Abstract
Objectives
The aim of this study was to compare the accuracy of genetic tables and formal pharmacogenetic algorithms for warfarin dosing.
Background
Pharmacogenetic algorithms based on regression equations can predict warfarin dose, but they require detailed mathematical calculations. A simpler alternative, recently added to the warfarin label by the U.S. Food and Drug Administration, is to use genotype-stratified tables to estimate warfarin dose. This table may potentially increase the use of pharmacogenetic warfarin dosing in clinical practice; however, its accuracy has not been quantified.
Methods
A retrospective cohort study of 1,378 patients from 3 anticoagulation centers was conducted. Inclusion criteria were stable therapeutic warfarin dose and complete genetic and clinical data. Five dose prediction methods were compared: 2 methods using only clinical information (empiric 5 mg/day dosing and a formal clinical algorithm), 2 genetic tables (the new warfarin label table and a table based on mean dose stratified by genotype), and 1 formal pharmacogenetic algorithm, using both clinical and genetic information. For each method, the proportion of patients whose predicted doses were within 20% of their actual therapeutic doses was determined. Dosing methods were compared using McNemar’s chi-square test.
Results
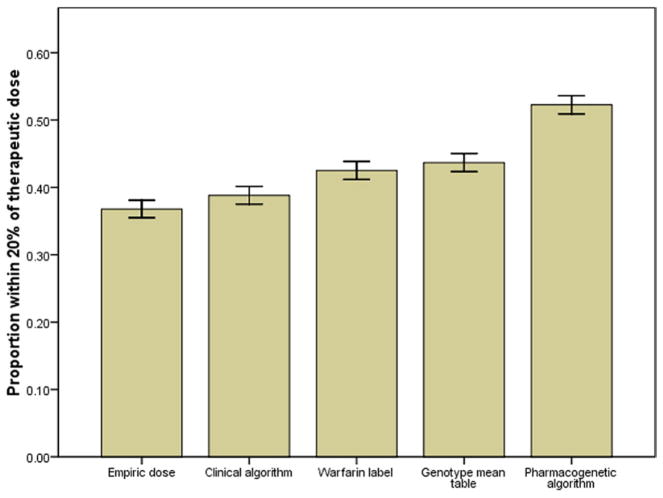
Warfarin dose prediction was significantly more accurate (all p < 0.001) with the pharmacogenetic algorithm (52%) than with all other methods: empiric dosing (37%; odds ratio [OR]: 2.2), clinical algorithm (39%; OR: 2.2), warfarin label (43%; OR: 1.8), and genotype mean dose table (44%; OR: 1.9).
Conclusions
Although genetic tables predicted warfarin dose better than empiric dosing, formal pharmacogenetic algorithms were the most accurate.
Keywords: coumarins, dose prediction, dosing algorithms, FDA label, genetic tables, pharmacogenetics, warfarin
In August 2007, the U.S. Food and Drug Administration updated the warfarin (Coumadin, Bristol-Myers Squibb, Princeton, New Jersey) product label to add pharmacogenetic information (1). In January 2010, the agency added specific instructions on how to use genotype to predict individualized doses: the new label provides a concise table of dosing recommendations, stratified by genotype (2). In contrast, most research on pharmacogenetic warfarin dosing has focused on developing and validating complex predictive algorithms (3,4), which integrate clinical and genetic factors to predict dose on the basis of regression equations. These algorithms accurately predict warfarin dose (4–8), although they may not be as accurate in African Americans (9).
Formal predictive algorithms require the use of a computer or web-based application (10) to conduct mathematical calculations and are difficult to summarize in written form. Providing an estimated dose in a table, such as the one in the new warfarin label, renders a genotype-specific dose readily accessible and easier to implement in clinical practice. However, no published study has quantified the accuracy of the new table. As a result, clinicians prescribing warfarin might increase their use of pharmacogenetic dosing without a clear understanding of its accuracy.
