Abstract
Epigenetics is the study of the transmission of cell memory through mitosis or meiosis that is not based on the DNA sequence. At the molecular level the epigenetic memory of a cell is embedded in DNA methylation, histone post-translational modifications, RNA interference and histone isoform variation. There is a tight link between histone post-translational modifications (the histone code) and DNA methylation, as modifications of histones contribute to the establishment of DNA methylation patterns and vice versa. Interestingly, proteins have recently been identified that can simultaneously read both methylated DNA and the histone code. UHRF1 ful-fills these requirements by having unique structural domains that allow concurrent recognition of histone modifications and methylated DNA. Herein, we review our current knowledge of UHRF1 and discuss how this protein ensures the link between histone marks and DNA methylation. Understanding the molecular functions of this protein may reveal the physiological relevance of the linkage between these layers of epigenetic marks.
Keywords: DNMT1, epigenetic code inheritance, hemi-methylated DNA, histones, SRA domain, tudor domain, UHRF1
What is Epigenetics?
The term epigenetics derives from the Greek prefix “epi” which signifies “above” or “in addition to”, associated to the word “genetics”. The broadest definition of epigenetics is the study of the transmission and perpetuation of information through mitosis or meiosis that is not based on the DNA sequence.1–3 In other words epigenetics corresponds to the study of inherited phenotypic variations that are not caused by variations in the DNA sequence.2,3 At the molecular level, epigenetics includes the study of the gene expression regulation through DNA methylation, histone post-translational modifications and RNA interference.3–6 These layers of DNA modification result in a kind of sequence (an epigenomic sequence) that lies above the DNA substrate and is also inherited and susceptible to variation. The epigenome is the sum of both the chromatin structure and the DNA methylation patterns resulting from an interaction between the genome and the environment.1 The genomic and epigenomic sequences, together, determine when genes are expressed and by how much and provide a form of cell memory for the maintenance of cellular functions.3,7
The Reasons Why the Epigenetic Information must be Inherited and Replicated
Except during development, when a cell divides, there is no other goal for the mother cell than to generate two identical cells, i.e. with the same cell phenotype, that are called the daughter cells. This is true in adults when differentiated cells need to proliferate following injury or renewal of specific cells, e.g. cells of the hematopoietic lineages, but during development this rule does not apply since early embryos can reset epigenetic marks.8–10 The doubling of differentiated cells is ensured by the ability of the daughter cells to faithfully inherit the epigenetic code, i.e. DNA methylation patterns, histone code and histone variants at the right place, without any loss of epigenetic marks.2,11,12 Cellular memories define both specific cell lineages and cell types in which epigenetic marks are varying, giving rise to an epigenome that is specific for each cell type. A recent study showed that the epigenomes of H1 (human embryonic stem cells) and IMR90 (fetal lung fibroblasts) are quite different in terms of DNA methylation with 82.7% and 67.7% of all CpG being methylated in H1 and IMR90 cells, respectively.13
The cell phenotype results from gene expression patterns that are governed by epigenetic events. As a consequence, these epigenetic events must follow several rules. The first regards memory; an epigenetic template is copied in comparison with the DNA template. A second regards fidelity; information is transmitted to daughter cells without error or with silenced errors.
