Abstract
Ebolaviruses are the etiologic agents of severe viral hemorrhagic fevers in primates, including humans, and could be misused for the development of biological weapons. The ability to rapidly detect and differentiate these viruses is therefore crucial. Antibodies that can detect reliably the ebolavirus surface envelope glycoprotein GP1,2 or a truncated variant that is secreted from infected cells (sGP) are required for advanced development of diagnostic assays such as sandwich ELISAs or Western blots (WB). We used a GP1,2 peptide conserved among Bundibugyo, Ebola, Reston, Sudan, and Taï Forest viruses and a mucin-like domain-deleted Sudan virus GP1,2 (SudanGPΔMuc) to immunize mice or rabbits, and developed a panel of antibodies that either cross-react or are virus-specific. These antibodies detected full-length GP1,2 and sGP in different assays such as ELISA, FACS, or WB. In addition, some of the antibodies were shown to have potential clinical relevance, as they detected ebolavirus-infected cells by immunofluorescence assay and gave a specific increase in signal by sandwich ELISA against sera from mouse-adapted Ebola virus-infected mice over uninfected mouse sera. Rabbit anti-SudanGPΔMuc polyclonal antibody neutralized gammaretroviral particles pseudotyped with Sudan virus GP1,2, but not particles pseudotyped with other ebolavirus GP1,2. Together, our results suggest that this panel of antibodies may prove useful for both in vitro analyses of ebolavirus GP1,2, as well as analysis of clinically relevant samples.
Keywords: Ebola, ebolavirus, envelope glycoprotein, filovirus, sGP
1. Introduction
The family Filoviridae currently includes the two established genera, Ebolavirus and Marburgvirus, and one tentative genus, Cuevavirus. The Ebolavirus genus consists of five species, all of which have one virus member: Bundibugyo ebolavirus (Bundibugyo virus, BDBV), Reston ebolavirus (Reston virus, RESTV), Sudan ebolavirus (Sudan virus, SUDV), Taï Forest ebolavirus (Taï Forest virus, TAFV), and Zaire ebolavirus (Ebola virus, EBOV). The genus Marburgvirus contains one species (Marburg marburgvirus) with two member viruses, Marburg virus (MARV) and Ravn virus (RAVV) (Kuhn, et al, 2010). With the exception of RESTV, ebolaviruses have been associated with naturally occurring outbreaks of severe viral hemorrhagic fever in humans. EBOV causes the most severe disease outbreaks with lethality reaching up to 90% (Feldmann et al., 2003; Towner et al., 2008).
Ebolaviruses are enveloped viruses and cell entry is mediated by the envelope glycoprotein (GP1,2). Definitive identification of the ebolavirus receptor remains elusive, although several surface molecules have been shown to enhance infection (Dolnik et al., 2008). The full-length GP1,2 on the surface of ebolavirions is translated from an mRNA that is the product of cotranscriptional editing and contains an additional non-template adenosine residue within a stretch of seven consecutive adenosine residues near the center of the coding region for the protein (Sanchez et al., 1996; Volchkov et al., 1995). Post-translational cleavage by a cellular protease yields the mature virion-associated form comprised of two protein subunits, the virion surface protein, GP1, and the transmembrane protein, GP2, linked together through a disulfide bond (GP1,2). GP1 contains the receptor binding site (RBS) (Kuhn et al., 2006) and a highly N- and O-glycosylated and diverse region at the C-terminus called the mucin-like domain. GP2 is anchored on the virus envelope and cell surface through the transmembrane domain (TMD). The extracellular domain (ECD) of GP2 contains the functional domains necessary for inducing the fusion of viral and cellular membranes upon GP1 binding to the ebolavirus receptor. Translation from the unedited GP gene (~80% of the time) gives rise to a much shorter soluble GP (sGP). The N-terminal 295 amino acid residues of sGP are identical to those of GP1. The C-terminus of sGP is unique and also highly diverse between sGP proteins from different ebolaviruses. One potential role of sGP is to serve as a decoy for ebolavirus-neutralizing antibodies (Feldmann et al., 1999; Volchkov, 1999; Volchkova et al., 1998).
There are no FDA-licensed therapies or vaccines for filoviruses. Neutralizing antibodies reacting with GP1,2 may provide passive immunity (Gupta et al., 2001; Jahrling et al., 1996; Parren et al., 2002; Takada et al., 2007). GP1,2-specific antibodies are also very useful reagents for research. For example, Lee and coworkers used an EBOV-neutralizing antibody, KZ52, to form a complex with ebolavirus GP1,2 and elucidated the crystal structure of GP1,2 (Lee et al., 2008). In addition, ebolavirus-specific antibodies are critical tools for detection and diagnosis of ebolavirus infection in the field and in the laboratory. Most anti-ebolavirus GP1,2 monoclonal antibodies reported to date are ebolavirus-specific, and many target the highly variable mucin-like domain, presumably because it contains immuno-dominant epitopes. In addition, most of the reported monoclonal antibodies are of mouse origin only (Shahhosseini et al., 2007; Wilson et al., 2000), which provides less flexibility in the development of some assays, for example, sandwich ELISAs.
This report describes the development and characterization of a new panel of ebolavirus-specific antibodies. Some of these antibodies are broadly cross-reactive to GP1,2 of different ebolaviruses. In addition, both mouse and rabbit monoclonal antibodies were generated that might allow for the development of complementary strategies in the establishment of ebolavirus-detection assays. Previous findings that the phenylalanine residue at position 88 (F88) of EBOV GP1 plays an important role in viral entry, is conserved among all currently known ebola- and marburgviruses, and regions flanking F88 are highly conserved among different ebolaviruses (Mpanju et al., 2006) suggested that antibodies raised against this highly conserved region would be broadly cross-reactive. A 38-mer peptide was synthesized corresponding to this region, designated as F88 peptide, and was used to develop rabbit polyclonal antibodies and mouse monoclonal antibodies. Additionally, SUDV GP1,2 with a deletion of 190 amino acid residues comprising the mucin-like domain (referred to as GP-S) was also used to develop rabbit polyclonal and monoclonal antibodies. Since most type-specific neutralizing antibodies reported to date recognize the highly glycosylated mucin-like domain that is also the most variable region of GP1,2, it is proposed that the use of mucin-like-domain deleted GP1,2 would induce more broadly-reactive antibodies. The mouse and rabbit polyclonal and monoclonal antibodies generated in this work will contribute to the development of detection assays for all the known ebolavirus species.
2. Materials and Methods
2.1. Cell culture
Human embryonic kidney (HEK) 293T (obtained from T. Dull, Cell Genesys, CA, now BioSante Pharmaceutical, Lincolnshire, IL), grivet kidney Vero E6 (ATCC, # CRL-1586, Manassas, VA) and grivet COS-7 cells (ATCC, #CRL-1651, Manassas, VA) were cultured at 37 °C in 5% CO2 and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Lonza, Walkersville, MD) supplemented with 10% heat-inactivated fetal bovine serum (FBS, HyClone, Logan, UT), 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Lonza, Walkersville, MD). Culture conditions used for mouse and rabbit hybridoma cells are described below.