In this study, we compared the accuracy of warfarin dose prediction in a formal pharmacogenetic algorithm with 2 genetic tables, the aforementioned warfarin label table (reproduced in Table 1) and a similar table constructed from the mean dose by genotype in a separate cohort (Table 2). We also compared these dosing methods with 2 nongenetic methods: a clinical predictive algorithm and standard empiric dosing of 35 mg/week (11).
Table 1.
FDA Label: Range of Expected Therapeutic Warfarin Doses (mg/day) Based on CYP2C9 and VKORC1 Genotypes
| VKORC1-1639 | CYP2C9 *1*1 | CYP2C9 *1*2 | CYP2C9 *1*3 | CYP2C9 *2*2 | CYP2C9 *2*3 | CYP2C9 *3*3 |
|---|---|---|---|---|---|---|
| GG | 5–7 | 5–7 | 3–4 | 3–4 | 3–4 | 0.5–2 |
| GA | 5–7 | 3–4 | 3–4 | 3–4 | 0.5–2 | 0.5–2 |
| AA | 3–4 | 3–4 | 0.5–2 | 0.5–2 | 0.5–2 | 0.5–2 |
Reproduced from updated warfarin (Coumadin, Bristol-Myers Squibb, Princeton, New Jersey) product label (2).
CYP2C9 = cytochrome P450 2C9; FDA = U.S. Food and Drug Administration; VKORC1 = vitamin K epoxide reductase complex subunit 1.
Table 2.
IWPC Cohort: Mean Therapeutic Warfarin Dose (mg/day) by CYP2C9 and VKORC1 Genotype in 2,858 Patients
| VKORC1-1639 | CYP2C9 *1*1 | CYP2C9 *1*2 | CYP2C9 *1*3 | CYP2C9 *2*2 | CYP2C9 *2*3 | CYP2C9*3*3 |
|---|---|---|---|---|---|---|
| GG | 6 | 5 | 4 | 4 | 4 | 3 |
| GA | 5 | 4 | 3 | 3 | 2 | 1 |
| AA | 3 | 3 | 2 | 2 | 1 | 2 |
IWPC = International Warfarin Pharmacogenetics Consortium; other abbreviations as in Table 1.
Methods
Patient population
The cohort comprised 1,378 patients taking warfarin at the University of Pennsylvania, the University of Florida, and Washington University. Most of these patients were originally recruited as part of prior prospective cohort studies (3,12–14), and many were also later included in the International Warfarin Pharmacogenetics Consortium cohort (accessed via PharmGKB accession number PA162355460) (4,15). Participants were required to have achieved a stable therapeutic dose of warfarin, defined as the dose that led to therapeutic international normalized ratio (INR) values on 2 or 3 consecutive visits (4), and to have complete data for dose prediction by all dosing methods. Because of institutional review board restrictions on access to individual, patient-level data for clinical and genetic variables for some of the patients in our cohort, we could not determine the overall frequency of the clinical and genetic variables discussed in the following; however, these frequencies are summarized online for the subset of our patients who were part of the International Warfarin Pharmacogenetics Consortium cohort (n = 1,025) (Online Table 1). Of note, predicted and actual warfarin doses were available at the patient level for all patients in our cohort.
Dose prediction methods
EMPIRIC DOSE
Patients were assigned the population average dose of 35 mg/week.
CLINICAL ALGORITHM
Patients were assigned a dose on the basis of a formal regression equation (3). The factors included in this validated algorithm are age, body surface area (BSA), African American race, amiodarone usage, target INR, smoking status, and warfarin indication.
WARFARIN LABEL
Patients were assigned a dose equal to the midpoint of the daily dose range provided in the table in the newly revised warfarin label (Table 1) (2). The exact methods used by the Food and Drug Administration to derive this table are not publicly available. Doses were multiplied by 7 to provide consistent units of milligrams per week.
GENOTYPE MEAN TABLE
Patients were assigned the mean dose in those with the same genotype, on the basis of the CYP2C9 *2, CYP2C9 *3, and VKORC1 (−1639 G>A) genetic variants. Mean dose was determined in a separate population, consisting of the International Warfarin Pharmacogenetics Consortium cohort minus those patients used in the analysis cohort described previously (final n = 2,858) (Table 2).
