T., Shanafelt A.B. FGF-21/FGF-21 receptor interaction and activation is determined by betaKlotho. J. Cell. Physiol. 2008;215:1–7. doi: 10.1002/jcp.21357. [DOI] [PubMed] [Google Scholar]
- 32.Hotta Y., Nakamura H., Konishi M., Murata Y., Takagi H., Matsumura S., Inoue K., Fushiki T., Itoh N. Fibroblast growth factor 21 regulates lipolysis in white adipose tissue but is not required for ketogenesis and triglyceride clearance in liver. Endocrinology. 2009;150:4625–4633. doi: 10.1210/en.2009-0119. [DOI] [PubMed] [Google Scholar]
- 33.Tomiyama K., Maeda R., Urakawa I., Yamazaki Y., Tanaka T., Ito S., Nabeshima Y., Tomita T., Odori S., Hosoda K., Nakao K., Imura A., Nabeshima Y. Relevant use of Klotho in FGF19 subfamily signaling system in vivo. Proc. Natl Acad. Sci, USA. 2010;107:1666–1671. doi: 10.1073/pnas.0913986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang P., Stern M.J. FGF signaling in flies and worms: more and more relevant to vertebrate biology. Cytokine Growth Factor Rev. 2005;16:151–158. doi: 10.1016/j.cytogfr.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 35.Satou Y., Imai K.S., Satoh N. Fgf genes in the basal chordate Ciona intestinalis. Dev. Genes Evol. 2002;212:432–438. doi: 10.1007/s00427-002-0266-8. [DOI] [PubMed] [Google Scholar]
- 36.Miyakawa K., Hatsuzawa K., Kurokawa T., Asada M., Kuroiwa T., Imamura T. A hydrophobic region locating at the center of fibroblast growth factor-9 is crucial for its secretion. J. Biol. Chem. 1999;274:29352–29357. doi: 10.1074/jbc.274.41.29352. [DOI] [PubMed] [Google Scholar]
- 37.Revest J.M., DeMoerlooze L., Dickson C. Fibroblast growth factor 9 secretion is mediated by a non-cleaved amino-terminal signal sequence. J. Biol. Chem. 2000;275:8083–8090. doi: 10.1074/jbc.275.11.8083. [DOI] [PubMed] [Google Scholar]
- 38.Miyakawa K., Imamura T. Secretion of FGF-16 requires an uncleaved bipartite signal sequence. J. Biol. Chem. 2003;278:35718–3524. doi: 10.1074/jbc.M300690200. [DOI] [PubMed] [Google Scholar]
- 39.Miller D.L., Ortega S., Bashayan O., Basch R., Basilico C. Compensation by fibroblast growth factor 1 (FGF1) does not account for the mild phenotypic defects observed in FGF2 null mice. Mol. Cell. Biol. 2000;20:2260–2268. doi: 10.1128/mcb.20.6.2260-2268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raballo R., Rhee J., Lyn-Cook R., Leckman J.F., Schwartz M.L., Vaccarino F.M. Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J. Neurosci. 2000;20:5012–5023. doi: 10.1523/JNEUROSCI.20-13-05012.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou M., Sutliff R.L., Paul R.J., Lorenz J.N., Hoying J.B., Haudenschild C.C., Yin M., Coffin J.D., Kong L., Kranias E.G., Luo W., Boivin G.P., Duffy J.J., Pawlowski S.A., Doetschman T. Fibroblast growth factor 2 control of vascular tone. Nat. Med. 1998;4:201–207. doi: 10.1038/nm0298-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dono R., Texido G., Dussel R., Ehmke H., Zeller R. Impaired cerebral cortex development and blood pressure regulation in FGF-2-deficient mice. EMBO J. 1998;17:4213–4225. doi: 10.1093/emboj/17.15.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.House S.L., Bolte C., Zhou M., Doetschman T., Klevitsky R., Newman G., Schultz J.E.J. Cardiac-specific overexpression of fibroblast growth factor-2 protects against myocardial dysfunction and infarction in a murine model of low-flow ischemia. Circulation. 2003;108:3140–3148. doi: 10.1161/01.CIR.0000105723.91637.1C. [DOI] [PubMed] [Google Scholar]
- 44.Virag J.A., Rolle M.L., Reece J., Hardouin S., Feigl E.O., Murry C.E. Fibroblast growth factor-2 regulates myocardial infarct repair: effects on cell proliferation, scar contraction, and ventricular function. Am. J. Pathol. 2007;171:1431–1440. doi: 10.2353/ajpath.2007.070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mansour S.L., Goddard J.M., Capecchi M.R. Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development. 1993;117:13–28. doi: 10.1242/dev.117.1.13. [DOI] [PubMed] [Google Scholar]