2.2. Plasmids
Expression plasmids encoding the GP1,2 of EBOV (Mayinga variant), SUDV (Gulu variant), and TAFV (Côte d’Ivoire variant), pVR1012-ZaireGP, pVR1012-SudanGP and pVR1012-TaiForestGP, respectively, were provided by Dr. Gary Nabel (Vaccine Research Center, NIH, Bethesda, MD) and Dr. Anthony Sanchez (CDC, Atlanta, GA) (Yang et al., 1998). Each of the plasmids had been mutated at the GP gene editing site, resulting in predominant expression of full-length GP1,2 rather than sGP. Plasmid pGP-S, which encodes SUDV (Gulu variant) GP1,2 amino acid residues (aa) 1-315, aa 506-650, a short linker (GG) and a His6 tag (Fig. 2C), was obtained by over-lapping PCR using pVR1012-SudanGP as template. Primers used to amplify the region that encodes aa 1-315 were designated GP-S-#1 (5′-GATCTCGAGCTCGCCACCATGGAGGGTCTTAGCCTACTCC-3′) and GP-S-#2 (5′-ACCCGTGGCCCTCTCGTTGAGCGATAAAGTTTCGAA-3′). Primers used to amplify the region that encodes aa 506-650 were designated GP-S-#3 (5′0TCGCTCAACGAGAGGGCCACGGGTAAATGCAATCCC-3′) and GP-S-#4 (5′-CGGGCCCGCGGTTAGTGATGGTGATGGTGATGGCCACCCTGTCTCCAGCCCG TCCACCAATTATC-3′). The coding sequence for the His6 tag (underlined) is contained within primer GP-S-#4. After gel purification, these two DNA fragments were linked by overlapping PCR using primers GP-S-#1 and GP-S-#4 and then ligated into pIRES2-EGFP (Clontech, Mountain View, CA) that had been restriction digested with Nhe I and Sac II (New England BioLabs, Ipswich, MA). The sequence of the resulting plasmid pGP-S was confirmed using the ABI Prism 3100 Sequence Detection System using the BigDye® Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). pBundiGP encodes the full-length BDBV (Bundibugyo variant) GP1,2. The coding sequence was designed according to the deposited sequence information (GenBank Accession No. FJ217161), codon-optimized for optimal expression in mammalian cells, synthesized, and inserted into pcDNA3.1(+) (Invitrogen, Carlsbad, CA, USA) by a commercial gene-synthesizing company (DNA2.0, Menlo Park, CA, USA). pReston-sGP encodes the wild-type unedited gene for RESTV (Pennsylvania variant) GP1,2 and expresses predominantly sGP (a gift from Michael Farzan, Harvard Medical School, Boston, MA, USA). Plasmid pMARV-Mus GP1,2, encodes Marburg virus (MARV) GP1,2 and a C9 epitope tag at its C-terminus, was described previously (Kuhn et al., 2006).
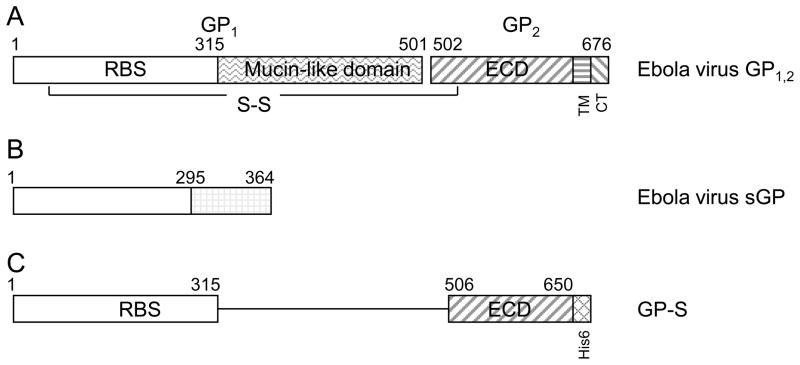
Fig. 2. Diagrammatic depiction of ebolavirus GP1,2 and antigen GP-S used for immunization.
A. full-length ebolavirus GP1,2, B. sGP; C. GP-S chimeric protein that includes aa1-315 and aa506-650 of SUDV GP1,2. The mucin-like domain, GP1/GP2 cleavage site, transmembrane domain (TM), and cytoplasmic tail (CT) were deleted, and a His6 epitope was added to the C-terminus. RBS: receptor-binding site; ECD: extracellular domain.
2.3. Peptide and protein immunogens used for antibody development
F88 peptide (Fig. 1B) includes 38 amino acid residues of EBOV GP1,2, a short linker peptide of serine-glycine-serine residues (SGS) and biotin at the N-terminus. It was synthesized and purified to ≥95% homogeneity at PolyPeptide Laboratories (San Diego, CA). To increase its immunogenicity, F88 peptide was conjugated to Keyhole Limpet Hemacyanin (KLH) carrier protein using the Imject Maleimide Activated mcKLH kit from Pierce (Rockford, IL) and inoculated at Spring Valley Laboratories, Inc. (Woodline, MD). The chimeric fusion protein GP-S includes SUDV GP1,2 aa 1-315, aa 506-650, a short linker (GG) and a His6 tag. The mucin-like domain, cleavage site between GP1 and GP2, transmembrane domain, and cytoplasmic tail of SUDV GP1,2 were deleted in GP-S (Fig. 2C). The fusion protein was expressed and purified at Chesapeake PERL (Savage, MD) using the PERLXpress System, a baculovirus based system to express proteins in cabbage looper (Trichoplusia ni) larvae (Kovaleva et al., 2009). The purity of GP-S was estimated as higher than 90% homogeneity by Coomassie blue staining.
Fig. 1. F88 peptide used for antibody development.
A. Alignment of a region of GP1,2 that flanks the phenylalanine residue at position 88 (F88) (amino acids 72-109) from five ebolaviruses and Marburg virus. The GenBank Accession Numbers for these sequences from top to bottom are ACI28624.1, AAB37093.1, AAC24346.1, AAU43887.1, AAD14585.1 and ABA87127.1, respectively. Conserved residues are shown in black and non-conserved residues in gray. B. F88 peptide used for immunization is derived from Ebola virus GP1,2 (GenBank No. AAD14585.1). The linker sequence SGS (underlined) and biotin were added to the N-terminus of the peptide to facilitate detection and antibody purification.
2.4. Development of rabbit anti-F88 polyclonal antibody
Each of the two rabbits was primed subcutaneously once with 250 μg F88-KLH conjugate protein mixed with equal volume of complete Freund’s adjuvant and boosted subcutaneously four times with 250 μg F88-KLH conjugate protein mixed with an equal volume of incomplete Freund’s adjuvant in a total volume of 1 ml peptide-adjuvant mixture for each injection. Peptide-KLH conjugation and injections were performed by Epitomics, Inc (Burlingame, CA) following the schedule illustrated in Fig. 3A. The specific anti-F88 antibody was purified from the terminal bleed of the second rabbit serum using F88 peptide captured on streptavidin magnetic beads (MyOne Streptavidin T1, Invitrogen, Carlsbad, CA) through its N-terminal biotin moiety, following the manufacturer’s instructions. The antibodies were eluted using 0.1 M glycine-HCl (pH 2.7) buffer, which was neutralized immediately with 1 M Tris-HCl (pH 8.0) and then exchanged to phosphate-buffered saline (PBS) using Amicon filters (Ultracel-10k, Millipore, Billerica, MA).
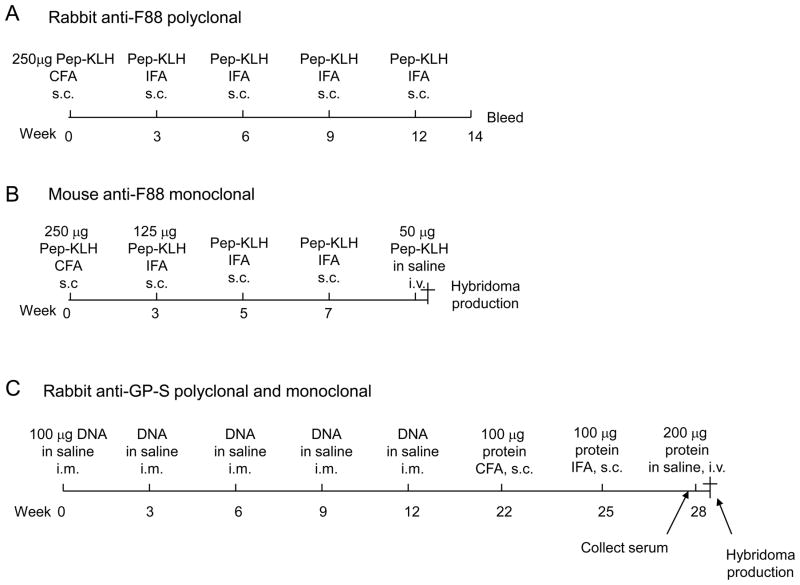
Fig. 3. Immunization schedules for raising rabbit and mouse anti-ebolavirus GP1,2 antibodies.