PHARMACOGENETIC ALGORITHM
Patients were assigned a dose according to a previously validated formal regression equation (3). This is similar to the algorithm available at www.warfarindosing.org; however, the online version has been expanded since the original validation studies to also accommodate newer single-nucleotide polymorphisms that have minor effects on dose. Factors included in this algorithm are CYP2C9 and VKORC1 genotype, age, BSA, African American race, amiodarone use, target INR, smoking status, and warfarin indication.
Data analysis
The accuracy of each dosing method was based on whether the predicted dose fell within a clinically meaningful range of within 20% of the stable therapeutic dose, as used in other studies (4). Because this is a matched dataset, the proportion of patients whose predicted doses fell within this range was compared between each method using McNemar’s chi-square test. The resultant odds ratio (OR) in this test reflects the odds of patients’ being dosed within range by one method versus the other. Comparisons of all possible pairwise combinations of the 5 dosing methods were made, with no correction for multiple comparisons.
Additionally, to quantify the extent to which each dosing method was either overestimating or underestimating warfarin dose, we examined the proportion of patients whose predicted doses were above or below the stable therapeutic dose by at least 20%. Finally, we examined the proportion of patients predicted at therapeutic dose ± 20% on the basis of whether the patient required a low (≤21 mg/week), intermediate (>21 and ≤49 mg/week), or high (>49 mg/week) therapeutic dose of warfarin.
To explore how clinical judgment may affect the accuracy of the warfarin label, we conducted a secondary analysis in which patients older than age 60 years (the cut point in the warfarin label at which patients “appear to exhibit greater than expected [prothrombin time]/INR response to … warfarin” [2]) were assigned the lower estimate of the dose range, and patients age 60 years or younger were given the upper estimate of the dose range. All principal analyses were then repeated. Additionally, we conducted a similar secondary analysis incorporating more of the clinical variables used in the algorithms in the subset of patients for which we had access to all of these variables at the individual level. Patients were given the upper estimate of the dose range in the warfarin label if they were age ≤60 years, had BSAs >2.25 m2 (>1 SD above the mean), were African American, or had target INRs >2.0 to 3.0. Patients were given the lower estimate of the dose range in the warfarin label if they were age >60 years, had BSAs <1.75 m2, or used amiodarone. If patients had factors that both raised and lowered their predicted warfarin doses, they were assigned the midpoint of the dose range in the warfarin label.
To examine the effect of race on dosing method accuracy, a secondary analysis was conducted in which the cohort was stratified by race and all principal analyses were repeated.
Analyses were performed using SPSS Statistics 17.0 (SPSS, Inc., Chicago, Illinois) and SAS 9.1 (SAS Institute Inc., Cary, North Carolina).
Results
The percents of patients whose predicted doses were within 20% of their stable therapeutic doses for the empiric dose (37%), clinical algorithm (39%), warfarin label (43%), genotype mean table (44%), and pharmacogenetic algorithm (52%) dosing methods are shown in Figure 1. The observed mean therapeutic doses by genotype compared with the doses predicted by the warfarin label and genotype mean table are shown in Online Tables 2 and 3, respectively.
Figure 1. Dosing Method Accuracy.
The proportion dosed within 20% of the stable therapeutic dose was calculated for each dosing method. Having a predicted dose within 20% of the therapeutic dose was considered a clinically meaningful degree of accuracy. Error bars indicate ±1 SE.