- 46.Alvarez Y., Alonso M.T., Vendrell V., Zelarayan L.C., Chamero P., Theil T., Bosl M.R., Kato S., Maconochie M., Riethmacher D., Schimmang T. Requirements for FGF3 and FGF10 during inner ear formation. Development. 2003;130:6329–6338. doi: 10.1242/dev.00881. [DOI] [PubMed] [Google Scholar]
- 47.Tekin M., Hismi B.O., Fitoz S., Ozdag H., Cengiz F.B., Sirmaci A., Aslan I., Inceoglu B., Yüksel-Konuk E.B., Yilmaz S.T., Yasun O., Akar N. Homozygous mutations in fibroblast growth factor 3 are associated with a new form of syndromic deafness characterized by inner ear agenesis, microtia, and microdontia. Am. J. Hum. Genet. 2007;80:338–344. doi: 10.1086/510920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charles C., Lazzari V., Tafforeau P., Schimmang T., Tekin M., Klein O., Viriot L. Modulation of Fgf3 dosage in mouse and men mirrors evolution of mammalian dentition. Proc. Natl. Acad. Sci. USA. 2009;106:22364–22368. doi: 10.1073/pnas.0910086106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feldman B., Poueymirou W., Papaioannou V.E., DeChiara T.M., Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- 50.Sun X., Meyers E.N., Lewandoski M., Martin G.R. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 1999;13:1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hébert J.M., Rosenquist T., Götz J., Martin G.R. FGF5 as a regulator of the hair growth cycle: evidence from targeted and spontaneous mutations. Cell. 1994;78:1017–1025. doi: 10.1016/0092-8674(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 52.Floss T., Arnold H.H., Braun T. A role for FGF-6 in skeletal muscle regeneration. Genes Dev. 1997;11:2040–2051. doi: 10.1101/gad.11.16.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo L., Degenstein L., Fuchs E. Keratinocyte growth factor is required for hair development but not for wound healing. Genes Dev. 1996;10:165–175. doi: 10.1101/gad.10.2.165. [DOI] [PubMed] [Google Scholar]
- 54.Qiao J., Uzzo R., Obara-Ishihara T., Degenstein L., Fuchs E., Herzlinger D. FGF-7 modulates ureteric bud growth and nephron number in the developing kidney. Development. 1999;126:547–554. doi: 10.1242/dev.126.3.547. [DOI] [PubMed] [Google Scholar]
- 55.Min H., Danilenko D.M., Scully S.A., Bolon B., Ring B.D., Tarpley J.E., DeRose M., Simonet W.S. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sekine K., Ohuchi H., Fujiwara M., Yamasaki M., Yoshizawa T., Sato T., Yagishita N., Matsui D., Koga Y., Itoh N., Kato S. Fgf10 is essential for limb and lung formation. Nat. Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 57.Ohuchi H., Hori Y., Yamasaki M., Harada H., Sekine K., Kato S., Itoh N. FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem. Biophys. Res. Commun. 2000;277:643–649. doi: 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- 58.Sakaue H., Konishi M., Ogawa W., Asaki T., Mori T., Yamasaki M., Takata M., Ueno H., Kato S., Kasuga M., Itoh N. Requirement of fibroblast growth factor 10 in development of white adipose tissue. Genes Dev. 2002;16:908–912. doi: 10.1101/gad.983202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colvin J.S., Green R.P., Schmahl J., Capel B., Ornitz D.M. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell. 2001;104:875–889. doi: 10.1016/s0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- 60.Colvin J.S., White A.C., Pratt S.J., Ornitz D.M. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- 61.Ohbayashi N., Shibayama M., Kurotaki Y., Imanishi M., Fujimori T., Itoh N., Takada S. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16:870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Z., Xu J., Colvin J.S., Ornitz D.M. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 2002;16:859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Usui H., Shibayama M., Ohbayashi N., Konishi M., Takada S., Itoh N. Fgf18 is required for embryonic lung alveolar development. Biochem. Biophys. Res. Commun. 2004;322:887–892. doi: 10.1016/j.bbrc.2004.07.198. [DOI] [PubMed] [Google Scholar]
