Abstract
Discovery of EX1 kinetics in hydrogen exchange (HX) mass spectrometry (MS) experiments is rare. Proteins follow the EX1 kinetic regime when cooperative unfolding events simultaneously expose multiple residues to solvent such that they all become deuterated together before the region is able to refold. A number of factors can contribute to what we call “false EX1” in which it appears as though EX1 occurs in a protein when it probably does not. One of the contributors to false EX1 is peptide carryover between chromatographic runs. In this work, we explore the origins of peptide carryover in HX MS, describe how carryover causes mass spectra to indicate false EX1 kinetics and then describe an optimized washing protocol that can be used to eliminate peptide carryover. A series of solvent injections was developed and found to efficiently eliminate carryover signatures such that analysis of deuterium incorporation could be reliably followed for two proteins prone to high carryover.
Keywords: Peptide carryover, EX1 kinetics, hydrogen exchange, mass spectrometry, deuterium
1. Introduction
In hydrogen exchange mass spectrometry (HX MS), unique spectral signatures are generated depending on whether the protein under investigation displays all EX2 kinetics, all EX1 kinetics or a mixture of the two [1–4]. In EX1 kinetics, a percentage of the molecular population being labeled undergoes an unfolding event such that a number of residues are simultaneously exposed to deuterium in the labeling solution [5–10]. Exchange occurs at all the exposed residues before the exposed region refolds. Therefore, in the mass spectra, there are two isotope distributions (a bimodal pattern): 1) a lower mass distribution that represents the population that did not undergo partial unfolding and was not labeled by deuterium, and 2) a higher mass distribution that represents the population of molecules that did undergo partial unfolding and was exposed to deuterium (see Figure 1a). Because most proteins under native conditions follow EX2 kinetics rather than EX1 kinetics [4–5, 7, 11], discovering a protein or region of a protein that follows EX1 kinetics is a rare event which can reveal important clues concerning protein function. However, other factors can lead to so-called “false EX1” kinetic signatures, or mass spectra in which it appears that EX1 kinetics are occurring in the protein as a natural phenomenon when in reality they are not. When ascribing EX1 kinetics to a protein, it is important to be sure that they are in fact real. Therefore, being aware of these false EX1 patterns and their causes is valuable.
Figure 1.
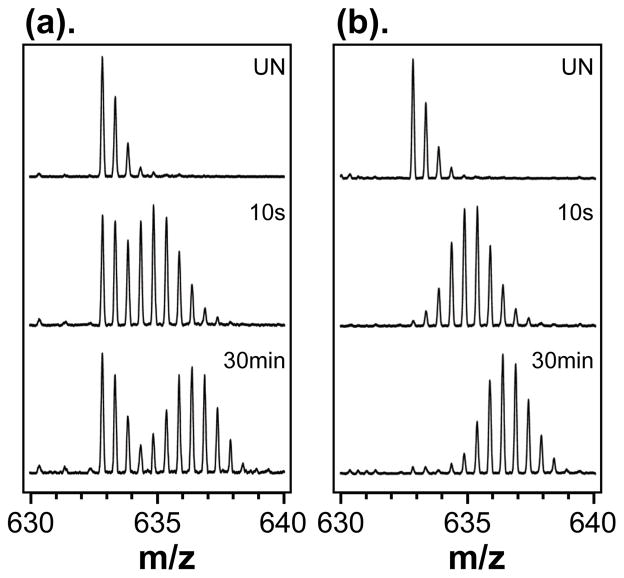
Schematic depicting typical real versus false EX1 signatures in HX MS. Real EX1 (a) involves two populations: a lower mass species (peak 1, red) that has not yet unfolded to become deuterated and a higher mass species (peak 2, green) that has undergone cooperative unfolding and has become deuterated. In False EX1 (b), there are two distributions, but the lower mass distribution (1) is the result of some phenomena other than EX1 kinetic unfolding, including aggregation, abnormal backexchange or sample carryover. If part of the protein population exists as an aggregate, that part of the population may not become deuterated (peak 1, red) while the rest of the population will (peak 2, blue). Abnormal total-backexchange [13] can result when all the deuterium from a portion of the population (peak 1, red) is backexchanged during analysis. False EX1 as a result of carryover occurs when some of the sample does not elute from the LC column, but elutes later, typically with the solvent gradient for the next injected sample. Because the protein from the first injection (indicated here with a *) has been backexchanging longer than that from the second injection (indicated with a ▲), an artifactual lower mass distribution (peak 1, red) appears. Note that * and ▲ represent the same protein/peptides but are indicated here with different symbols to distinguish them in injection 2. This figure is meant to be illustrative and does not address all the types of intensities, mass differences and peaks shapes that are possible.