Duplication of the DNA Methylation Patterns
DNA methylation patterns regulate tissue-specific expression of genes and chromatin state via different mechanisms dependent on the developmental stage of the cell.5,14–16 In differentiated cells, DNA is methylated only on cytosine that are 5′ of guanines, i.e. in CpG dinucleotides. In stem cells methylated cytosine can also be found at CHG and CHH trinucleotides (H represents A, T or C).13 In differentiated cells, DNA methylation is symmetric, i.e. occurs on both DNA strands; this represents the basis for mitotic duplication. In vertebrates, approximately 80% of all CpG dinucleotides are subject to methylation; exceptions are “CpG islands”, which correspond to short regions of DNA with elevated density of CpG dinucleotides. That these CpG islands are often found at promoters has provided a possible etiology of cancer.17 For example, promoters of tumor suppressor genes in cancer cells are frequently hyper-methylated and transcriptionally silent with subsequent unpaired apoptosis.17 Also, sites that vary normally in tissue differentiation are hot-spots of differential DNA methylation in cancer cells. These sites have been termed “CpG island shores” and have been found up to 2 kbp from the proximal promoter. It has been proposed that methylation of these shores is responsible for the expression of oncogenes and is consistent with an epigenetic progenitor model of cancer: epigenetic alterations affecting tissue-specific differentiation are the predominant mechanism by which epigenetic changes cause cancer.18
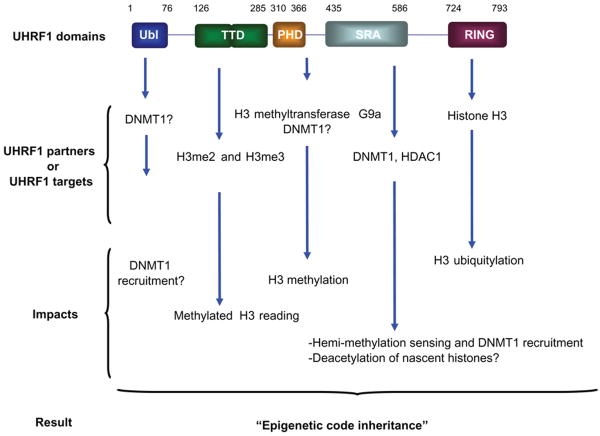
As with lower species the human set of DNA methyl transferases (DNMT1, and to a lesser extent DNMT3A and DNMT3B) likely catalyzes replication of the parental-strand pattern onto the newly synthesized strand via a semi-conservative mechanism.19–22 With only a few exceptions,16 which will not be discussed here, DNA methylation is an irreversible process; once methylated, CpG dinucleotides are stable. If maintenance methylation is not conducted on the newly-synthesized strand, the methyl mark will be diluted in subsequent cycles of cell division. In differentiated cells, because both daughter DNA strands exhibit the same DNA methylation patterns after DNA duplication, DNA duplication likely acts in concert with maintenance DNA methylation. A major scientific challenge is to understand how epigenetic marks are propagated during cell division. Major breakthroughs have been achieved, especially regarding the role and mechanism of action of UHRF1 (Ubiquitin-like PHD RING Finger 1). Although containing multiple domains (Fig. 1), UHRF1's singularity is conferred by its SRA domain,23,24 a 150–170 amino-acid long domain characterized by the conservation of up to 13 evenly spaced glycine residues and a VRV(I/V)RG motif.
Figure 1.
Role of the structural domains of UHRF1. The targets and the partners of each domain of UHRF1 are indicated. when a question mark is used, it means that it still requires further data to support this evidence.
Abbreviations: UBL, ubiquitin-like domain; TTD, tandem tudor domain; PHD, plant homeo domain; SRA, set and ring associated; RING, really interesting new gene; DNMT1, dna methyltransferase 1; HDAC1, Histone DeAcetylase 1.
UHRF1 exhibits affinity for hemi-methylated DNA through the SRA domain25–28 and is thought to be required by DNMT1 for proper localization.25 To our knowledge the only human proteins known to have preference for hemi-methylated DNA on their own, are UHRF1 and DNMT1.25–28
UHRF1 interacts with DNMT1 (see Fig. 1), but the mechanism remains unclear. We have observed that the human SRA domain of UHRF1 interacts with a poorly-characterized region of DNMT1 encompassing AA215 to AA415 of DNMT1;29 others found that the PHD or the UBL domain of mouse UHRF1 is involved in DNMT1 interaction.25,30 Three regions of DNMT1 have been shown to be implicated, namely amino-acid residues from 1–446, 1081–140825 and 401–615.29 Although direct interaction of UHRF1 and DNMT1 is incontestable, the reason for these discrepancies is not yet clear. Species-specific differences might be one. The possibility that during the catalytic reaction of DNA methylation, UHRF1 and DNMT1 interacts with different domains, may be another one.