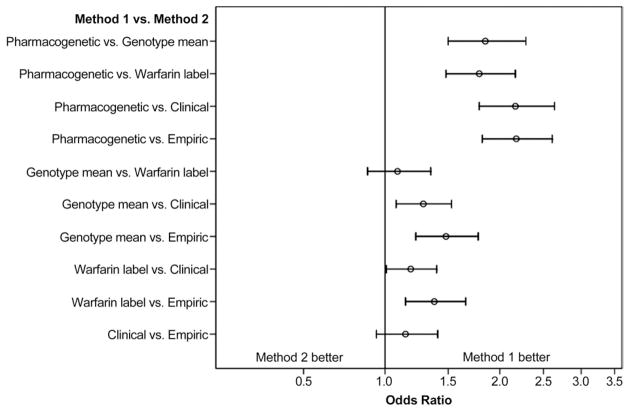
As shown in Figure 2, the pharmacogenetic algorithm was significantly more accurate compared with each of the other dosing methods: empiric dose (OR: 2.18; 95% confidence interval [CI]: 1.82 to 2.61), clinical algorithm (OR: 2.17; 95% CI: 1.79 to 2.64), warfarin label (OR: 1.79; 95% CI: 1.48 to 2.17), and genotype mean table (OR: 1.85; 95% CI: 1.50 to 2.29). The 2 genetic tables were more accurate than either the empiric dose or clinical algorithm. The difference between the warfarin label and genotype mean tables was not statistically significant.
Figure 2. Two-Way Comparisons of Dosing Methods.
The proportion dosed within 20% of stable therapeutic dose was compared using McNemar’s chi-square test. The resultant odds ratio reflects the odds of a patient’s being properly dosed by the first method versus the second. Error bars represent exact 95% confidence intervals.
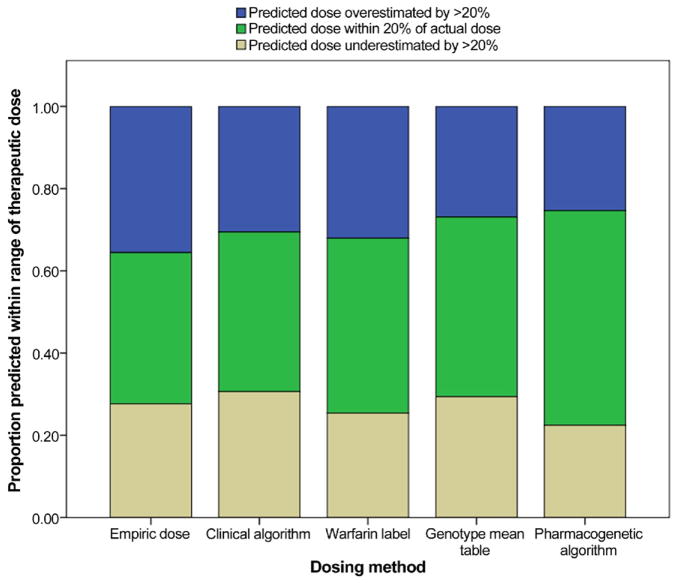
The extent of overestimation and underestimation of warfarin dose by each dosing method is shown in Figure 3, and both were lowest with the pharmacogenetic algorithm. Additionally, all methods performed best in patients requiring intermediate warfarin doses. The pharmacogenetic algorithm performed better than all other methods in predicting dose in those requiring higher (>49 mg/week) and lower (≤21 mg/week) doses (Online Fig. 1).
Figure 3. Dosing Method Accuracy.
The proportions dosed below 80%, within 20%, and above 120% of the stable therapeutic dose were calculated for each dosing method.
The age-adjusted warfarin label was minimally better than the warfarin label based on the table midpoint (OR: 1.16; 95% CI: 0.95 to 1.42), but the pharmacogenetic algorithm was still significantly better than the former method (OR: 1.59; 95% CI: 1.31 to 1.93). The warfarin label adjusted for several clinical factors was minimally better than the warfarin label based on the table midpoint (OR: 1.12; 95% CI: 0.86 to 1.47) as well as the age-adjusted warfarin label (OR: 1.21; 95% CI: 0.75 to 1.96), but it was significantly worse than the pharmacogenetic algorithm (OR: 1.62; 95% CI: 1.29 to 2.05).
The percents of patients predicted at therapeutic dose ±20% for the empiric dose, clinical algorithm, warfarin label, genotype mean table, and pharmacogenetic algorithm dosing methods were 36%, 42%, 41%, 38%, and 42%, respectively, in African Americans and 37%, 38%, 43%, 45%, and 54% in non–African Americans.