- 64.Hung I.H., Yu K., Lavine K.J., Ornitz D.M. FGF9 regulates early hypertrophic chondrocyte differentiation and skeletal vascularization in the developing stylopod. Dev. Biol. 2007;307:300–313. doi: 10.1016/j.ydbio.2007.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hotta Y., Sasaki S., Konishi M., Kinoshita H., Kuwahara K., Nakao K., Itoh N. Fgf16 is required for cardiomyocyte proliferation in the mouse embryonic heart. Dev. Dyn. 2008;237:2947–2954. doi: 10.1002/dvdy.21726. [DOI] [PubMed] [Google Scholar]
- 66.Lu S.Y., Sheikh F., Sheppard P.C., Fresnoza A., Duckworth M.L., Detillieux K.A., Cattini P.A. FGF-16 is required for embryonic heart development. Biochem. Biophys. Res. Commun. 2008;373:270–274. doi: 10.1016/j.bbrc.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu S.Y., Jin Y., Li X., Sheppard P., Bock M.E., Sheikh F., Duckworth M.L., Cattini P.A. Embryonic survival and severity of cardiac and craniofacial defects are affected by genetic background in fibroblast growth factor-16 null mice. DNA Cell Biol. 2010;29:407–415. doi: 10.1089/dna.2010.1024. [DOI] [PubMed] [Google Scholar]
- 68.Xu J., Liu Z., Ornitz D.M. Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development. 2000;127:1833–1843. doi: 10.1242/dev.127.9.1833. [DOI] [PubMed] [Google Scholar]
- 69.Terauchi A., Johnson-Venkatesh E.M., Toth A.B., Javed D., Sutton M.A., Umemori H. Distinct FGFs promote differentiation of excitatory and inhibitory synapses. Nature. 2010;465:783–787. doi: 10.1038/nature09041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanders E.J., Harvey S. Peptide hormones as developmental growth and differentiation factors. Dev. Dyn. 2008;237:1537–1552. doi: 10.1002/dvdy.21573. [DOI] [PubMed] [Google Scholar]
- 71.Vincentz J.W., McWhirter J.R., Murre C., Baldini A., Furuta Y. Fgf15 is required for proper morphogenesis of the mouse cardiac outflow tract. Genesis. 2005;41:192–201. doi: 10.1002/gene.20114. [DOI] [PubMed] [Google Scholar]
- 72.Badman M.K., Koester A., Flier J.S., Kharitonenkov A., Maratos-Flier E. Fibroblast growth factor 21-deficient mice demonstrate impaired adaptation to ketosis. Endocrinology. 2009;150:4931–4940. doi: 10.1210/en.2009-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ben-Dov I.Z., Galitzer H., Lavi-Moshayoff V., Goetz R., Kuro-o M., Mohammadi M., Sirkis R., Naveh-Many T., Silver J. The parathyroid is a target organ for FGF23 in rats. J. Clin. Invest. 2007;117:4003–4008. doi: 10.1172/JCI32409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang W.J., Wang L.F., Xu X.Y., Zhou Y., Jin W.F., Wang H.F., Gao J. Autocrine/paracrine action of vitamin D on FGF23 expression in cultured rat osteoblasts. Calcif. Tissue Int. 2010;86:404–410. doi: 10.1007/s00223-010-9355-2. [DOI] [PubMed] [Google Scholar]
- 75.Fon Tacer K., Bookout A.L., Ding X., Kurosu H., John G.B., Wang L., Goetz R., Mohammadi M., Kuro-O M., Mangelsdorf D.J., Kliewer S.A. Research resource: comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Mol. Endocrinol. 2010;24:2050–2064. doi: 10.1210/me.2010-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bohm F., Speicher T., Hellerbrand C., Dickson C., Partanen J.M., Ornitz D.M., Werner S. FGF receptors 1 and 2 control chemically induced injury and compound detoxification in regenerating livers of mice. Gastroenterology. 2010;139:1385–1396. doi: 10.1053/j.gastro.2010.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang J., Meyer M., Müller A.K., Böhm F., Grose R., Dauwalder T., Verrey F., Kopf M., Partanen J., Bloch W., Ornitz D.M., Werner S. Fibroblast growth factor receptors 1 and 2 in keratinocytes control the epidermal barrier and cutaneous homeostasis. J. Cell Biol. 2010;188:935–952. doi: 10.1083/jcb.200910126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin Y., Wang F. FGF signalling in prostate development, tissue homoeostasis and tumorigenesis. Biosci. Rep. 2010;30:285–291. doi: 10.1042/BSR20100020. [DOI] [PubMed] [Google Scholar]