Three main sources of false EX1 signatures that we have routinely encountered in our laboratory are: aggregation/multimerization, abnormal complete backexchange, and carryover (Figure 1b). In cases of protein aggregation, amyloids for instance [12], the aggregate itself can prevent exchange of deuterium as many sites are protected, protein motions are restricted and it is generally difficult for deuterium to access labile positions in the aggregate. The population of molecules that is not aggregated exchanges normally and produces the higher mass isotope distribution. Another type of false EX1 arises from abnormal complete backexchange, as described previously [13], results from accelerated backexchange that occurs during the online digestion step of the HX MS experiment. This phenomenon causes nearly all peptic peptides to have false EX1 signatures, as shown in the example in Figure 2. In those experiments, and in others, false EX1 kinetics are revealed when all peptides appear to undergo EX1 kinetics, an event that would be exceedingly rare for a protein. Finally, as we illustrate in this paper, peptide carryover between chromatographic runs can also be a source of false EX1 kinetic signatures.
Figure 2.
Comparison of deuterium levels in a dihydrofolate reductase (DHFR) peptic peptide under (a) conditions that caused accelerated backexchange during analysis or (b) conditions that did not cause accelerated backexchange. The peptide shown is: m/z = 632.85 (+2), TTSSVEGKQNLV, DHFR residues 81–92, either undeuterated (UN) or deuterated for 10 seconds or 30 minutes prior to digestion. This Figure reprinted with permission from Ref. [13], Copyright 2006 American Chemical Society.
Carryover is the general phenomenon whereby analyte from one injection is undesirably retained in an LC system such that it elutes in subsequent injections. Carryover can be a common problem in any LC MS system (e.g., Refs. [14–15]) but in HX MS experiments the problem of carryover can be amplified by the fact that chromatographic separations must occur at 0 °C wherein the kinetics of analyte association/dissociation with the stationary phase are slower. The ever-increasing sensitivity of mass spectrometers has probably contributed in some fashion to better detection of carryover, as has variability in chromatographic devices and stationary phases. For example, we have noticed that carryover can become a greater concern in ultra-high pressure separations, those occuring at pressures greater than 8000–8500 psi or so, wherein small diameter particles (1.5–1.7 micron) are utilized for the separation. Perhaps this phenomena is related to the specific type of stationary phase particle engineered to handle such high pressures, but much more experimentation will be required to determine the exact reason behind the phenomenon. The is an additional reason why carryover is more problematic in HX MS experiments than in regular kinds of chromatography. In many chromatographic measurements where carryover could occur, isotopes are not involved and the mass of the carried over species remains constant. However, in HX MS experiments, labile deuterium at backbone amide positions is present, and the mass of ions can change depending on how long they are retained in the chromatographic system (i.e., backexchange of deuterium in a peptide/protein to hydrogen is greater the longer something sticks or carries-over in an LC column). Figure 1b illustrates the phenomena. Deuterated species (protein or peptides) are injected into the system and mass analysis is performed. Upon a second injection, material that was retained from the first injection carries-over and has now been exposed to the 100% H2O environment of the LC system for much longer than the deuterated protein in the second injection. As there has been more time for backexchange to occur, the material from the first injection that then elutes during the gradient in the second injection has a lower mass while the material from the second injection has a higher mass. The result is a bimodal isotope pattern similar to what is observed in real EX1 kinetics.
As part of any HX MS experiment, it is wise to monitor carefully if carryover between injections occurs and then to minimize it as much as possible. We present here a description of the carryover phenomena as it applies to HX MS experiments for two proteins [UmuD and the kinase domain of epidermal growth factor receptor (EGFR)] that have a large number of “sticky” peptides prone to carryover. A solution to the carryover problem is shown which involves multi-solvent washing. By applying a customized washing method, a significant reduction in carryover can be achieved, thereby permitting correct analysis of the HX mass spectra and reliable data interpretation of protein behavior in solution.
2. Experimental
2.1 Chemicals
Solvents used included: Formic acid (FA, Sigma-Aldrich, reagent grade, ≥95%), trifluoroethanol (TFE, Sigma-Aldrich, ≥99.5%), methanol (MeOH, Fisher Scientific, HPLC grade), acetonitrile (ACN, Fisher Scientific, HPLC grade), 2-isopropanol (IPA, Fisher Scientific, HPLC grade), trifluoroacetic acid (TFA, Sigma-Aldrich, reagent grade, ≥95%), dimethyl sulfoxide (DMSO, Sigma-Aldrich, reagent grade, ≥95%) and deuterium oxide (99.9%, Cambridge Isotope Laboratories, Inc.). All other reagents were analytical grade and used without further purification. HPLC grade water was purchased from Fisher Scientific. Porcine pepsin was obtained from Sigma-Aldrich (P6887).