Immunization schedule, doses and routes for developing (A) rabbit anti-F88 polyclonal antibodies; (B) mouse anti-F88 monoclonal antibodies; and (C) rabbit anti-GP-S polyclonal antibodies. The same rabbit that was immunized to make anti-GP-S polyclonal antibodies was used to make monoclonal. Pep-KLH, peptide and Keyhole Limpet Hemacyanin protein conjugate; CFA, complete Freund’s adjuvant; IFA, incomplete Freund’s adjuvant; s.c., subcutaneously; i.v., intravenously; i.m., intramuscularly.
2.5. Development of mouse anti-F88 monoclonal antibodies
Each of five mice was primed subcutaneously once with 250 μg F88-KLH conjugate protein in complete Freund’s adjuvant, boosted subcutaneously three times with 125 μg F88-KLH conjugate protein in incomplete Freund’s adjuvant, and finally one more time intravenously with 50 μg F88-KLH conjugate protein in saline solution. Peptide-KLH conjugation and injection were performed by Promab Biotechnologies (Richmond, CA) following the schedule illustrated in Fig. 3B. Splenocytes isolated from the mouse that produced the highest antibody response, which was measured by EBOV GP1,2 sandwich ELISA (below), were fused with SP2/0 myeloma cells for monoclonal antibody production. Three thousand clones were screened against F88-BSA using ELISA assay and 30 positive clones were obtained. Five clones that produced the highest specific signal on the F88 ELISA were subcloned by limiting dilution and sent to us for confirmation and characterization using ELISA, FACS and/or Western blot (WB, below). The hybridoma clones were propagated in MegaCell DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with 10% heat-inactivated FBS, 1 mM glutamine (Invitrogen, Carlsbad, CA), and 50 μg/ml kanamycin (Invitrogen, Carlsbad, CA). For antibody production, the expanded hybridoma cells were washed twice with PBS and grown in Opti-MEM medium (Invitrogen, Carlsbad, CA) for 3 days before harvesting the antibody-containing supernatant. Antibodies were purified from the supernatant using a protein G column (GE Healthcare, Piscataway, NJ), eluted using 0.1 M glycine-HCl (pH 2.7) buffer, which was immediately neutralized with 1 M Tris-HCl (pH 8.0) and then exchanged to PBS using an Amicon filter (Ultracel-10k Millipore, Billerica, MA). The isotype of these monoclonal antibodies was determined using the IsoStrip Mouse Monoclonal Antibody Isotyping Kit from Roche Applied Sciences (Indianapolis, IN) following the manufacturer’s manual.
2.6. Development of rabbit anti-GP-S polyclonal and monoclonal antibodies
Each of two rabbits was first immunized with five intramuscular injections of 100 μg plasmid pGP-S in 100 μl saline solution every three weeks, followed by one additional subcutaneous injection with 100 μg purified GP-S in complete Freund’s adjuvant, one subcutaneous injection with 100 μg purified GP-S in incomplete Freund’s adjuvant, and one final intravenous injection with 200 μg purified protein in saline solution. The immunizations were performed by Spring Valley Laboratory (Sykesville, MD) following the schedule illustrated in Fig. 3C. Sera were collected prior to the last intravenous (i.v.) injection and were used to analyze and subsequently purify polyclonal antibodies. GP-S-specific antibodies were isolated from the rabbit sera using GP-S captured on AminoLink Plus beads (Pierce, Rockford, IL) following the manufacturer’s instructions. The antibody was eluted using 0.1 M glycine-HCl (pH 2.7) buffer, which was immediately neutralized with 1 M Tris-HCl (pH 8.0) and then exchanged to PBS using an Amicon filter (Ultracel-10k, Millipore, Billerica, MA).
Rabbit sera were screened by GP-S sandwich ELISA. Rabbit #1 had the highest antibody response and was euthanized 4 days after the last i.v. boost and the spleen was harvested and sent to Epitomics, Inc. (Burlingame, CA). Splenocytes were isolated and used to generate hybridomas through fusion with their proprietary myeloma-like tumor cell line 240E-W2 (Huang et al., 2007). Hybridoma clones (a total of 3,840) were screened by Epitomics, Inc. (Burlingame, CA) using ELISA with GP-S protein captured on 96-well plates that were coated with anti-His6 antibody. Sixty positive clones were obtained. The three hybridoma clones with highest scores in this assay were sent to us for confirmation by ELISA, FACS and/or WB (below). Upon confirmation they were subcloned by limiting dilution and expanded in RPMI 1640 medium supplemented with 0.05 mM 2-mercaptoethanol (Invitrogen, Carlsbad, CA), RabMAb Supplement A (Epitomics Inc Burlingame, CA), 2 mM GlutaMAX-1 (Invitrogen, Carlsbad, CA), and 10% heat-inactivated fetal bovine serum. For antibody production, the expanded hybridoma cells were washed twice with PBS and grown in CD Hybridoma Medium (Invitrogen, Carslbad, CA) for 3 days before harvesting the antibody-containing supernatant. The antibodies were purified from the supernatant using a protein A column (GE Healthcare, Piscataway, NJ), eluted using 0.1 M glycine-HCl (pH 2.7) buffer, that was immediately neutralized with 1 M Tris-HCl (pH 8.0) and then exchanged to PBS using an Amicon filter (Ultracel-10k, Millipore, Billerica, MA).
2.7. Coomassie blue staining
Two micrograms of purified antibodies were boiled in 1× NuPAGE lithium dodecylsulfate (LDS) sample buffer and 1× NuPAGE sample reducing agent (Invitrogen, Carlsbad, CA) or dithiothreitol (DTT) at 50 or 100 mM for 5 min and resolved on a pre-cast NuPAGE 4–12% Bis-Tris Gel. The gel was stained using the SimplyBlue SafeStain (Invitrogen, Carlsbad, CA) and destained following the manufacturer’s manual.
2.8. Western blot analysis
Confirmation of bands observed by Coomassie Blue stained SDS-PAGE was performed by Western blot analysis. Two micrograms of purified R.F88-2, R.GP-S-1, R.GP-S-46-12, R.GP-S-53-7, R.GP-S-55-1 and rabbit IgG, as a control, were boiled in 1× NuPAGE LDS sample buffer and 1× NuPAGE sample reducing agent (Invitrogen, Carlsbad, CA) for 5 min, resolved on pre-cast NuPAGE 4–12% Bis-Tris gels, and transferred to PVDF membranes (Invitrogen, Carlsbad, CA). Membranes were blocked with 3% BSA overnight at 4 °C, incubated with horseradish peroxidase-conjugated anti-rabbit IgG (goat anti-rabbit; 80ng/mL, Pierce, Rockford, IL) for 45 min at room temperature on a rocker, and washed 4 times with TBST buffer (25 mM Tris buffer, pH8.0, KD Medical, Columbia, MD, 125 mM sodium chloride, Promega, Madison, WI, 0.1% Tween 20, Sigma-Aldrich, St. Louis, MO). Western Lightning™ Plus Chemiluminescence Reagent (PerkinElmer, Waltham, MA) was used to develop films (Kodak’s BioMax MR, Rochester, NY).