It was proposed that UHRF1 is a fidelity factor of DNMT1. The SRA-DNA interaction may serve as an anchor to keep UHRF1 at a hemi-methylated CpG site where it recruits DNMT1 for DNA methylation maintenance.30 According to structures of the SRA-DNA complex, it seems unlikely that, in the presence of DNA bound UHRF1, DNMT1 catalyzes its reaction on the unmethylated target cytosine that is specifically recognized by the NRK finger of the SRA domain. An attractive model would have the UHRF1 to slide along the DNA in order to detect the hemimethylated CpG and once it had, a “message” would be transmitted to DNMT1 through a yet unidentified mechanism. Indeed, it has been suggested that UHRF1 might slide along the DNA,28 and that this sliding is hindered by the base flipping mechanism mediated by the SRA domain.26–28 Thus, when DNMT1 enters into action, UHRF1 has dissociated from its CpG site. Possibly, different sets of interactions between UHRF1 and DNMT1 come into play depending on who is bound to the hemimethylated CpG and would be consistent with studies showing different domains of DNMT1 associate with either PHD or SRA domains of UHRF1.25,29,31 This hypothesis is compatible with the model proposed by Arita and co-workers.26 It remains to be investigated what domains of UHRF1 and DNMT1 interact with each other in the presence or absence of hemi-methylated and fully methylated DNA.
Consistent with the “Epigenetic Code REplication Machinery” (ECREM) hypothesis,4 which proposes the existence of a machinery that duplicates epigen-etic marks, DNMT1 and UHRF1 could be localized in a single macro-molecular complex in a constitutive manner. Moreover, the fact that these two proteins readily co-immunoprecipitate25,29 strongly suggests that DNMT1 and UHRF1 do not dissociate from each other during the catalytic cycle. We further believe that the UHRF1 and DNMT1 macro-molecular complex moves along DNA during epigenetic code replication at DNA replication forks. UHRF1 and DNMT1 likely use different interacting domains with each other and with other partners in a spatio-temporal fashion that may be determined by whether UHRF1 is anchored or not to a flipped methylated cytosine.
A Link between Histone H3 Methylation and Cytosine Methylation, Ensured by UHRF1, may be the Basis of the Primary Events for Epigenetic Code Replication
When compared with DNA methylation, much less is known regarding the replication and inheritance of histone post-translational modifications. At least 150 different modifications are known, the most being ensured by specific enzymes.4 Of note not all modifications are conserved through mitosis. Inherited marks are special in that they require binding by specific proteins in a manner that is stable through mitosis, i.e. remain bound on mitotic chromosomes. Histone H3 methylation is one of those marks and because it is intimately related with DNA methylation will be the focus of this section.
DNA methylation and histone modifications act together to influence chromatin structure and gene expression.30,32–36 This synergy is most evident in gene silencing, which is associated with H3 hypo-acetylation, H3K9 methylation, and DNA methylation. Kinetically, it was observed that these events occur in the listed order,37 and that DNA methylation, being a late event, may serve to irrevocably silence genes.38 UHRF1 has been implicated in all of these events4,24,25,39–43 and likely coordinates a macro-molecular complex able to serially catalyze all of these events, i.e. histone deacetylation (via HDAC1), histone methylation (via G9) and DNA methylation (via DNMT1). A better understanding of the structure of this complex particularly in relation to the replication fork will help not only reveal mechanisms of epigenetic replication but also evaluate the current chronology model of epigenetic mark inheritance.