2.2 Sample preparation
UmuD was overexpressed in E. coli BL21(DE3) strain and purified as described previously [16]. EGFR kinase domain was obtained from Prof. M. J. Eck at DFCI/Harvard Medical School, Boston, MA. The purity and mass of both proteins were verified by electrospray mass spectrometry (LCT premier, Waters) and the theoretical masses matched the measured mass to within 0.4 Da. For UmuD, a stock solution (50 μM, monomer concentration) was equilibrated in 100% H2O buffer of 20 mM HEPES, 0.1 mM EDTA, 50 mM NaCl, and 1 mM DTT, pH 7.5. To initiate deuterium exchange, the stock was diluted 17-fold (v/v) with D2O buffer (20 mM HEPES, 0.1 mM EDTA, 50 mM NaCl, and 10 mM DTT, pD 7.5 and exchange allowed to proceed at 25 °C for 30 minutes. Exchange was quenched by lowering the pH to 2.5 with phosphate buffer containing 6M Guanidine HCl, the quenched samples were immediately frozen on dry ice and stored at −80 °C until analysis. For analysis, each labeled and quenched UmuD sample was thawed in ice and pepsin was added at a 1:10 ratio (protein: pepsin) by mass and allowed to incubate for 5 min at 0 °C before injection. EGFR protein was labeled in a similar fashion except the buffer system was 20 mM Tris, 150 mM NaCl, pH 7.4. EGFR was labeled with a 15-fold dilution and incubated for 12 hours at 37 °C, and digested with a protein:pepsin ratio (by mass) of 1:2.
2.3 Chromatography and mass spectrometry
Approximately 20 pmol of peptic digested protein (for both UmuD and EGFR) were injected into a custom nanoACQUITY UPLC system (Waters Corp, Milford, MA) as described previously [17]. The entire system post injector was kept at 1.0 °C. Peptides were trapped with a VanGuard Pre-Column (2.1 mm × 5 mm, ACQUITY UPLC BEH C18, 1.7 μm) for 4 min of desalting at 100 μL/min (essentially no reduction in carryover was seen when a C8 trap was used). The trap was then placed in line with an analytical column (1.0 mm × 100 mm, ACQUITY UPLC BEH C18, 1.7 μm), see Figure 3 inset. A 8–40% gradient of acetonitrile over 6 min at a flow rate of 40 μL/min was used to separate the peptides and elute them into the mass spectrometer. At the end of the gradient, the ACN concentration was held at 40% for 1 minute and increased to 85% and held for 1 minute. The UPLC mobile phases contained 0.05% formic acid for a pH of 2.5 [18]. The volume of the injector system from the mixer to the head of the trap column was ~110 μL, which included an ~100 μL sampling loop.
Figure 3.
Timeline and solvent conditions for UPLC column washing. The solvent programs for the trap column (blue line) and analytical column (red dash line) are shown. Multiple combinations of four solvents (see Table 1) were injected onto the trap and allowed to flow through it for the time indicated. Then two saw-tooth gradients of 8–85% acetonitrile were used to wash the analytical column. The inset shows the plumbing scheme of the UPLC. In our system, pump A+B was switched to 85% ACN prior to the first injection and then brought back down to 8% for component injection. This step is optional, but introduces another wash of the analytical column.
The eluant from the UPLC analytical column was directed into a QTof-Premier mass spectrometer (Waters Corp.) equipped with a standard ESI source. The QTof-premier instrument settings were as follows: 3.5kV cone and 40 V capillary voltages; source and desolvation temperature were 80 and 175 °C, respectively; desolvation gas flow of 600 L/h. All spectra were collected in ESI (+) and V mode. The deuterium levels were not corrected for backexchange during analysis and are therefore reported as relative [19].
2.4 Wash procedure and data processing
After each injection, carryover was checked by running a blank in which the identical solvent and mass acquisition programs were used but 0.1% formic acid in water or quenched sample buffer without protein was injected. When carryover was observed, as described below, a wash was applied to remove peptides between runs. The washing procedure was composed of two steps. In the first step, peptides stuck to the trap column were removed with pure or combinations of several organic solvents, inorganic acids and their mixtures, such as TFE, MeOH, IPA, FA, Mix1 (IPA:MeOH:ACN, 1:1:1 by volume) or Mix2 (1.0% DMSO, 60% ACN, 30% IPA, 8.9% H2O, 0.1% TFA, from Ref. [20]) etc. as described below. Each solvent component was injected, in order (Table 1), as shown in Figure 3. In the second step, peptides that made it past the trap and were stuck to the analytical column were removed. This second step involved performing a saw-tooth (8–85%) gradient of acetonitrile as shown in Figure 3. The flowrate for this gradient was the same as the flowrate for normal separation (40 μL/min).