Analysis of antibody reactivity properties by Western blot (WB) was performed using lysates of HEK 293T cells after transient transfection with plasmids expressing either ebolavirus or marburgvirus GP1,2. To prepare cell lysates, HEK 293T cells were seeded in 10-cm dishes at a density of 5 × 106 cells per dish, and transfected the next day with 24 μg of one of the GP1,2-expressing plasmids (described in 2.2) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) following the manufacturer’s manual. Forty-eight hours after transfection, cells were washed twice in PBS, detached by gentle pipetting and centrifuged at 500×g for 5 min. Cell pellets were frozen on dry ice, thawed once, and then lysed in RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris pH8.0) supplemented with the Complete Mini Protease Inhibitor Cocktail (1 tablet/7 ml, Roche Applied Science, Indianapolis, IN) for 30 min on ice, vortexing once every 10 min. Lysates were cleared by centrifugation for 10 min at 18,000 x g at 4 °C in a bench top microcentrifuge. The GP1,2-containing lysates were resolved by SDS-PAGE, and transferred to PVDF membranes. After incubation with primary antibodies as indicated above, the membranes were incubated with a horseradish peroxidase conjugated-secondary antibody pre-diluted to 50 ng/ml (Pierce, Rockford, IL) for 45 minutes at room temperature on a rocker, and washed 4 times with PBST buffer. Western Lightning™ Plus Chemiluminescence Reagent (PerkinElmer, Waltham, MA) was used as substrate, sensitive film (Kodak’s BioMax MR, Rochester, NY) was exposed to the membranes for different times and developed.
2.9. Sandwich ELISA
Maxisorp 96-well ELISA plates were coated with 50 μl capturing antibody (2 μg/ml in PBS) overnight at 4 °C and blocked with 100 μl 3% BSA in PBS for 1 h at 37 °C. Fifty microliters of cell lysate, which was prepared in the same way as for WB (above), was added to the blocked ELISA plate. After 2 h of incubation at 37 °C, the plate was washed twice with PBST buffer (0.05% Tween 20 in PBS) and incubated with primary antibody for 1 h at 37 °C. Each of the primary antibodies was used at 0.2 μg/ml (primary antibodies used for these experiments: R.F88-2, R.GP-S-1, R.GP-S-46-12, R.GP-S-53-4, R.GP-S-55-1, M.F88-H3D5, M.F88-H3C8, 13C6-1-1. The last mouse monoclonal antibody was a kind gift of Gene Olinger, USAMRIID, Ft. Detrick, MD (Wilson et al., 2000). Plates were washed twice in PBST and incubated with horseradish peroxidase conjugated anti-mouse IgG or anti-rabbit IgG antibody pre-diluted to 50 ng/ml for 30 min at 37 °C. After four washes in PBST, one hundred microliters of ABTS peroxidase substrate (KPL, Gaithersburg, MD) was added to each well, and 10 min later the plate was read at 405 nm on a Victor3 V microplate reader (Perkin Elmer, Waltham, MA). Serum samples from infected and control mice were analyzed using a similar procedure with the exception that the plates were washed two times prior to adding ABTS peroxidase substrate, not four times.
2.10. Analysis of antibody binding affinity
Binding of M.F88-H3C8 and M.F88-H3D5 monoclonal antibodies to EBOV GP1,2 was analyzed with a Biacore 3000 surface Plasmon resonance spectrometer (GE Healthcare). All samples were analyzed in 10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, and 0.005% surfactant P-20 buffer. ZEBOVGP-Fc, a fusion protein derived from EBOV GP1,2 with the transmembrane domain and cytoplasmic tail replaced with human IgG1 Fc, or control protein Fc with a FLAG epitope tag at its N-terminus (FLAG-Fc) (Konduru et al., 2011) was immobilized by covalent attachment to a carboxymethyl dextrate-coated biosensor chip (CM5) using amino coupling to yield a surface density of 1,000 response units. The H3C8 or H3D5 antibody was injected over the coupled sensor chip with concentrations of 25, 50, 100, and 200 nM at a flow rate of 30 μl/min for 3 min at 25 °C. The dissociation rate of the complexes was monitored for 15 min at a flow rate of 30 μl/min. To account for background effects, the results obtained using the FLAG-Fc-containing flow cell were subtracted from the values obtained from the flow cells coated with ZEBOVGP-Fc. BIAevaluation version 4.1 was used to analyze the association and dissociation data, and a 1:1 Langmuir binding model was used to fit data globally to extract kinetic parameters of interaction with ZEBOVGP-Fc.
2.11. FACS analysis
Antibody binding to GP1,2 expressed on the cell surface was determined by fluorescence-activated cell sorting (FACS) analysis. To that end, COS-7 cells were seeded in 10-cm dishes at the density of 3×106 cells per dish and transfected with 12 μg of one of each of the ebolavirus GP1,2-expressing plasmids (as described in section 2.2) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. Thirty-six hours after transfection, cells were washed twice in PBS, detached using 5 mM EDTA in PBS, centrifuged at 500×g for 5 min, and resuspended in staining solution (CO2-independent Medium of Invitrogen supplemented with 2.5 mM EDTA and 2.5% FBS) at a concentration of 107 cells/ml. For each sample, 0.1 ml of cells was mixed with the corresponding primary antibody pre-diluted to 1 μg/ml in staining solution and incubated at 4°C for 1 h. After washing in 3 ml PBS, cells were resuspended in 0.1 ml staining solution and incubated with Alexa 647-conjugated anti-mouse IgG (Invitrogen, Carlsbad, CA) or Alexa 647-conjugated anti-rabbit IgG (Invitrogen, Carlsbad, CA) pre-diluted to 1 μg/ml in staining solution for 45 min at 4°C. After washing twice in 3 ml PBS each time, cells were resuspended in 0.25 ml staining solution. Immediately prior to analysis using a FACS Calibur (BD, Franklin Lakes, NJ), 0.25 ml staining solution supplemented with 0.5 μl cell viability dye YO-PRO-1 (Invitrogen, Carlsbad, CA) was added. FACS data were analyzed using FlowJo software (TreeStar, Ashland, OR).
2.12. Filovirus infections
Vero or HeLa cells were infected with EBOV-Kik, RESTV-Pen, SUDV-Bon, TAFV-CI, or MARV-Ci67 at a multiplicity of infection (MOI) of 0.3. Viruses were adsorbed to cells for 1 h, the inocula were removed, and cells were washed three times with PBS and supplemented with fresh growth media. Two days later, cells were fixed in 10% buffered formalin (Val Tech Diagnostics, Pittsburg, PA) for 72 h and stained for quantitative image-based analysis with the murine monoclonal antibody M.F88-H3D5 or the rabbit antibodies R.GP-S-46-12 or R.GP-S-1, followed by Alexa Fluor 488 goat anti-mouse IgG (Invitrogen, Carlsbad, CA) or by DyLight 488 goat anti-rabbit IgG (Thermo Fisher Scientific, Rockford, IL). All infected cells were stained with Hoechst 33342 and HCS CellMask Red (Invitrogen, Carlsbad, CA). Fluorescence images of stained cells were acquired and analyzed on an Opera confocal reader (model 3842-Quadruple Excitation High Sensitivity (QEHS), Perkin Elmer, Waltham, MA) using a 10X air objective. Analysis of the images was accomplished within the Opera environment using standard Acapella scripts. All experiments involving infectious filoviruses were performed under biosafety level 4 containment at the United States Army Medical Research Institute for Infectious Diseases.
2.13. Pseudotype virus neutralization
Production of gammaretroviral (Moloney murine leukemia virus, MoMLV) pseudotypes bearing BDBV, EBOV, SUDV, TAFV GP1,2 or VSIV G protein was performed as described previously (Ou et al., 2010). Briefly, Vero E6 cells were seeded in 24-well plates at the density of 3.5×104 cells per well one day prior to the experiment. Pseudotypes were mixed with testing antibodies at the indicated concentrations, incubated for 45 min at 37°C, and another 15 min at 37 °C after adding polybrene (American Bioanalytical, Natick, MA) to a final concentration of 8 μg/ml. After cell culture media were removed from cells, 200 μl of pseudotype/antibody/polybrene mixture was added to each well, and the cells were returned to the CO2 incubator at 37 °C. Eight hours later, the pseudotype/antibody/polybrene mixture was removed and 500 μl of regular culture medium was added to each well. After another 40 h of culture, cells were washed, fixed and stained for LacZ expression as previously described (Ou et al., 2010). Pseudotype virus titers were calculated according to the number of the blue-forming units per ml and normalized to 100% for the control that did not contain antibody.