- 79.Gecz J., Baker E., Donnelly A., Ming J.E., McDonald-McGinn D.M., Spinner N.B., Zackai E.H., Sutherland G.R., Mulley J.C. Fibroblast growth factor homologous factor 2 (FHF2): gene structure, expression and mapping to the Börjeson-Forssman-Lehmann syndrome region in Xq26 delineated by a duplication breakpoint in a BFLS-like patient. Hum. Genet. 1999;104:56–63. doi: 10.1007/s004390050910. [DOI] [PubMed] [Google Scholar]
- 80.van Swieten J.C., Brusse E., de Graaf B.M., Krieger E., van de Graaf R., de Koning I., Maat-Kievit A., Leegwater P., Dooijes D., Oostra B.A., Heutink P. A mutation in the fibroblast growth factor 14 gene is associated with autosomal dominant cerebellar ataxia. Am. J. Hum. Genet. 2003;72:191–199. doi: 10.1086/345488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brusse E., de Koning I., Maat-Kievit A., Oostra B.A., Heutink P., van Swieten J.C. Spinocerebellar ataxia associated with a mutation in the fibroblast growth factor 14 gene (SCA27): a new phenotype. Mov. Disord. 2006;21:396–401. doi: 10.1002/mds.20708. [DOI] [PubMed] [Google Scholar]
- 82.Misceo D., Fannemel M., Baroy T., Roberto R., Tvedt B., Jaeger T., Bryn V., Stromme P., Frengen E. SCA27 caused by a chromosome translocation: further delineation of the phenotype. Neurogenetics. 2009;10:371–374. doi: 10.1007/s10048-009-0197-x. [DOI] [PubMed] [Google Scholar]
- 83.Trarbach E.B., Abreu A.P., Silveira L.F., Garmes H.M., Baptista M.T., Teles M.G., Costa E.M., Mohammadi M., Pitteloud N, Mendonca B.B., Latronico A.C. Nonsense mutations in FGF8 gene causing different degrees of human gonadotropin-releasing deficiency. J. Clin. Endocrinol. Metab. 2010;95:3491–3496. doi: 10.1210/jc.2010-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Riley B.M., Mansilla M.A., Ma J., Daack-Hirsch S., Maher B.S., Raffensperger L.M., Russo E.T., Vieira A.R., Dodé C., Mohammadi M., Marazita M.L., Murray J.C. Impaired FGF signaling contributes to cleft lip and palate. Proc. Natl Acad. Sci. USA. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Entesarian M., Dahlqvist J., Shashi V., Stanley C.S., Falahat B., Reardon W., Dahl N. FGF10 missense mutations in aplasia of lacrimal and salivary glands (ALSG) Eur. J. Hum. Genet. 2007;15:379–382. doi: 10.1038/sj.ejhg.5201762. [DOI] [PubMed] [Google Scholar]
- 86.Rohmann E., Brunner H.G., Kayserili H., Uyguner O., Nürnberg G., Lew E.D., Dobbie A., Eswarakumar V.P., Uzumcu A., Ulubil-Emeroglu M., Leroy J.G., Li Y., Becker C., Lehnerdt K., Cremers C.W., Yüksel-Apak M, Nürnberg P., Kubisch C., Schlessinger J., van Bokhoven H., Wollnik B. Mutations in different components of FGF signaling in LADD syndrome. Nat. Genet. 2006;38:414–417. doi: 10.1038/ng1757. [DOI] [PubMed] [Google Scholar]
- 87.Ohmachi S., Mikami T., Konishi M., Miyake A., Itoh N. Preferential neurotrophic activity of fibroblast growth factor-20 for dopaminergic neurons through fibroblast growth factor receptor-1c. J. Neurosci. Res. 2003;72:436–443. doi: 10.1002/jnr.10592. [DOI] [PubMed] [Google Scholar]
- 88.Gao X., Scott W.K., Wang G., Mayhew G., Li Y.J., Vance J.M., Martin E.R. Gene-gene interaction between FGF20 and MAOB in Parkinson disease. Ann. Hum. Genet. 2008;72:157–162. doi: 10.1111/j.1469-1809.2007.00418.x. [DOI] [PubMed] [Google Scholar]