Table 1.
The components and order of different wash protocols
| Wash Protocol | Components and order |
|---|---|
| 1 | 100% IPA, 80% ACNa, 100% IPA, 80% ACN |
| 2 | 100% MeOH, 80% ACN, 100% MeOH, 80% ACN |
| 3 | Mix1, 80% ACN, Mix1, 80% ACN |
| 4 | Mix2, 80% ACN, Mix2, 80% ACN |
| 5 | 10% FA, 80% ACN, 10% FA, 80% ACN |
| 6 | 100% TFE, 80% ACN, Mix1, 80% ACN |
| 7* | 10% FA, 50% TFE, 80% MeOH, 80% ACN |
| 8 | 5% FA, 50% TFE, 80% MeOH, 80% ACN |
| 9 | 10% FA, 50% MeOH, 80% MeOH, 80% ACN |
any solvent not listed as 100% was made in water, e.g. 80% ACN means 80% ACN, 20% water.
To process the data and quantify carryover, the relative mass spectral intensity (peak height) of each peptide was compared in a blank run performed right after a sample analysis run (no washing) or in a blank run performed with an intermediate washing step (i.e., sample run, washing step, blank). The resulting value reported on the amount of material retained in the system: 0% indicated no peptide was retained and 100% indicated that the intensity of peptides eluted in the blank run was identical to the signal observed in the actual sample analysis run.
3. Results and Discussion
3.1 Possible locations of peptide carryover
Carryover in a chromatographic system can be caused by residual material from a previously injected sample that was either absorbed to, or trapped within the injection system. Material can be trapped in a number of potential sites, including: the injection syringe barrel, void/dead volumes in the system, the rotor seal, the tubing, the tubing connections, and finally on the analytical and/or trapping columns [15, 20–22]. We have found that in a well-controlled UPLC HX MS system, carryover in most of the potential places can be minimized except for carryover occurring on the columns. Column carryover can be very compound-dependent and is related mainly to analyte:stationary phase interactions. Strong interactions between analyte and the stationary phase can prevent peptides from being eluted by the solvent gradient. Not surprisingly, material sticking to the trap column (as sample interacts with the trap before the analytical column) is more prone to carryover to the next run. Depending on the protein under investigation, these “sticky peptides” may be unavoidable and in the case of HX MS experiments, can show up in subsequent runs as false EX1 signatures.
3.2 HX MS analysis of UmuD and EGFR proteins
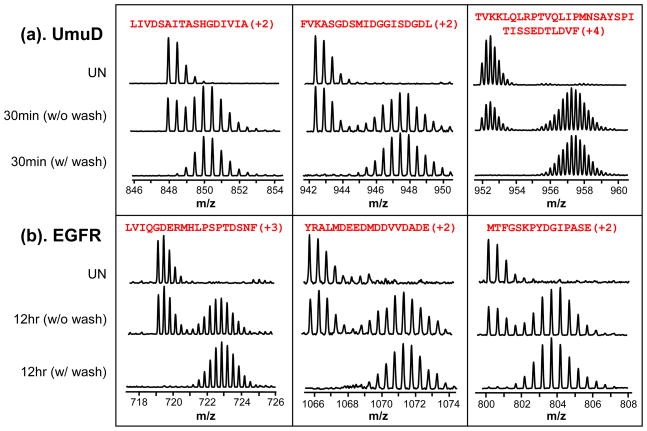
Two proteins with an uncharacteristically high proportion of sticky peptides were chosen to develop the washing protocol. The DNA repair protein UmuD [16] and the epidermal growth factor receptor kinase domain (here called EGFR) both had a number of sticky peptides that produced false EX1 signatures. Figure 4 illustrates typical spectra observed for three representative peptides from each of these proteins. These example peptides represent different sequences and charge states. As shown later in Figures 5 and S1, length did not correlate with stickyness and there was no strong correlation between hydrophobicity of the peptides, from calculations using Bull and Breese parameters for example, with the degree of carryover. In any case, without considering carryover, one may be falsely led to think many of these peptides were undergoing EX1 kinetics as obvious bimodal isotopic distributions were observed for many of the peptides from both proteins (Figure 4a,b, middle row of each panel). Again it should be noted that if EX1 is observed for the majority of peptides in an HX MS experiment, it is very unlikely that these EX1 signatures represent true EX1 kinetic events in the protein. Rather, false EX1, perhaps as a result of carryover, is a much more likely explanation. To determine if sticky peptides are being carried over, blank runs with the identical solvent program should be performed between samples, particularly when starting to work on a new protein for which carryover has not yet been established. Adding a washing step between runs can significantly reduce or eliminate false EX1 signatures (see bottom row of each panel, Figure 4a,b). The choice and manner of washing depends somewhat on the sample but some recommended solvent systems are shown below based on our observations.