All animal experiments were performed according to international standards for animal welfare.
3. Results
3.1. Development of rabbit and mouse monoclonal and polyclonal antibodies
Following the immunization regimens depicted in Fig. 3 and the antibody production and purification methods described above, one rabbit polyclonal antibody against the conserved F88 peptide (R.F88-2), two mouse monoclonal antibodies against the F88 peptide (M.F88-H3D5, M.F88-H3C8), one rabbit polyclonal antibody (R.GP-S-1) and four rabbit monoclonal antibodies (R.GP-S-46-12, R.GP-S-53-4, R.GP-S-53-7, R.GP-S-55-1) against the GP-S protein were developed. R.GP-S-53-4 and R.GP-S-53-7 are different subclones derived from the same positive clone, and therefore should have the same characteristics. The isotype of the murine monoclonal antibodies M.F88-H3D5 and M.F88-H3C8 was found to be IgG1 with a kappa light chain (data not shown). To evaluate the purity of the antibodies, each antibody preparation was analyzed on an SDS PAGE gel stained with Coomassie blue. The purity of all the antibodies was estimated as higher than 90% except for R.GP-S-1, which had an unexpected band of ≈70 kDa (Fig. 4). Since R.GP-S-1 polyclonal antibody was purified by affinity to solid phase GP-S, the 70 kDa is an unknown rabbit serum protein that binds to GP-S. The heavy chain of R.GP-S-55-1 was a smear. To confirm the identity of the bands as immunoglobulin, the proteins were analyzed by Western blot,. An approximately 55 kDa band was detected with horseradish peroxidase-conjugated anti-rabbit IgG by Western blot analysis, similar to control rabbit immunoglobulin loaded onto the same immunoblot (data not shown).
Fig. 4. Coomassie blue staining of rabbit and mouse anti-ebolavirus GP1,2 antibodies.
Each of the polyclonal antibodies was purified from sera using the corresponding antigens conjugated to beads and monoclonal antibodies were purified from the supernatant of hybridoma cells grown in serum-free medium using protein A (for rabbit monoclonals) or protein G (for mouse monoclonals) beads. Two micrograms of each antibody were resolved on SDS-PAGE gel and stained.
3.2. Detection of filovirus GP1,2 by Western blot
To assess whether the panel of antibodies can detect the GP1,2 of various filoviruses by WB, 293T cells were transfected with plasmids that encode EBOV, SUDV and TAFV full-length GP1,2, RESTV sGP, or MARV full-length GP1,2 that contains a C9 epitope tag at its C-terminus, and used the cell lysate for WB analysis. As shown in Fig. 5, R.F88-2, M.F88-H3D5, M.F88-H3C8, and R.GP-S-1 detected all the ebolavirus GP1,2 tested. However, R.GP-S-46-12, R.GP-S-53-4 and R.GP-S-55-1 detected SUDV GP1,2 only. None of the antibodies detected MARV GP1,2,. The expression of MARV GP1,2 was confirmed by WB with anti-C9 antibody (Fig. 5, lower right panel). Since MARV GP1,2 was not detected by any of the antibodies tested by Western blot analysis, MARV GP1,2 was not included in any further analyses.
Fig. 5. Detection of various Filovirus GP1,2 by anti-ebolavirus GP1,2 antibodies using WB.
Lysate of cells that were transiently transfected to express the full-length GP1,2 of EBOV (E), SUDV (S), TAFV (TF) or the MARV (M), RESTV sGP (R), or without plasmid (Mock) were resolved on a 4-12% NuPAGE gel, and transferred to a PVDF membrane. Shown here are the bands detected after incubation with the indicated mouse or rabbit primary antibodies (indicated above or below the blots) and the corresponding horseradish peroxidase-conjugated secondary antibodies. Anti-C9 was used for detecting MARV GP1,2, which had the C9 epitope attached to its C-terminus. The size bands expected for GP1,2, sGP, and the GP-C9 fusion protein are shown on the right.
3.3. Detection of ebolavirus GP1,2 by sandwich ELISA
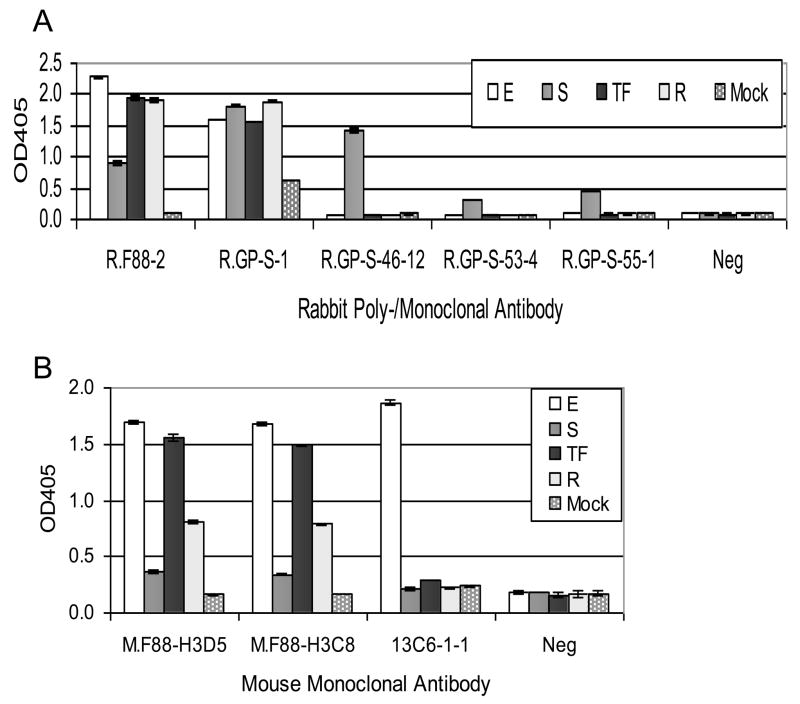
The utility of the antibodies for detecting ebolavirus GP1,2 was evaluated by sandwich ELISA. To assess the detection properties of rabbit polyclonal and monoclonal antibodies, M.F88-H3D5 was used to capture GP1,2. To evaluate the mouse monoclonal antibodies, GP1,2 was captured using R.F88-2. Consistent with the WB results, R.F88-2, R.GP-S-1, M.F88-H3D5 and M.F88-H3C8 detected all the ebolavirus GP1,2 tested, while R.GP-S-46-12, R.GP-S-53-4 and R.GP-S-55-1 detected SUDV GP1,2 only (Fig. 6). The mouse monoclonal antibody 13C6-1-1 served as a control for the assay format, since it is known to detect EBOV GP1,2 (Wilson et al., 2000).
Fig. 6. Detection of various ebolavirus GP1,2 by anti-GP1,2 antibodies using sandwich ELISA.
A. To detect GP1,2 using rabbit antibodies, mouse anti-F88 monoclonal M.F88-H3D5 (2 μg/ml) was used to coat the plate and capture the indicated GP1,2 in cell lysate of transiently transfected cells. B. To detect GP1,2 by mouse antibodies, rabbit anti-F88 polyclonal R.F88-2 (2 μg/ml) was used to coat the plate and capture the indicated GP1,2. The lysates were the same as those used for WB (Fig. 5). E, EBOV GP1,2; S, SUDV GP1,2, TF, TAFV GP1,2; R, RESTV sGP. For negative controls (Neg), PBS was used in place of primary antibody, and the same horseradish peroxidase conjugated anti-rabbit IgG (A) or anti-mouse IgG (B) secondary antibody was used. Antibody 13C6-1-1 served a positive control for the assay design, by detection of EBOV GP1,2. The average OD405 ± standard deviation of triplicate samples is shown.