Discussion
Recent research on warfarin dose prediction has focused on the use of formal pharmacogenetic algorithms based on complex regression equations. However, in January 2010, the Food and Drug Administration updated the warfarin label to include pharmacogenetic dosing information in the form of a more straightforward table of estimated warfarin dose, stratified by genotype. This change may potentially increase the use of pharmacogenetic dose prediction by clinicians prescribing warfarin; thus, it is critical to determine the accuracy of this method. In this analysis, we compared the accuracy of these various dosing methods on the basis of whether the predicted dose fell within a clinically meaningful range of within 20% of the stable therapeutic dose, as done previously (4).
In our study, dosing based on genetic tables was somewhat more accurate than that based on nongenetic methods. Using the warfarin label or the genotype mean tables, accurate dosing could be predicted 43% or 44% of the time, respectively, compared with 39% using a clinical algorithm and 37% using an empiric 5 mg/day dose. However, the pharmacogenetic algorithm was superior to all other methods, including these tables. Using a pharmacogenetic algorithm, accurate dose prediction was achieved 52% of the time. Furthermore, the pharmacogenetic algorithm had lower rates of both dose overestimation and underestimation than all other dosing methods (Fig. 3) as well as better dosing prediction in those requiring high and low warfarin doses (Online Fig. 1).
Both genetic tables were found to be an improvement over empiric dosing and a clinical algorithm, affirming the importance of genetics in determining warfarin dose. Because both genetic tables performed similarly, it seems likely that their suboptimal performance relative to the pharmacogenetic algorithm is based on inherent limitations of using tables, rather than the result of inaccurate estimates. Formal algorithms would be expected to more precisely quantify the effects of multiple genetic factors on dose, compared with the average effects that are used in dosing tables. Moreover, age, BSA, target INR, drug interactions, and other clinical factors have all previously been shown to affect warfarin dose (11). Thus, it is unsurprising that the combination of clinical factors with genotype in the context of a formal pharmacogenetic algorithm seemed to be the most beneficial for improving the accuracy of dose prediction.
The warfarin label does in fact state that clinical factors should be accounted for; however, it does not give guidelines on how both clinical and genetic factors should be combined to determine an appropriate dose. As a result, there was no way to formally test the impact of clinical factors on the use of the genetic table in the warfarin label. Analyses that approximated how clinicians may account for clinical factors led to minimal improvement in the accuracy of the warfarin label; however, the formal pharmacogenetic algorithm remained superior.
One possible reason that adjusting for clinical factors did not result in more improvement in dosing accuracy is that the magnitude of the effect on warfarin dose varies from factor to factor and that level of complexity is difficult to account for in a simple table. Additionally, another limitation of the warfarin label is that it gives a wide relative dosing range for those most sensitive to the drug. Thus, any simplified table most likely will not be able to capture the full complexity of personalized warfarin dosing, because it will not be able to easily and accurately quantify the effects of both genetic and clinical factors.
Nonetheless, even with the pharmacogenetic algorithm, 48% of patients were ultimately not dosed accurately. There are several reasons why such a substantial proportion of patients may not be dosed correctly. First, clinical factors other than those included in our study (and, to our knowledge, not included in well-validated dosing algorithms) can affect warfarin dose (16–22). Second, genes other than CYP2C9 and VKORC1 may affect warfarin dose, such as CYP4F2, GGCX, EPHX1, and ApoE (14,23–25), although their effects on warfarin dose require confirmation (26–28). These and other genes could be incorporated into future pharmacogenetic algorithms or tables to improve their accuracy; however, studies are needed to assess their clinical utility.
Finally, as expected on the basis of previous results (9,29,30), both the pharmacogenetic algorithm and the genetic tables were less accurate in African Americans. In fact, the clinical algorithm seemed to perform just as well as these methods in African Americans, although our analysis cohort did not have sufficient power to formally compare dosing methods within this group. To show their greatest benefit, pharmacogenetic algorithms will likely have to incorporate genetic information from studies done in African Americans. In short, it is clear that despite the extensive amount of research that has been done on warfarin dosing variability, a substantial percent of that variability remains to be explained (9,29–34).