Figure 4.
Examples of false EX1 kinetics and effective removal with washing. Mass spectra of three selected peptides each from UmuD (a) and EGFR (b) are shown. Both proteins were analyzed after digesting with pepsin in solution when undeuterated (UN) or after labeling with deuterium for either 30 min (UmuD) or 12 hr (EGFR). Material was injected, a regular HX MS separations gradient was performed (8–40% ACN in 6 minutes) and then another acquisition made of the same sample without an intermediate washing step (middle row of each panel) or with a washing step (bottom row of each panel) using wash protocol 7, Table 1. The bimodal pattern in the sample with no intermediate wash step resembles real EX1 kinetics, but is in fact false (see also Figure 1). The sequence and charge state of each peptide are shown at the top of each panel.
Figure 5.
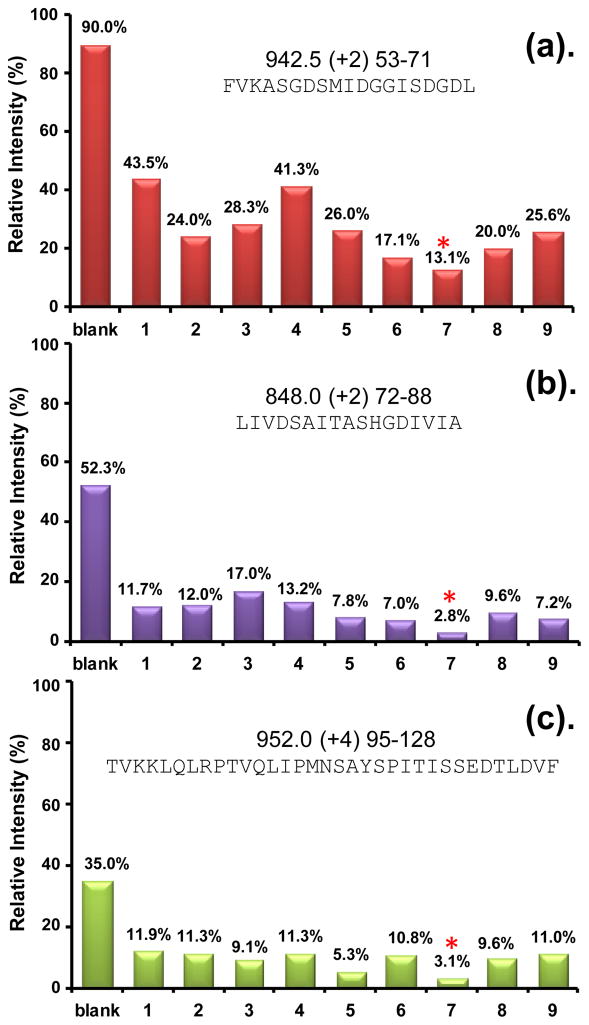
Effectiveness of different wash protocols in eliminating peptide carryover. Three of the most sticky peptides from UmuD are shown (see Figure S1 for other peptides). The m/z, charge state, and sequence of each peptide ion are shown at the top of each graph. The far left column indicates carryover without washing (e.g., a value of 90% means that the intensity of the peptide ion signal in a blank run following a sample run was 90% of the signal intensity in the sample run). Columns 1–9 show the relative intensity of the peptide ion in a blank run after an intermediate washing step using the washing protocols labeled 1–9, respectively, in Table 1. For example, the intensity of the peptide ion in panel (a) fell to 13.1% in a blank run after wash protocol 7 was used immediately post sample run. The most effective wash protocol overall was number 7, indicated with an *.