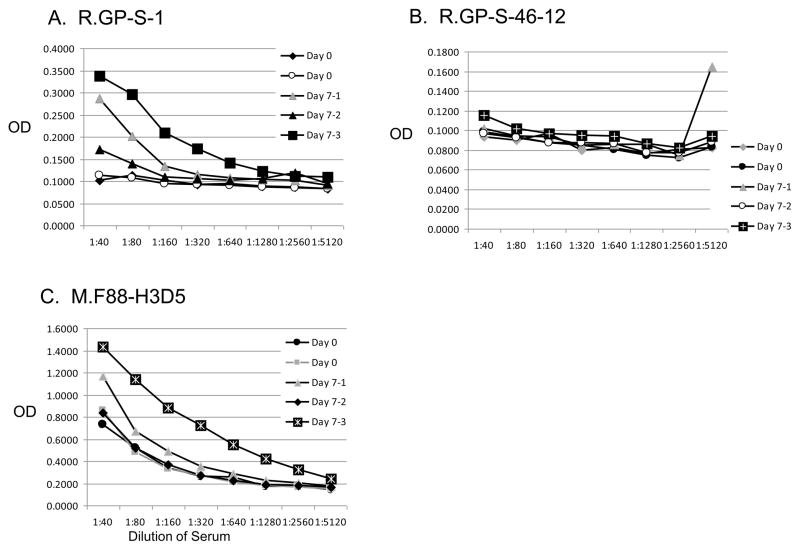
In addition, the ability of the antibodies to detect EBOV GP1,2 in serum samples from mice infected with mouse-adapted EBOV at various time points after injection was evaluated using the sandwich ELISA format. R.GP-S-1 or M.F88-H3C8 detected EBOV GP12 in sera from mice infected with EBOV at 7 days postinfection compared to uninfected mice at 0 day (Fig. 7). As expected, R.GP-S-46.12 did not detect EBOV GP1,2, since it only reacts with SUDV-GP1,2.
Fig. 7. Detection of mouse-adapted Ebola virus in murine serum samples by sandwich ELISA.
Sera from mice before (day 0) and 7 days after injection with mouse-adapted Ebola virus were serially diluted and then evaluated for presence of EBOV GP1,2 using the polyclonal antibody R.GP-S-1 or monoclonal antibodies R-GP-S-46-12 or M.F88-H3D5.
3.4. Antibody binding affinity
Binding affinity of M.F88-H3D5 and M.F88-H3C8 to ZEBOVGP-Fc was measured using surface plasmon resonance technology. ZEBOVGP-Fc is a recombinant protein composed of the extracellular regions of EBOV GP1,2 fused to the Fc domain of human IgG1 (Konduru et al., 2011). The dissociation constants (KD) for M.F88-H3C8 and M.F88-H3D5 were 148 nM and 361 nM, respectively (data not shown).
3.5. Detection of ebolavirus GP1,2 by FACS
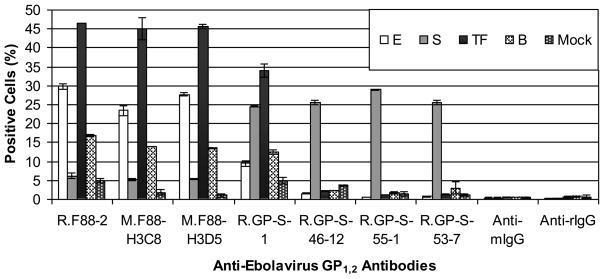
Flow cytometry was used to evaluate whether the antibodies could detect native GP1,2 exposed at the cell surface. Consistent with WB and ELISA results, R.GP-S-46-12, R.GP-S-53-7 and R.GP-S-55-1 bound to cells expressing SUDV GP1,2 but not the other ebolavirus GP1,2. In contrast, R.F88-2, M.F88-H3D5 and M.F88-H3C8 bound to cells expressing BDBV, EBOV, and TAFV but reacted poorly with cells expressing SUDV GP1,2. R.GP-S-1 detected the GP1,2 of all tested ebolaviruses, although detection of BDBV and EBOV GP1,2-expressing cells was lower than detection of cells expressing SUDV or TAFV GP1,2 (Fig. 8).
Fig. 8. Detection of various ebolavirus GP1,2 by anti-GP1,2 antibodies using flow cytometry.
COS-7 cells were transiently transfected with a plasmid that encodes GP1,2 of either EBOV (E), SUDV (S), TAFV (TF), or BDBV (B), or β-galactosidase (Mock), stained with the indicated primary antibodies (first five groups) or without the primary antibody (last two groups) and the corresponding Alexa 647-conjugated anti-rabbit or mouse IgG. Dead cells, detected positively with a stain for cell viability (YO-PRO-1), were excluded from analysis. The average percentage of positively stained cells ± standard deviation of duplicate samples is shown.
3.6. Detection of ebolavirus-infected cells by fluorescent microscopy
To investigate whether the antibodies could detect cells infected with authentic filoviruses and not just GP-1,2, Vero or HeLa cells were infected with EBOV, SUDV, TAFV, RESTV, or MARV were stained with M.F88-H3D5, R.GP-S-46-12 or R.GP-S-1 antibodies followed by a fluorescent-labeled secondary antibody (Alexa Fluor 488 goat anti-mouse IgG or by DyLight 488 goat anti-rabbit IgG). Mock-infected cells treated in a similar manner to infected cells, were included as negative controls.. As shown in Fig. 9, fluorescence microscopy analysis revealed that cells infected with EBOV, SUDV, TAFV, and RESTV that were stained with M.F88-H3D5 contained green fluorescent foci characteristic of filovirus infected cells. Similarly, R.SGP-1 and R.SGP-46-12 antibodies stained cells infected with SUDV, TAV, and RESTV. The panel of EBOV infected cells were not analyzed with R.SGP-1 and R.SGP-46-12 antibodies. None of the antibodies detected MARV-infected cells.
Fig. 9. Detection of filovirus-infected cells using immunofluorescence.
As described in Materials and Methods, Vero or Hela cells were exposed to different filoviruses and two days later fixed and stained with the indicated antibodies. HCS CellMask Red was for nuclear/cytoplasmic staining of all cells (shown in red). Cells that are positive for virus staining are shown in green (10X objective).
3.7. Neutralization of retroviral vector pseudotypes bearing ebolavirus GP1,2
Retroviral vector pseudotypes bearing ebolavirus GP1,2 or VSIV G as a specificity control, were used to assess the neautralization potential of the antibodies. R.GP-S-1 neutralized approximately 50% of the SUDV GP1,2 pseudotype at 10 μg/ml and approximately 60% at 25 μg/ml whereas 10 μg/ml of R.F88-2, M.F88-H3D5, M.F88-H3C8, R.GP-S-46-12, or R.GP-S-53-7 did not have a significant effect on the titers of the the EBOV, SUDV, TAFV of BDBV compared to the VSIV G negative control pseudotypes.
4. Discussion
The aim of this study was to raise antibodies with broad cross-reactivity by using antigens that were either highly conserved (F88 peptide) or had highly divergent sequences deleted (Sudan GP with mucin-like domain deleted). The observation that rabbit polyclonal antibodies (R.F88-2 and R.GP-S-1) and mouse monoclonal antibodies (M.F88-H3D5, M.F88-H3C8) could be raised to detect recombinant or Filovirus produced GP1,2 representing all known ebolaviruses in selected assays suggests that this strategy was effective. However, the rabbit monoclonals derived from splenocytes of the GP-S-mucin-deleted-immunized rabbit, showed differences in specificity, in that those are specific only for SUDV GP1,2 (Figs. 5, 6, and 8; Table 1), perhaps due to clonal bias. Interestingly, in a prior study by Yu et al., use of SUDV-GP1,2-derived peptides generated polyclonal antibodies with more restricted ability to detect ebolaviruses (i.e., only SUDV or SUDV plus one other ebolavirus) (Yu et al., 2006). Lucht et al., isolated antibodies with more restricted reactivity when full-length EBOV GP1,2 was used as the immunogen (Lucht et al., 2004), suggesting again the use of the mucin-deleted form of GP1,2 as an improved strategy to induce broadly reactive antibodies.