The importance of UHRF1 in coordinating H3 methylation and DNA methylation was most convincingly revealed recently by the serendipitous discovery of a cryptic Tandem Tudor Domain (TTD) in UHRF1 that specifically and tightly bind H3K9me2 or H3K9me3.44 In one scenario, UHRF1 may be laying down DNA methylation only where histones are modified with H3K9me2/3. Thus, with TTD and SRA domains, UHRF1 guides its associated epigenetic enzymes to maintain a relationship between histone methylation and DNA methylation. A better understanding of the biological role and molecular functions of UHRF1 will reveal the details of the relationship between the histone code and DNA methylation.
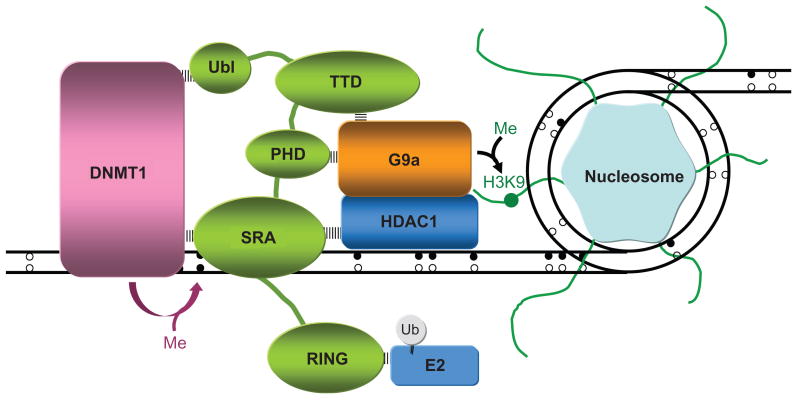
A central question in the epigenetic field is: how are histone marks replicated? Recently, UHRF1 and G9a (a histone methyltransferase that catalyses the synthesis of di- and tri- methylated H3 at lysine 9) have been found in the same macro-molecular complex.45 Perhaps, UHRF1 reads H3K9me2/3 in the parental histone molecule and recruits or directs G9a to write H3K9me2/3 in another, newly-deposited histone. This mechanism would be consistent with the basic features of replicative machinery, namely that they can read a specific code and can write that same code. Here, we propose a model in which UHRF1, localized at the interface of DNA and histones (Fig. 2), senses the presence of hemi-methylated DNA and H3K9 methylation via the SRA and the TTD domains, respectively and signals its enzymatic partners such as DNMT1, HDAC1, and G9a to exert their catalytic activities on the newly synthesized DNA strand and on newly assembled histones. This model is the first to be proposed for the histone code replication and thus inheritance.
Figure 2.
Hypothetical model of the spatial organization of UHRF1 at the interface between DNA and histones. The interactions of the domains of UHRF1 with different partners are illustrated. Unmethylated and methylated cytosines are symbolized by open circles and filled circles, respectively. Methyl groups are represented by “Me” and lysine 9 of histone H3 by “H3K9”. E2 is an Ubiquitin-conjugating enzyme that interacts with the RING domain of UHRF1.
If the enzymes act on the same nucleosome that is being read, this would fit a semi-conservative model of the epigenetic code inheritance.4 If the TTD and SRA domains bind their ligands simultaneously, this would suggest UHRF1 serves as a guide for G9a and DNMT1 to localize their targets at the same time. This is consistent with a report showing that G9a and DNMT1 are in the same complex.46 Another intriguing question is what is the role of the ubiquitylation activity of UHRF1?47 Does UHRF1 preferentially ubiquitylate histone H3 in its methylated form or unmethylated form? The answer to this question may reveal a link between histone ubiquitylation, DNA methylation and histone H3 methylation. An interesting analogy is that of NSPc1-mediated H2A ubiquitylation and DNA methylation, both being directed by EZH2 and are interdependent in long-term target gene silencing within cancer cells.48
In summary, we propose that a part of UHRF1 is working at the histone interface and that another part is working at the DNA interface (Fig. 2), a model that would be compatible with a role at the replication fork.