3.3 Wash method development and optimization
An ideal wash protocol will be as thorough as possible and add as little additional time to the length of an experiment as possible. We strived for maximal efficiency of washing with minimal time consumption. Different types of solvent or solvent mixtures were injected through the injector and trap column. For these experiments (see Figure 3 inset), no online pepsin digestion column [23] was used (see also below) and digestion was done in solution offline. In the system we used, each injection was 100 μL and the flowrate from the C pump was 100 μL/min. Four components were used in sequence (see Figure 3, blue line) in each washing protocol. In terms of timing, in our system, upon the start of the wash program, pumps A and B switched to 85% ACN for 1 minute and returned to 8% when the first wash component was injected. The injected solvent, driven by pump C pumping 0.1% FA in water, flowed over the trap column for 1 minute. The second solvent was loaded, injected, allowed to flow over the trap for another minute etc., until all four solvents had been injected for a total trap washing protocol time of 4 minutes. During trap washing, the analytical column was exposed to 8% ACN. At the end of the trap washing with four components, the analytical column was washed using gradients of acetonitrile (Figure 3, red line). Total time for the entire wash program was 11 minutes. Variations on this timing can readily be envisioned, depending on the system in question.
Several factors were taken into consideration in the choice of the washing solvents: 1) potential power to solubilize or denature sticky peptides, 2) potential to release strong hydrophobic interactions between peptide and column, 3) solvents not detrimental to mass spectrometry analysis later-on (i.e., no detergents), 4) solvents that generally do not harm the lifetime of the separations columns and do not alter separation efficiency, and 5) easy solvents to deal with. Trifluoroethanol (TFE) can effectively solubilize both peptides and proteins and it is sometimes used as a co-solvent in protein folding studies [24–25]. Acetonitrile (70–90%) is empirically known to be a good choice for removing peptides absorbed by hydrophobic interactions in C18 columns, hence its wide use as the organic solvent for HPLC. Asakawa and co-workers [26] tested the effectiveness of 5–80% acetonitrile in removing carryover. As expected, their results showed that a higher percent organic solvent resulted in better washing and 80% ACN was their final choice. As an alternative, a protic solvent, such as methanol or 80–90% methanol, can sometimes be more effective for more polar hydrophobic compounds. Acids, or acidified acetonitrile, isopropanol/methanol/water solutions can also be quite efficient and universally used to dissociate analyte adsorption caused by dipole-dipole and ionic interactions. Low pH can influence the charge state of analytes thereby affecting the efficiency of washing; some charged compounds may more easily dissolve in water solutions while having more affinity towards pure organic solutions in an unchanged state [21]. However, very concentrated acid could damage the stationary phase and high concentrations may increase back-pressure and lead to noisy mass spectra. After considering all these factors, the solvents that were chosen were: TFE, methanol, isopropanol, acetonitrile, formic acid, and two mixtures: Mix1 (IPA:MeOH:ACN, 1:1:1 by volume) and Mix2 (1.0% DMSO, 60% ACN, 30% IPA, 8.9% H2O, 0.1% TFA [20]), see Table 1.
To determine the washing efficiency in experiments with both the small UmuD protein and the much larger EGFR protein, representative sticky peptides of both proteins were selected. Three peptides selected for UmuD were: FVKASGDSMIDGGISDGDL, m/z 942.5, +2 (Figure 5a, red); LIVDSAITASHGDIVIA, m/z 848.0, +2 (Figure 5b, purple); and TVKKLQLRPTVQLIPMNSAYSPITISSEDTLDVF, 952.0, +4 (Figure 5c, green). The EGFR peptides that were studied, as well as others from UmuD that were followed, are shown in Supporting Information Figure S1. Washing efficiency was determined by comparing the amount of carryover of each peptide with washing to that of the same peptide without washing. Because the absolute signal for a given peptide will vary with the concentration of the injection, efficiency of ionization, etc. and may vary from day to day for the same sample depending on multiple factors including the sensitivity of the mass spectrometer, the relative intensity of sticky peptide was used to determine washing efficiency. To determine carryover, the amount of injected sample was fixed at 20 pmol and we measured the peak height of each peptide ion in the sample run and the subsequent blank run(s). The ratio of peak heights represents the amount of carryover, or stickiness, as shown in the far left column of each peptide in Figure 5 (the error of determining the intensity for n=3 is shown in Supplementary Figure S2). A value of 90% means that 90% of the signal remained in the subsequent blank injection. We made the same intensity measurement after each of the washing protocols shown in Table 1 was applied and compared the effectiveness of the different washing protocols.