- 89.Schaap F.G., van der Gaag N.A., Gouma D.J., Jansen P.L. High expression of the bile salt-homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis. Hepatology. 2009;49:1228–1235. doi: 10.1002/hep.22771. [DOI] [PubMed] [Google Scholar]
- 90.Reiche M., Bachmann A., Lössner U., Blüher M., Stumvoll M., Fasshauer M. Fibroblast growth factor 19 serum levels: relation to renal function and metabolic parameters. Horm. Metab. Res. 2010;42:178–181. doi: 10.1055/s-0029-1243249. [DOI] [PubMed] [Google Scholar]
- 91.Schreuder T.C., Marsman H.A., Lenicek M., van Werven J.R., Nederveen A.J., Jansen P.L., Schaap F.G. The hepatic response to FGF19 is impaired in patients with nonalcoholic fatty liver disease and insulin resistance. J. Physiol. Gastrointest. Liver Physiol. 2010;298:G440–G445. doi: 10.1152/ajpgi.00322.2009. [DOI] [PubMed] [Google Scholar]
- 92.Mraz M., Bartlova M., Lacinova Z., Michalsky D., Kasalicky M., Haluzikova D., Matoulek M., Dostalova I., Humenanska V., Haluzik M. Serum concentrations and tissue expression of a novel endocrine regulator fibroblast growth factor-21 in patients with type 2 diabetes and obesity. Clin. Endocrinol. 2009;71:369–375. doi: 10.1111/j.1365-2265.2008.03502.x. [DOI] [PubMed] [Google Scholar]
- 93.Durovcová V., Marek J., Hána V., Matoulek M., Zikán V., Haluzíková D., Kaválková P., Lacinová Z., Kršek M., Haluzík M. Plasma concentrations of fibroblast growth factors 21 and 19 in patients with Cushing's syndrome. Physiol. Res. 2010;59:415–422. doi: 10.33549/physiolres.931801. [DOI] [PubMed] [Google Scholar]
- 94.Li H., Fang Q., Gao F., Fan J., Zhou J., Wang X., Zhang H., Pan X., Bao Y., Xiang K., Xu A., Jia W. Fibroblast growth factor 21 levels are increased in nonalcoholic fatty liver disease patients and are correlated with hepatic triglyceride. J. Hepatol. 2010;53:934–940. doi: 10.1016/j.jhep.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 95.Dostálová I., Kaválková P., Haluzíková D., Lacinová Z., Mráz M., Papezová H., Haluzík M. Plasma concentrations of fibroblast growth factors 19 and 21 in patients with anorexia nervosa. J. Clin. Endocrinol. Metab. 2008;3:3627–3632. doi: 10.1210/jc.2008-0746. [DOI] [PubMed] [Google Scholar]
- 96.Razzaque M.S. The FGF23-Klotho axis: endocrine regulation of phosphate homeostasis. Nat. Rev. Endocrinol. 2009;5:611–619. doi: 10.1038/nrendo.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.ADHR Consortium. Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat. Genet. 2000;26:345–348. doi: 10.1038/81664. [DOI] [PubMed] [Google Scholar]
- 98.White K.E., Carn G., Lorenz-Depiereux B., Benet-Pages A., Strom T.M., Econs M.J. Autosomal-dominant hypophosphatemic rickets (ADHR) mutations stabilize FGF-23. Kidney Int. 2001;60:2079–2086. doi: 10.1046/j.1523-1755.2001.00064.x. [DOI] [PubMed] [Google Scholar]
- 99.Benet-Pagès A., Orlik P., Strom T.M., Lorenz-Depiereux B. An FGF23 missense mutation causes familial tumoral calcinosis with hyperphosphatemia. Hum. Mol. Genet. 2005;14:385–390. doi: 10.1093/hmg/ddi034. [DOI] [PubMed] [Google Scholar]
- 100.Shimada T., Mizutani S., Muto T., Yoneya T., Hino R, Takeda S., Takeuchi Y., Fujita T., Fukumoto S., Yamashita T. Cloning and characterization of FGF23 as a causative factor of tumor-induced osteomalacia. Proc. Natl Acad. Sci. USA. 2001;98:6500–6505. doi: 10.1073/pnas.101545198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Larsson T., Nisbeth U., Ljunggren O., Jüppner H., Jonsson K.B. Circulating concentration of FGF-23 increases as renal function declines in patients with chronic kidney disease, but does not change in response to variation in phosphate intake in healthy volunteers. Kidney Int. 2003;64:2272–2279. doi: 10.1046/j.1523-1755.2003.00328.x. [DOI] [PubMed] [Google Scholar]