Epigenetic code Inheritance by UHRF1: Always Faithful?
In lower organisms, where cell division occurs at a high rate and phenotypic variance is undetectable, duplication of the epigenetic code likely approaches perfection. In multicellular organisms, where a single pluripotent cell generates many cell types, the problem is much more complex. Development is, by definition, epigenetic.16 Multicellular organisms have evolved to modify the epigenetic code (not the underlying DNA code) as the basis of cellular differentiation. Considering that changes in the DNA sequence can be ruled out, it is likely that during development, the epigenetic marks inherited by the daughter cells are not identical to those of the mother cells. It is likely that changes from one generation to the next are hardly perceptible but over a range of divisions becomes visible.
An intriguing possibility is that UHRF1 plays an important role in development to determine which loci will be maintained and inherited and which loci will not. With this power, UHRF1 may direct/control/regulate development of multicellular organisms. Once differentiated, cells must maintain their specific epigenetic code; faithful maintenance of methylation patterns is then essential for their survival. Multifunctional UHRF1 in differentiated cells plays this critical maintenance role.
Interestingly, alteration of the epigenetic code is the basis of many diseases. For example, disruption of DNA methylation patterns in imprinted genes is linked to carcinogenesis.49 Altered epigenetic mechanisms have been observed in neurological disorders such as Alzheimer's disease, Huntington's disease, Rett Syndrome, Fragile X syndrome, Rubinstein-Taybi Syndrome, in psychiatric disorders such as schizophrenia, addiction and depression and in disease susceptibility.50 Among them, obesity, glucose intolerance and especially cancer are the most prominent.
Hemi-methylated CpG sites were seen in both carcinomas and controls but, importantly, in carcinoma DNA molecules, they were significantly more likely to occur in clusters displaying the same orientation (the same strand methylated).51 A recent study suggests that hemimethylated CpG dyads are intermediates in active demethylation during carcinogenesis and not just due to a failure of maintenance methylation during replicative DNA synthesis.51 However, considering that UHRF1 is able to sense the presence of hemi-methylated DNA and subsequently to recruit DNMT1,25–28 it is surprising to observe that hemi-methylated DNA sites persist in cells considering that: (i) UHRF1 is supposed to detect them with subsequent recruitment of DNMT1; (ii) UHRF1 greatly increases the preference of DNMT1 for hemimeth-ylated DNA and reconciles the faithful transmission of genomic methylation patterns with a small (5- to 30-fold) inherent preference of DNMT1 for hemimethylated DNA; (iii) UHRF1 and DNMT1 are over-expressed in cancer cells, which should reduce the number of hemi-methylated CpG sites. One interesting track is that in cancer cells, altered interactions of UHRF1 and DNMT1 have been observed.52 Indeed, in this study the authors have observed less UHRF1/DNMT1 complexes in cancer cells when compared with non-cancerous cells which could be a plausible mechanism to explain the hypo-methylation status of cancer cells. However, this remains to be confirmed. Consequently, it is possible that the epigenetic code transmission governed by UHRF1 is likely to be unfaithful during pathogenesis. In the same order, it would be interesting to investigate whether altered interactions occur between UHRF1 and histone modifying enzymes, such as G9a and HDAC1. However, so far to our knowledge no alteration in the H3K9 methylation inheritance has yet been reported in cancer cells.
Concluding Remarks
UHRF1 behaves as an epigenetic integration platform involved in the cross-talk between H3K9 methylation and DNA methylation as suggested previously and now confirmed by our recent studies.20,24–30,44 Considering that UHRF1 is able to detect hemi-methylated DNA and hypothetically hemi-methylated histone H3 (i.e. only one histone monomer is methylated), and that UHRF1 recruits and/or interacts with the DNA and histone methylation enzymes, undoubtedly UHRF1 plays a key role in the epigenetic code inheritance. Hence, we propose that UHRF1 is in a spatio-temporal way the first intervening protein to read and to induce the epigenetic code duplication.