The injection order of each component was optimized. The first and third wash components of each wash protocol were varied, such as 100% MeOH, IPA etc. whereas 80% acetonitrile was consistently used as the second and fourth component to both remove sticky peptides as well as all other previous washing solvent(s) that may have remained in the injection/trapping system. Ten percent formic acid was chosen as the best acid for washing, although 5% FA was tested (Wash 8) but not found to be as effective as 10% FA. While formic acid was efficient in removing sticky peptides, it introduced too many inorganic ions into subsequent chromatographic and mass spectrometry steps and caused a much higher back pressure in UPLC. To reduce this problem, we washed the FA away with another solvent (Wash 5,7,8,9). We found 50% TFE was optimal as the beneficial peptide removal aspects of TFE were preserved while also efficiently removing FA from the prior injection. When 100% TFE was used as the first component (wash 6), peptide mass spectral intensity was dramatically reduced in later runs. It was important to remove residual TFE from the injection path and from the stationary phase prior to a new HX MS sample being injected. We therefore used 50% TFE instead. In wash protocol 9, 50% TFE was replaced with 50% MeOH but there was not an improvement in the removal of most sticky peptides (i.e., wash 9 was not better than wash 7). Wash protocol 7 was found to be the best combination of FA and TFE tested. FA, methanol, and TFE were the most efficient solvents overall and as mentioned above, ACN was the perfect choice for the last component.
The wash protocol found ideal for removing the majority of sticky peptides from the trap was protocol 7: 10% FA, 50% TFE, 80% MeOH, 80% ACN. As shown in Figure 5, with this washing protocol, the intensity of the most sticky UmuD peptide was reduced from 90% intensity in subsequent runs without washing to 13% with wash protocol 7. The intensity of many of the sticky peptides of both UmuD and EGFR were reduced to less than 5% of the intensity observed without washing (Figures 5 and S1). Two uses of wash protocol 7 back to back removed all traces of sticky peptides. After the use of wash protocol 7, the false EX1 kinetic signatures of either UmuD or EGFR disappeared, as shown in Figure 4, bottom rows. All of these experiments were reproduced more than 3 times.
4. Conclusions
Falsely attributing EX1 kinetics to proteins and peptides that do not behave in the EX1 kinetic regime can be a problem. By being aware of how real EX1 kinetics arise and the typical sources of false EX1 signatures, ascribing EX1 kinetics incorrectly can greatly be reduced and/or eliminated. As we have shown here, testing for peptide carryover is essential in any HX MS experiment. We have demonstrated the reduction of carryover by a time-efficient washing procedure that allows washing of the trap column with multiple solvents in an optimized order. Although this method adds to the acquisition time required for an HX MS experiment, it converts useless data into useful data that can be processed as a typical HX MS data set would be. Such conversion is well worth the additional time.
We note that in the system described here, online digestion [23] was not implemented. An online digestion column can become another source of carryover and other methods must be used to eliminate carryover occurring in online digestion. Primarily, solvents that destroy the enzymatic activity of the immobilized acid protease cannot be used to wash the column and other strategies must be adopted. We will describe such strategies and methods in a future report.
Supplementary Material
Acknowledgments
We are pleased to acknowledge Prof. M. J. Eck for providing EGFR kinase domain. This work was supported by the NIH (R01-GM086507 to JRE), the NSF (CAREER Award, MCB-0845033 to PJB), a New Faculty Award from the Camille & Henry Dreyfus Foundation (PJB), the NU Office of the Provost (PJB), The Danish Natural Science Research Council (Grant 272-07-0276 to KDR) and the Waters Corporation (JRE). PJB is a Cottrell Scholar of the Research Corporation for Science Advancement. This work is contribution number 968 from the Barnett Institute.