Table 1.
Reactivity of anti-ebolavirus GP1,2 antibodies assessed by different assays
| Anti-F88 Peptide | Anti-GP-S | ||||
|---|---|---|---|---|---|
| Assay | GP1,2 | Rabbit polyclonal1 | Mouse monoclonals2 | Rabbit polyclonal3 | Rabbit monoclonals4 |
| WB | EBOV | + | + | + | − |
| SUDV | + | + | + | + | |
| TAFV | + | + | + | − | |
| RESTV (sGP) | + | + | + | − | |
| MARV | − | − | − | − | |
|
| |||||
| ELISA | EBOV | + | + | + | − |
| SUDV | + | +/− | + | + | |
| TAFV | + | + | + | − | |
| RESTV (sGP) | + | + | + | − | |
|
| |||||
| FACS | EBOV | + | + | +/− | − |
| SUDV | +/− | +/− | + | + | |
| TAFV | + | + | + | − | |
| BDBV | + | + | + | − | |
R.F88-2
M.F88-H3D5 and M.F88-H3C8
R.GP-S-1
R.GP-S-46-12, R.GP-S-53-4, R.GP-S-53-7 and R.GP-S-55-1
+, positive; −, negative; +/−, barely above background
In the initial analysis, the heavy chain of the rabbit monoclonal R.GP-S-55-1 was difficult to reduce and/or denature (Fig. 4). Even when the concentration of DTT was increased to 100 mM during the preparation of the sample, the heavy chain was observed to form a smear, rather than a distinct band. Scientists at Epitomics, Inc (Burlingame, CA) also observed a similar phenomenon for other rabbit monoclonal antibodies, but regarding light chains, not heavy chains (personal communications). The smear is likely to be derived from the rabbit immunoglobulin heavy chain for three reasons. First, the hybridoma cells were grown in serum-free medium for production and the antibody was purified using protein A beads. Second, the purified antibody can detect SUDV GP1,2 in different assays, which involved the use of horseradish peroxidae/Alexa647-conjugated anti-rabbit IgG. Third, the light chain size was comparable to that of others. Similarly, the bands for R.GP-S-46-12 light chain and R.GP-S-53-7 heavy chain were not sharp either.
Both the rabbit polyclonal and the mouse monoclonal anti-F88 antibodies were generally broadly reactive with the ebolavirus GP1,2 tested with the exception of SUDV GP1,2 (Table 1). In both ELISA (Fig. 6) and FACS (Fig. 8) assays, but not in WB (Fig. 5) assay, binding to SUDV GP1,2 was lower compared to the other ebolavirus GP1,2 tested (EBOV, TAFV, RESTV in ELISA and BDBV, EBOV, and TAFV in FACS). Interestingly, alignment of the F88 38-mer peptide sequences from these five ebolaviruses reveals a striking diversity within the N-terminal eight amino acid residues (Fig. 1). In the case of SUDV GP1,2, three of these eight residues are distinct, whereas the BDBV, EBOV, and TAFV residues are identical and the RESTV peptide differs from those in only a single residue. This observation suggests indirectly that the anti-F88 antibodies may be directed primarily towards the N-terminal region or that this portion of the peptide influences the conformation resulting in a different tertiary conformation of this region in SUDV GP1,2. Notably, the N-terminal region of MARV in this 38 amino acid region is almost completely distinct from the corresponding region in EBOV GP1,2, sharing only a single glycine residue at the N-terminus. Therefore, perhaps not surprisingly, none of the anti-F88 antibodies used could detect MARV GP1,2.
Of the detection assays used, the FACS analysis gave the most variability compared to ELISA and WB. When comparing across the different antibodies, there is sufficient data to suggest that each of the GP1,2 was expressed at comparable levels based on the percentage of positive cells detected. While 5% of pVR1012-SudanGP-transfected cells were detected as positive when evaluated using the anti-F88 panel of antibodies, 25% or more of the same cells were detected as positive when evaluated with the anti-GP-S panel of antibodies, demonstrating that the reduced detection level with the anti-F88 antibodies was due to reduced reactivity as opposed to poor expression levels (Fig. 8). Reduced detection of the cells expressing EBOV and BDBVGP1,2 by the rabbit polyclonal anti-GP-S antibody (R.GP-S-1) was also due to reduced reactivity to these native proteins (Fig. 8). Notably, in the sandwich ELISA or WB assays, detection of EBOV and BDBV GP1,2 by this same polyclonal antibody was not decreased, suggesting that the epitopes that this polyclonal antibody recognizes are presented in a different manner in the ELISA or WB assay compared to cell surface-expressed GP1,2. The ability of the R.GP-S-1, R-GP-S-46-12, and M.F88-H3D5 antibodies to recognize authentic ebolavirus-infected cells by immunofluorescence assay supports the assertion that these antibodies recognize native GP1,2.
R.GP-S-1 neutralized weakly SUDV GP1,2-pseudotyped MoMLV, whereas none of the other antibodies demonstrated neutralization activity against the tested pseudotypes (Fig. 10). Of note, the affinity study of the two monoclonal anti-F88 antibodies exhibited a fairly weak binding to EBOV GP-Fc with KD values greater than 100 nM (148 nM and 361 nM for H3C8 and H3D5, respectively). This low affinity to native GP1,2 may provide one explanation for the lack of neutralization activity of these antibodies.
Fig. 10. Neutralization of gammaretroviral pseudotypes bearing various ebolavirus GP1,2.
MoMLV pseudotypes bearing ebolavirus GP1,2 of indicated strain (x axis) or the G protein of VSIV were pre-incubated with or without anti-GP1,2 antibodies at the indicated concentrations (μg/ml), as indicated, in the presence of 8 μg/ml polybrene. Forty-eight h post-exposure to the antibody-vector mixtures, Vero E6 cells were stained for LacZ activity. Blue-forming units were counted and normalized to 100% for the control that had no antibody. The average ± standard deviation of triplicate samples is shown.
All anti-F88 antibodies (R.F88-2, M.F88-H3D5, and M.F88-H3C8) could detect RESTV sGP by WB (Fig. 5). Given that the F88 peptide sequence is identical in both sGP and full-length GP1,2 and that these antibodies could detect EBOV, SUDV and TAFV full-length GP1,2, these antibodies should detect the sGP of EBOV, SUDV and TAFV as well. The ability to detect sGP is significant as sGP accounts for about 80% of the ebolavirus GP gene transcripts in natural infections. Consequently, these cross-reactive antibodies may be useful for detection and diagnosis of ebolavirus infections (Sanchez et al., 1996; Volchkova et al., 1998). While the polyclonal R,GP-S-1 was also able to detect RESTV sGP by WB, it’s notable that the rabbit anti-GP-S monoclonal antibodies (R.GP-S-46-12, 53-4, 53-7, 55-1) were specific for SUDV GP1,2 only. The specificity of the rabbit monoclonals may provide useful reagents for ebolavirus differentiation with respect to SUDV only (Figs. 5, 6 and 8). The ELISA results with sera from mice injected with mouse-adapted Ebola virus demonstrated that the R.GP-S-46.12 antibody could not detect this virus specifically. However, analysis using either the R.GP-S polyclonal or the M.F88-H3D5 antibodies yielded specific responses above levels detected for control sera. These results support the further development of these antibodies in combinations as described here. Alternatively, pairing with other antibodies proven useful for detection of ebolaviruses by ELISA (Lucht et al., 2004) (Yu et al., 2006) may provide tools for detection and potentially differential diagnosis of ebolavirus infections in animal studies and/or clinical samples.
In summary, a panel of cross-reactive or ebolavirus-specific rabbit and mouse antibodies was developed and analyzed in a variety of different assay formats. The development of antibodies with similar specificity but different host origins may allow for more versatile pairing in the development of diagnostic assays, such as sandwich ELISA, using one for capture and the other one for detection. Using such sandwich ELISA for detecting ebolavirus GP1,2, may allow for increased assay specificity and sensitivity. In addition, these antibodies could also be useful for research of GP1,2 structure and function.