Perhaps most important, although we know that pharmacogenetic algorithms lead to more accurate warfarin dose prediction, it remains unknown whether using a formal pharmacogenetic algorithm will actually improve laboratory or clinical outcomes. Although some observational research suggests that this may be the case (35), the answer to these questions awaits the results of large randomized clinical trials that are currently under way (36–38).
The average cost of warfarin-related genetic testing is currently almost $400 per patient (39). This cost could fall in the near future, making genotyping more feasible for routine clinical practice. However, given the substantial number of patients started on warfarin each year, the added cost of genotyping over the current standard of care will still be significant. Thus, it is critical to use the information gained from genetic testing in the most effective way possible.
Our results suggest that the genetic table provided in the updated warfarin label is an improvement over empiric dosing and thus may be appropriate under certain circumstances. However, tables are less accurate than a formal pharmacogenetic algorithm for warfarin dose prediction. As a result, it seems advisable for clinicians to use a formal pharmacogenetic algorithm instead of a genetic dosing table when feasible. Should dosing algorithms prove effective at improving clinical outcomes in ongoing randomized trials, methods to improve access to these algorithms could include publication in the warfarin label, web-based access, and incorporation into handheld devices.
Supplementary Material
Acknowledgments
This work was supported by Medical Scientist Training Program grant T32-GM07170 from the National Institutes of Health, institutional funds from the University of Pennsylvania School of Medicine, and grant R01HL066176 from the National Institutes of Health. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript. Dr. Gage is funded by the grant R01HL097036 from the National Institutes of Health for warfarin-related research and owns the nonprofit domain name www.warfarindosing.org. Dr. Johnson is an advisory board member for Medco Health Solutions, Inc. Ms. Brensinger has consulted for a law firm representing Pfizer, Inc., unrelated to warfarin. Dr. Kimmel has received research funding from GlaxoSmithKline and Pfizer, Inc.; has served as a consultant to several drug companies, including Bayer AG, Novartis, and Pfizer, Inc., all unrelated to warfarin; has received an honorarium from Ortho-McNeil for a talk on warfarin; and receives funding from the National Institutes of Health and the Aetna Foundation for warfarin-related research.
Abbreviations and Acronyms
- BSA
body surface area
- CI
confidence interval
- CYP2C9
cytochrome P450 2C9
- INR
international normalized ratio
- OR
odds ratio
- VKORC1
vitamin K epoxide reductase complex subunit 1
APPENDIX
For supplementary tables and figures and their legends, please see the online version of this article.
Footnotes
All other authors have reported that they have no relationships to disclose.
References
- 1.Gage B, Lesko L. Pharmacogenetics of warfarin: regulatory, scientific, and clinical issues. J Thromb Thrombolysis. 2008;25:45–51. doi: 10.1007/s11239-007-0104-y. [DOI] [PubMed] [Google Scholar]
- 2.Brisotol-Meyers Squibb Company. [Accessed February 2, 2010.];Coumadin® tablets (warfarin sodium tablets, USP) crystalline; Coumadin® for injection (warfarin sodium for injection, USP) Available at: http://www.accessdata.fda.gov/drugsatfda_docs/label/2010/009218s108lbl.pdf.
- 3.Gage B, Eby C, Johnson J, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–31. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein T, Altman R, Eriksson N, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–64. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voora D, Eby C, Linder M, et al. Prospective dosing of warfarin based on cytochrome P-450 2C9 genotype. Thromb Haemost. 2005;93:700–5. doi: 10.1160/TH04-08-0542. [DOI] [PubMed] [Google Scholar]
- 6.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008;83:460–70. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 7.Anderson J, Horne B, Stevens S, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116:2563–70. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 8.Wen M, Lee M, Chen J, et al. Prospective study of warfarin dosage requirements based on CYP2C9 and VKORC1 genotypes. Clin Pharmacol Ther. 2008;84:83–9. doi: 10.1038/sj.clpt.6100453. [DOI] [PubMed] [Google Scholar]
- 9.Schelleman H, Chen Z, Kealey C, et al. Warfarin response and vitamin K epoxide reductase complex 1 in African Americans and Caucasians. Clin Pharmacol Ther. 2007;81:742–7. doi: 10.1038/sj.clpt.6100144. [DOI] [PubMed] [Google Scholar]
- 10.Gage BF. [Accessed February 16, 2010.];Warfarin dosing. Available at: http://www.warfarindosing.org/Source/Home.aspx.