ABBREVIATIONS
- IPA
Isopropanol
- ACN
Acetonitrile
- MeOH
Methanol
- FA
Formic acid
- TFE
Trifluoroethanol
- TFA
Trifluoroacetic acid
- DMSO
Dimethyl sulfoxide
- HX MS
hydrogen exchange mass spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miranker A, Robinson CV, Radford SE, Aplin RT, Dobson CM. Detection of transient protein folding populations by mass spectrometry. Science. 1993;262:896–900. doi: 10.1126/science.8235611. [DOI] [PubMed] [Google Scholar]
- 2.Arrington CB, Teesch LM, Robertson AD. Defining protein ensembles with native-state NH exchange: kinetics of interconversion and cooperative units from combined NMR and MS analysis. J Mol Biol. 1999;285:1265–1275. doi: 10.1006/jmbi.1998.2338. [DOI] [PubMed] [Google Scholar]
- 3.Engen JR, Smith DL. Investigating the higher order structure of proteins. Hydrogen exchange, proteolytic fragmentation, and mass spectrometry. Methods Mol Biol. 2000;146:95–112. doi: 10.1385/1-59259-045-4:95. [DOI] [PubMed] [Google Scholar]
- 4.Weis DD, Wales TE, Engen JR, Hotchko M, Ten Eyck LF. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J Am Soc Mass Spectrom. 2006;17:1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 6.Arrington CB, Robertson AD. Kinetics and thermodynamics of conformational equilibria in native proteins by hydrogen exchange. Meth Enzymol. 2000;323:104–124. doi: 10.1016/s0076-6879(00)23363-6. [DOI] [PubMed] [Google Scholar]
- 7.Sivaraman T, Robertson AD. Kinetics of conformational fluctuations by EX1 hydrogen exchange in native proteins. Methods Mol Biol. 2001;168:193–214. doi: 10.1385/1-59259-193-0:193. [DOI] [PubMed] [Google Scholar]
- 8.Kaltashov IA, Eyles SJ. Studies of biomolecular conformations and conformational dynamics by mass spectrometry. Mass Spectrom Rev. 2002;21:37–71. doi: 10.1002/mas.10017. [DOI] [PubMed] [Google Scholar]
- 9.Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu Rev Biophys Biomol Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 10.Wales TE, Engen JR. Partial unfolding of diverse SH3 domains on a wide timescale. J Mol Biol. 2006;357:1592–1604. doi: 10.1016/j.jmb.2006.01.075. [DOI] [PubMed] [Google Scholar]
- 11.Englander SW. Protein folding intermediates and pathways studied by hydrogen exchange. Annu Rev Biophys Biomol Struct. 2000;29:213–238. doi: 10.1146/annurev.biophys.29.1.213. [DOI] [PubMed] [Google Scholar]
- 12.Kheterpal I, Wetzel R. Hydrogen/deuterium exchange mass spectrometry--a window into amyloid structure. Acc Chem Res. 2006;39:584–593. doi: 10.1021/ar050057w. [DOI] [PubMed] [Google Scholar]
- 13.Wu Y, Kaveti S, Engen JR. Extensive deuterium back-exchange in certain immobilized pepsin columns used for H/D exchange mass spectrometry. Anal Chem. 2006;78:1719–1723. doi: 10.1021/ac0518497. [DOI] [PubMed] [Google Scholar]
- 14.Haeckel R. Recommendations for definition and determination of carry-over effects. J Automat Chem. 1988;10:181–183. doi: 10.1155/S1463924688000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng W, Musson DG, Fisher AL, Wang AQ. A new approach for evaluating carryover and its influence on quantitation in high-performance liquid chromatography and tandem mass spectrometry assay. Rapid Commun Mass Spectrom. 2006;20:635–640. doi: 10.1002/rcm.2353. [DOI] [PubMed] [Google Scholar]
- 16.Fang J, Rand KD, Silva MC, Wales TE, Engen JR, Beuning PJ. Conformational dynamics of the Escherichia coli DNA polymerase manager proteins UmuD and UmuD’. J Mol Biol. 2010;398:40–53. doi: 10.1016/j.jmb.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z. Doctoral Dissertation. Purdue University; 1995. Protein hydrogen exchange determined by mass spectrometry: a new tool for probing protein high-order structure and structural changes; p. 223. [Google Scholar]
- 19.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 20.Mitulovic G, Stingl C, Steinmacher I, Hudecz O, Hutchins JR, Peters JM, Mechtler K. Preventing carryover of peptides and proteins in nano LC-MS separations. Anal Chem. 2009;81:5955–5960. doi: 10.1021/ac900696m. [DOI] [PubMed] [Google Scholar]
- 21.Hughes NC, Wong EY, Fan J, Bajaj N. Determination of carryover and contamination for mass spectrometry-based chromatographic assays. Aaps J. 2007;9:E353–360. doi: 10.1208/aapsj0903042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vallano PT, Shugarts SB, Woolf EJ, Matuszewski BK. Elimination of autosampler carryover in a bioanalytical HPLC-MS/MS method: a case study. J Pharm Biomed Anal. 2005;36:1073–1078. doi: 10.1016/j.jpba.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics. 2002;1:132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 24.Sadek PC. The HPLC solvent guide. John Wiley and Sons, Inc; New York: 2002. [Google Scholar]
- 25.Aguilar M-I. HPLC of peptides and proteins: methods and protocols. Humana press Inc; New Jersey: 2004. [Google Scholar]
- 26.Asakawa Y, Ozawa C, Osada K, Kaneko S, Asakawa N. Reduction of carry-over in column-switching HPLC/MS system with automated system washing procedure for highly sensitive direct analysis of donepezil in dog plasma. J Pharm Biomed Anal. 2007;43:683–690. doi: 10.1016/j.jpba.2006.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.