Acknowledgments
We are grateful to Seton Pariser, Bertram W. Maidment III, and Evan Ewers for their technical assistance. This research was partially supported by CBER/NIAID Interagency Agreement (IAG) and the Biomedical Advanced Research and Development Authority (BARDA). This research was partially supported by the Joint Science and Technology Office Transformational Medical Technologies (proposal # TMTI0048_09_RD_T) and Defense Threat Reduction Agency (proposal #4.10022_08_RD_B) to SB. JHK performed this work as an employee of Tunnell Consulting, Inc., a subcontractor to Battelle Memorial Institute under its prime contract with NIAID, under Contract No. HHSN272200200016I. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the US Army, the US Department of Health and Human Services or of the institutions and companies affiliated with the authors. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Dolnik O, Kolesnikova L, Becker S. Filoviruses: Interactions with the host cell. Cell Mol Life Sci. 2008;65:756–76. doi: 10.1007/s00018-007-7406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H, Jones S, Klenk HD, Schnittler HJ. Ebola virus: from discovery to vaccine. Nat Rev Immunol. 2003;3:677–85. doi: 10.1038/nri1154. [DOI] [PubMed] [Google Scholar]
- Feldmann H, Volchkov VE, Volchkova VA, Klenk HD. The glycoproteins of Marburg and Ebola virus and their potential roles in pathogenesis. Arch Virol Suppl. 1999;15:159–69. doi: 10.1007/978-3-7091-6425-9_11. [DOI] [PubMed] [Google Scholar]
- Gupta M, Mahanty S, Bray M, Ahmed R, Rollin PE. Passive transfer of antibodies protects immunocompetent and imunodeficient mice against lethal Ebola virus infection without complete inhibition of viral replication. J Virol. 2001;75:4649–54. doi: 10.1128/JVI.75.10.4649-4654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Gu B, Wu R, Zhang J, Li Y, Zhang M. Development of a rabbit monoclonal antibody group against Smads and immunocytochemical study of human and mouse embryonic stem cells. Hybridoma (Larchmt) 2007;26:387–91. doi: 10.1089/hyb.2007.0517. [DOI] [PubMed] [Google Scholar]
- Jahrling PB, Geisbert J, Swearengen JR, Jaax GP, Lewis T, Huggins JW, Schmidt JJ, LeDuc JW, Peters CJ. Passive immunization of Ebola virus-infected cynomolgus monkeys with immunoglobulin from hyperimmune horses. Arch Virol Suppl. 1996;11:135–40. doi: 10.1007/978-3-7091-7482-1_12. [DOI] [PubMed] [Google Scholar]
- Konduru K, Bradfute SB, Jacques J, Manangeeswaran M, Nakamura S, Morshed S, Wood SC, Bavari S, Kaplan GG. Ebola virus glycoprotein Fc fusion protein confers protection against lethal challenge in vaccinated mice. Vaccine. 2011 doi: 10.1016/j.vaccine.2011.01.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaleva ES, O’Connell KP, Buckley P, Liu Z, Davis DC. Recombinant protein production in insect larvae: host choice, tissue distribution, and heterologous gene instability. Biotechnol Lett. 2009;31:381–6. doi: 10.1007/s10529-008-9883-2. [DOI] [PubMed] [Google Scholar]
- Kuhn JH, Becker S, Ebihara H, Geisbert TW, Johnson KM, Kawaoka Y, Lipkin WI, Negredo AI, Netesov SV, Nichol ST, Palacios G, Peters CJ, Tenorio A, Volchkov VE, Jahrling PB. Proposal for a revised taxonomy of the family Filoviridae - classification, names of taxa and viruses, and virus abbreviations. Arch Virol. 2010;155:2083–2103. doi: 10.1007/s00705-010-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JH, Radoshitzky SR, Guth AC, Warfield KL, Li W, Vincent MJ, Towner JS, Nichol ST, Bavari S, Choe H, Aman MJ, Farzan M. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J Biol Chem. 2006;281:15951–8. doi: 10.1074/jbc.M601796200. [DOI] [PubMed] [Google Scholar]
- Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–82. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucht A, Grunow R, Otterbein C, Moller P, Feldmann H, Becker S. Production of monoclonal antibodies and development of an antigen capture ELISA directed against the envelope glycoprotein GP of Ebola virus. Med Microbiol Immunol. 2004;193:181–7. doi: 10.1007/s00430-003-0204-z. [DOI] [PubMed] [Google Scholar]
- Mpanju OM, Towner JS, Dover JE, Nichol ST, Wilson CA. Identification of two amino acid residues on Ebola virus glycoprotein 1 critical for cell entry. Virus Res. 2006;121:205–14. doi: 10.1016/j.virusres.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Ou W, King H, Delisle J, Shi D, Wilson CA. Phenylalanines at positions 88 and 159 of Ebolavirus envelope glycoprotein differentially impact envelope function. Virology. 2010;396:135–42. doi: 10.1016/j.virol.2009.10.028. [DOI] [PubMed] [Google Scholar]
- Parren PW, Geisbert TW, Maruyama T, Jahrling PB, Burton DR. Pre- and postexposure prophylaxis of Ebola virus infection in an animal model by passive transfer of a neutralizing human antibody. J Virol. 2002;76:6408–12. doi: 10.1128/JVI.76.12.6408-6412.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc Natl Acad Sci U S A. 1996;93:3602–7. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahhosseini S, Das D, Qiu X, Feldmann H, Jones SM, Suresh MR. Production and characterization of monoclonal antibodies against different epitopes of Ebola virus antigens. J Virol Methods. 2007;143:29–37. doi: 10.1016/j.jviromet.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Takada A, Ebihara H, Jones S, Feldmann H, Kawaoka Y. Protective efficacy of neutralizing antibodies against Ebola virus infection. Vaccine. 2007;25:993–9. doi: 10.1016/j.vaccine.2006.09.076. [DOI] [PubMed] [Google Scholar]
- Towner JS, Sealy TK, Khristova ML, Albarino CG, Conlan S, Reeder SA, Quan PL, Lipkin WI, Downing R, Tappero JW, Okware S, Lutwama J, Bakamutumaho B, Kayiwa J, Comer JA, Rollin PE, Ksiazek TG, Nichol ST. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4:e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volchkov VE. Processing of the Ebola virus glycoprotein. Curr Top Microbiol Immunol. 1999;235:35–47. doi: 10.1007/978-3-642-59949-1_3. [DOI] [PubMed] [Google Scholar]
- Volchkov VE, Becker S, Volchkova VA, Ternovoj VA, Kotov AN, Netesov SV, Klenk HD. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology. 1995;214:421–30. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- Volchkova VA, Feldmann H, Klenk HD, Volchkov VE. The nonstructural small glycoprotein sGP of Ebola virus is secreted as an antiparallel-orientated homodimer. Virology. 1998;250:408–14. doi: 10.1006/viro.1998.9389. [DOI] [PubMed] [Google Scholar]
- Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL, Hart MK. Epitopes involved in antibody-mediated protection from Ebola virus. Science. 2000;287:1664–6. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- Yang Z, Delgado R, Xu L, Todd RF, Nabel EG, Sanchez A, Nabel GJ. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science. 1998;279:1034–7. doi: 10.1126/science.279.5353.1034. [DOI] [PubMed] [Google Scholar]
- Yu JS, Liao HX, Gerdon AE, Huffman B, Scearce RM, McAdams M, Alam SM, Popernack PM, Sullivan NJ, Wright D, Cliffel DE, Nabel GJ, Haynes BF. Detection of Ebola virus envelope using monoclonal and polyclonal antibodies in ELISA, surface plasmon resonance and a quartz crystal microbalance immunosensor. J Virol Methods. 2006;137:219–28. doi: 10.1016/j.jviromet.2006.06.014. [DOI] [PubMed] [Google Scholar]