- 11.Ansell J, Hirsh J, Hylek E, Jacobson A, Crowther M, Palareti G. Pharmacology and management of the vitamin K antagonists: American College of Chest Physicians evidence-based clinical practice guidelines (8th edition) Chest. 2008;133:160S–98S. doi: 10.1378/chest.08-0670. [DOI] [PubMed] [Google Scholar]
- 12.Aquilante C, Langaee T, Lopez L, et al. Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P450 2C9 gene polymorphisms on warfarin dose requirements. Clin Pharmacol Ther. 2006;79:291–302. doi: 10.1016/j.clpt.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Lenzini P, Grice G, Milligan P, et al. Laboratory and clinical outcomes of pharmacogenetic vs. clinical protocols for warfarin initiation in orthopedic patients. J Thromb Haemost. 2008;6:1655–62. doi: 10.1111/j.1538-7836.2008.03095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimmel S, Christie J, Kealey C, et al. Apolipoprotein E genotype and warfarin dosing among Caucasians and African Americans. Pharmacogenomics J. 2008;8:53–60. doi: 10.1038/sj.tpj.6500445. [DOI] [PubMed] [Google Scholar]
- 15.PharmGKB. [Accessed February 14, 2010.];IWPC Pharmacogenetic Dosing Algorithm. Available at: https://www.pharmgkb.org/do/serve?objCls=Dataset&objId=PA162372936.
- 16.Garcia D, Regan S, Crowther M, Hughes R, Hylek E. Warfarin maintenance dosing patterns in clinical practice: implications for safer anticoagulation in the elderly population. Chest. 2005;127:2049–56. doi: 10.1378/chest.127.6.2049. [DOI] [PubMed] [Google Scholar]
- 17.Gage B, Eby C, Milligan P, Banet G, Duncan J, McLeod H. Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin. Thromb Haemost. 2004;91:87–94. doi: 10.1160/TH03-06-0379. [DOI] [PubMed] [Google Scholar]
- 18.Hirsh J, Dalen J, Anderson D, et al. Oral anticoagulants: mechanism of action, clinical effectiveness, and optimal therapeutic range. Chest. 1998;114:445S–69S. doi: 10.1378/chest.114.5_supplement.445s. [DOI] [PubMed] [Google Scholar]
- 19.James A, Britt R, Raskino C, Thompson S. Factors affecting the maintenance dose of warfarin. J Clin Pathol. 1992;45:704–6. doi: 10.1136/jcp.45.8.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Booth S, Centurelli M. Vitamin K: a practical guide to the dietary management of patients on warfarin. Nutr Rev. 1999;57:288–96. doi: 10.1111/j.1753-4887.1999.tb01815.x. [DOI] [PubMed] [Google Scholar]
- 21.Cropp J, Bussey H. A review of enzyme induction of warfarin metabolism with recommendations for patient management. Pharmacotherapy. 1997;17:917–28. [PubMed] [Google Scholar]
- 22.Absher R, Moore M, Parker M. Patient-specific factors predictive of warfarin dosage requirements. Ann Pharmacother. 2002;36:1512–7. doi: 10.1345/aph.1C025. [DOI] [PubMed] [Google Scholar]
- 23.Wells P, Majeed H, Kassem S, et al. A regression model to predict warfarin dose from clinical variables and polymorphisms in CYP2C9, CYP4F2, and VKORC1: derivation in a sample with predominantly a history of venous thromboembolism. Thromb Res. 2010;125:e259–64. doi: 10.1016/j.thromres.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 24.Kimura R, Miyashita K, Kokubo Y, et al. Genotypes of vitamin K epoxide reductase, gamma-glutamyl carboxylase, and cytochrome P450 2C9 as determinants of daily warfarin dose in Japanese patients. Thromb Res. 2007;120:181–6. doi: 10.1016/j.thromres.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Pautas E, Moreau C, Gouin-Thibault I, et al. Genetic factors (VKORC1, CYP2C9, EPHX1, and CYP4F2) are predictor variables for warfarin response in very elderly, frail inpatients. Clin Pharmacol Ther. 2010;87:57–64. doi: 10.1038/clpt.2009.178. [DOI] [PubMed] [Google Scholar]