Acknowledgments
We apologize to authors whose work could not be cited owing to space limitations. Studies from our laboratories (CB & MM) concerning UHRF1 (ICBP90) have been supported by grants of the Ligue contre le Cancer, Comité du Haut-Rhin, France and by NIH grant CA060999.
Abbreviations
- DNMT1
DNA methyltransferase 1
- HAT
histone Acetyltransferase
- HDAC1
Histone DeAcetylase 1
- RING
Really Interesting New Gene
- SRA
Set and Ring Associated
- TTD
Tandem Tudor Domain
- UHRF1
Ubiquitin-like PHD RING Finger 1
Footnotes
Disclosures: The authors report no conflicts of interest.
Publisher's Disclaimer: This is an open access article. Unrestricted non-commercial use is permitted provided the original work is properly cited.
References
- 1.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–18. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- 2.Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol. 2007;19:266–72. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 4.Bronner C, Chataigneau T, Schini-Kerth VB, Landry Y. The “Epigenetic Code Replication Machinery”, ECREM: a promising drugable target of the epigenetic cell memory. Curr Med Chem. 2007;14:2629–41. doi: 10.2174/092986707782023244. [DOI] [PubMed] [Google Scholar]
- 5.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–94. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corpet A, Almouzni G. A histone code for the DNA damage response in mammalian cells? EMBO J. 2009;28:1828–30. doi: 10.1038/emboj.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–40. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 9.Dean W, Santos F, Reik W. Epigenetic reprogramming in early mammalian development and following somatic nuclear transfer. Semin Cell Dev Biol. 2003;14:93–100. doi: 10.1016/s1084-9521(02)00141-6. [DOI] [PubMed] [Google Scholar]
- 10.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–62. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 11.Nightingale KP, O'Neill LP, Turner BM. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev. 2006;16:125–36. doi: 10.1016/j.gde.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 12.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol. 2009;10:526–37. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 13.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009 Oct 14; doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buschhausen G, Wittig B, Graessmann M, Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci U S A. 1987;84:1177–81. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–43. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 16.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–32. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 17.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeltsch A. On the enzymatic properties of Dnmt1: specificity, processivity, mechanism of linear diffusion and allosteric regulation of the enzyme. Epigenetics. 2006;1:63–6. doi: 10.4161/epi.1.2.2767. [DOI] [PubMed] [Google Scholar]
- 20.Jeltsch A. Reading and writing DNA methylation. Nat Struct Mol Biol. 2008;15:1003–4. doi: 10.1038/nsmb1008-1003. [DOI] [PubMed] [Google Scholar]
- 21.Svedruzić ZM. Mammalian cytosine DNA methyltransferase Dnmt1: enzymatic mechanism, novel mechanism-based inhibitors, and RNA-directed DNA methylation. Curr Med Chem. 2008;15:92–106. doi: 10.2174/092986708783330700. [DOI] [PubMed] [Google Scholar]
- 22.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009 Sep 30; doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mousli M, Hopfner R, Abbady AQ, et al. ICBP90 belongs to a new family of proteins with an expression that is deregulated in cancer cells. Br J Cancer. 2003;89:120–7. doi: 10.1038/sj.bjc.6601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bronner C, Achour M, Arima Y, Chataigneau T, Saya H, Schini-Kerth VB. The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol Ther. 2007;115:419–34. doi: 10.1016/j.pharmthera.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–4. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 26.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–21. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 27.Avvakumov GV, Walker JR, Xue S, et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–5. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 28.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–9. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Achour M, Jacq X, Rondé P, et al. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene. 2008;27:2187–97. doi: 10.1038/sj.onc.1210855. [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto H, Horton JR, Zhang X, Cheng X. UHRF1, a modular multidomain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications. Epigenetics. 2009;4:8–14. doi: 10.4161/epi.4.1.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meilinger D, Fellinger K, Bultmann S, et al. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep. 2009 Oct 2; doi: 10.1038/embor.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, Feil R. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J. 2007;26:720–9. doi: 10.1038/sj.emboj.7601513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Epsztejn-Litman S, Feldman N, Abu-Remaileh M, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–83. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikegami K, Iwatani M, Suzuki M, et al. Genome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cells. Genes Cells. 2007;12:1–11. doi: 10.1111/j.1365-2443.2006.01029.x. [DOI] [PubMed] [Google Scholar]
- 35.Stancheva I. Caught in conspiracy: cooperation between DNA methylation and histone H3K9 methylation in the establishment and maintenance of heterochromatin. Biochem Cell Biol. 2005;83:385–95. doi: 10.1139/o05-043. [DOI] [PubMed] [Google Scholar]
- 36.Vaissière T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 2008;659:40–8. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Mutskov V, Felsenfeld G. Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J. 2004;23:138–49. doi: 10.1038/sj.emboj.7600013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 39.Arima Y, Hirota T, Bronner C, et al. Down-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transition. Genes Cells. 2004;9:131–42. doi: 10.1111/j.1356-9597.2004.00710.x. [DOI] [PubMed] [Google Scholar]
- 40.Macaluso M, Montanari M, Noto PB, Gregorio V, Bronner C, Giordano A. Epigenetic modulation of estrogen receptor-alpha by pRb family proteins: a novel mechanism in breast cancer. Cancer Res. 2007;67:7731–7. doi: 10.1158/0008-5472.CAN-07-1476. [DOI] [PubMed] [Google Scholar]
- 41.Hopfner R, Mousli M, Jeltsch JM, et al. ICBP90 a novel human CCAAT binding protein involved in the regulation of topoisomerase IIα expression. Cancer Res. 2000;60:121–8. [PubMed] [Google Scholar]
- 42.Jeanblanc M, Mousli M, Hopfner R, et al. The retinoblastoma gene and its product are targeted by ICBP90: a key mechanism in the G1/S transition during the cell cycle. Oncogene. 2005;24:7337–45. doi: 10.1038/sj.onc.1208878. [DOI] [PubMed] [Google Scholar]
- 43.Unoki M, Nishidate T, Nakamura Y. ICBP90 an E2F-1 target recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–10. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 44.Walker JR, Avvakumov GV, Nady N, et al. A cryptic Tandem Tudor Domain of UHRF1 mediates crosstalk between DNA methylation and the chromatin epigenetic codes. Submitted. [Google Scholar]
- 45.Kim JK, Estève PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Estève PO, Chin HG, Smallwood A, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Citterio E, Papait R, Nicassio F, et al. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol. 2004;24:2526–35. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu X, Gong Y, Yue J, Qiang B, Yuan J, Peng X. Cooperation between EZH2, NSPc1-mediated histone H2A ubiquitination and Dnmt1 in HOX gene silencing. Nucleic Acids Res. 2008;36:3590–9. doi: 10.1093/nar/gkn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ooi L, Wood IC. Regulation of gene expression in the nervous system. Biochem J. 2008;414:327–41. doi: 10.1042/BJ20080963. [DOI] [PubMed] [Google Scholar]
- 50.Gräff J, Mansuy IM. Epigenetic codes in cognition and behaviour. Behav Brain Res. 2008;192:70–87. doi: 10.1016/j.bbr.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 51.Shao C, Lacey M, Dubeau L, Ehrlich M. Hemimethylation footprints of DNA demethylation in cancer. Epigenetics. 2009;4:165–75. doi: 10.4161/epi.4.3.8277. [DOI] [PubMed] [Google Scholar]
- 52.Hervouet E, Lalier L, Debien E, et al. Tumor induction by disruption of the Dnmt1, PCNA and UHRF1 interactions. Nature Precedings. hdl:10101/npre.2008.2509.1:Posted 13 Nov 2008. [Google Scholar]