- 26.Carlquist J, Horne B, Mower C, et al. An evaluation of nine genetic variants related to metabolism and mechanism of action of warfarin as applied to stable dose prediction. J Thromb Thrombolysis. 2010;30:358–64. doi: 10.1007/s11239-010-0467-3. [DOI] [PubMed] [Google Scholar]
- 27.Lee M, Chen C, Chou C, et al. Genetic determinants of warfarin dosing in the Han-Chinese population. Pharmacogenomics. 2009;10:1905–13. doi: 10.2217/pgs.09.106. [DOI] [PubMed] [Google Scholar]
- 28.Cavallari L, Langaee T, Momary K, et al. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87:459–64. doi: 10.1038/clpt.2009.223. [DOI] [PubMed] [Google Scholar]
- 29.Schelleman H, Chen J, Chen Z, et al. Dosing algorithms to predict warfarin maintenance dose in Caucasians and African Americans. Clin Pharmacol Ther. 2008;84:332–9. doi: 10.1038/clpt.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schelleman H, Limdi N, Kimmel S. Ethnic differences in warfarin maintenance dose requirement and its relationship with genetics. Pharmacogenomics. 2008;9:1331–46. doi: 10.2217/14622416.9.9.1331. [DOI] [PubMed] [Google Scholar]
- 31.Kealey C, Chen Z, Christie J, et al. Warfarin and cytochrome P450 2C9 genotype: possible ethnic variation in warfarin sensitivity. Pharmacogenomics. 2007;8:217–25. doi: 10.2217/14622416.8.3.217. [DOI] [PubMed] [Google Scholar]
- 32.Loebstein R, Yonath H, Peleg D, et al. Interindividual variability in sensitivity to warfarin—nature or nurture? Clin Pharmacol Ther. 2001;70:159–64. doi: 10.1067/mcp.2001.117444. [DOI] [PubMed] [Google Scholar]
- 33.Limdi N, Goldstein J, Blaisdell J, Beasley T, Rivers C, Acton R. Influence of CYP2C9 genotype on warfarin dose among African American and European Americans. Per Med. 2007;4:157–69. doi: 10.2217/17410541.4.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cavallari L, Limdi N. Warfarin pharmacogenomics. Curr Opin Mol Ther. 2009;11:243–51. [PubMed] [Google Scholar]
- 35.van Schie R, Wadelius M, Kamali F, et al. Genotype-guided dosing of coumarin derivatives: the European Pharmacogenetics of Anticoagulant Therapy (EU-PACT) trial design. Pharmacogenomics. 2009;10:1687–95. doi: 10.2217/pgs.09.125. [DOI] [PubMed] [Google Scholar]
- 36.National Heart, Lung, and Blood Institute. [Accessed February 2, 2010];Clarification of Optimal Anticoagulation Through Genetics (COAG) Available at: http://www.clinicaltrials.gov/ct2/show/NCT00839657.
- 37.Washington University School of Medicine. [Accessed February 14, 2010.];Genetic Informatics Trial of Warfarin to Prevent Deep Vein Thrombosis. Available at: http://clinicaltrials.gov/ct2/show/NCT01006733?term=NCT01006733&.
- 38.Epstein R, Moyer T, Aubert R, et al. Warfarin genotyping reduces hospitalization rates: results from the MM-WES (Medco-Mayo Warfarin Effectiveness Study) J Am Coll Cardiol. 2010;55:2804–12. doi: 10.1016/j.jacc.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Eckman M, Rosand J, Greenberg S, Gage B. Cost-effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Ann Intern Med. 2009;150:73–83. doi: 10.7326/0003-4819-150-2-200901200-00005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.