Summary
The Arabidopsis thylakoid FtsH protease complex is composed of FtsH1/FtsH5 (type A) and FtsH2/FtsH8 (type B) subunits. Type A and type B subunits display a high degree of sequence identity throughout their mature domains, but no similarity in their amino-terminal targeting peptide regions. In chloroplast import assays, FtsH2 and FtsH5 were imported and subsequently integrated into thylakoids by a two-step processing mechanism that resulted in an amino-proximal lumenal domain, a single transmembrane anchor, and a carboxyl proximal stromal domain. FtsH2 integration into washed thylakoids was entirely dependent on the proton gradient, whereas FtsH5 integration was dependent on NTPs, suggesting their integration by Tat and Sec pathways, respectively. This was corroborated by in-organello competition and by antibody inhibition experiments. A series of constructs were made in order to understand the molecular basis for different integration pathways. The amino proximal domains through the transmembrane anchors were sufficient for proper integration as demonstrated with carboxyl-truncated versions of FtsH2 and FtsH5. The mature FtsH2 protein was found to be incompatible with the Sec machinery as determined with targeting peptide-swapping experiments. Incompatibility does not appear to be determined by any specific element in the FtsH2 domain as no single domain was incompatible with Sec transport. This suggests an incompatible structure that requires the intact FtsH2. That the highly homologous type A and type B subunits of the same multimeric complex use different integration pathways is a striking example of the notion that membrane insertion pathways have evolved to accommodate structural features of their respective substrates.
Keywords: chloroplast, protease, protein transport, twin arginine, SecY, Tat
Introduction
Thylakoidal FtsH proteins are bacterial-type metalloproteases that belong to the subfamily AAA+ (ATPases Associated with diverse cellular Activities). In general, FtsH proteases have two transmembrane domains followed by a cytosolic or stromal ATPase module with Walker A and B motifs, a Zn+ binding region, and a proteolytic domain (Ito and Akiyama, 2005). FtsH proteases are known to degrade short-lived membrane proteins in a number of organisms. Studies in prokaryotes show that FtsH proteins form hexameric rings where the ATP binding motif faces the center of the ring (Niwa et al., 2002). Although there are nine putative plastid FtsH family members in Arabidopsis, only one thylakoid FtsH complex has been detected in significant quantity, and this consists of FtsH1/FtsH5 (type A) and FtsH2/FtsH8 (type B) subunits (Sakamoto et al., 2003). Co-immunoprecipitation studies demonstrate that type A and type B subunits are members of the same complex (Sakamoto et al., 2003; Yu et al., 2004). Disruption of the FtsH5 gene results in a variegated phenotype (var1); disruption of the FtsH2 gene also results in a variegated phenotype (var2). The var1 and var2 phenotypes can be rescued by overexpression of FtsH1 and FtsH8, respectively (Yu et al., 2004; 2005). Thus, only one of each isomer is required for correct function of the thylakoid FtsH complex (Zaltsman et al., 2005).
All chloroplast FtsH proteins are encoded on nuclear genes and are synthesized in the cytosol. Cytosolically synthesized thylakoid proteins of chloroplasts are routed to their functional locations by several translocases (Cline and Dabney-Smith, 2008). Most are initially imported into the plastid stroma by the Toc and Tic complexes in the outer and inner envelope membranes. The Sec, Tat, SRP/Alb3, and an unassisted pathway integrate the stromal intermediates into the thylakoid bilayer or transport them to the lumen (Cline and Theg, 2007; Cline and Dabney-Smith, 2008). Of particular interest are the Tat pathway (for twin arginine translocation), which transports fully folded protein substrates and relies only on the proton gradient, and the Sec pathway, which transports unfolded proteins through a SecYE channel by virtue of ATP binding and hydrolysis by the SecA translocation motor.
Here we analyzed the targeting mechanisms of two Arabidopsis FtsH members, FtsH5 and FtsH2, as representative members of type A and type B families. Both isoforms have an amino terminal stroma-targeting transit peptide followed by a hydrophobic sequence, which we show functions as a cleavable hydrophobic signal peptide rather than a transmembrane domain. We show that the Tat pathway integrates FtsH2, whereas the Sec pathway integrates FtsH5. Carboxyl proximal truncations suggest that targeting specificity resides in the signal peptide of each isoform. Transit peptide swapping experiments indicated an incompatibility of the FtsH2 mature region with the Sec-pathway, explaining the need for two different integration systems to target two highly homologous subunits. This demonstrates an intriguing twist on the biogenesis membrane protein complexes, whereby two highly homologous subunits of the same multimeric complex are delivered to the membrane by different integration machineries.
Results
Type A and type B FtsH subunits differ by the presence/absence of the twin arginine motif
The two FtsH type A family members share a high degree of homology throughout their entire peptide sequence; this is also true for type B FtsH family members, suggesting recent gene duplication (Figure S1 b,c). By contrast, whereas FtsH type A and FtsH type B show identity in their stroma-exposed ATPase and protease domains, there is little homology between their chloroplast targeting peptides and the sequences flanking the amino proximal hydrophobic domain (Figure S1 a). In particular, the type B proteins FtsH2 and FtsH8 possess a twin-arginine motif just before, and an A-X-A signal peptidase cleavage site consensus (where X is any amino acid) just after the first hydrophobic domain, suggesting that they are Tat-directed cleavable signal peptides (Figure 1 b). For FtsH type A proteins the first hydrophobic domain also appears to be a cleavable signal peptide as evidenced by a relatively low hydrophobicity and an A-X-A cleavage site (Figure 1 a). This sequence analysis suggests that the mature proteins for type A and type B FtsH proteases consist of an amino terminal lumen-facing domain (L), a transmembrane anchor (T), and a large stroma-facing catalytic domain (S) (Figure 1 (c)). Because amino-acid sequence identity of family members within a type was very high, even in the signal peptide and transit peptide regions (Figure S1 b, c) we selected FtsH5 and FtsH2 for the type A and type B groups, respectively, for in vitro assays to determine their paths to the thylakoids.
Figure 1.
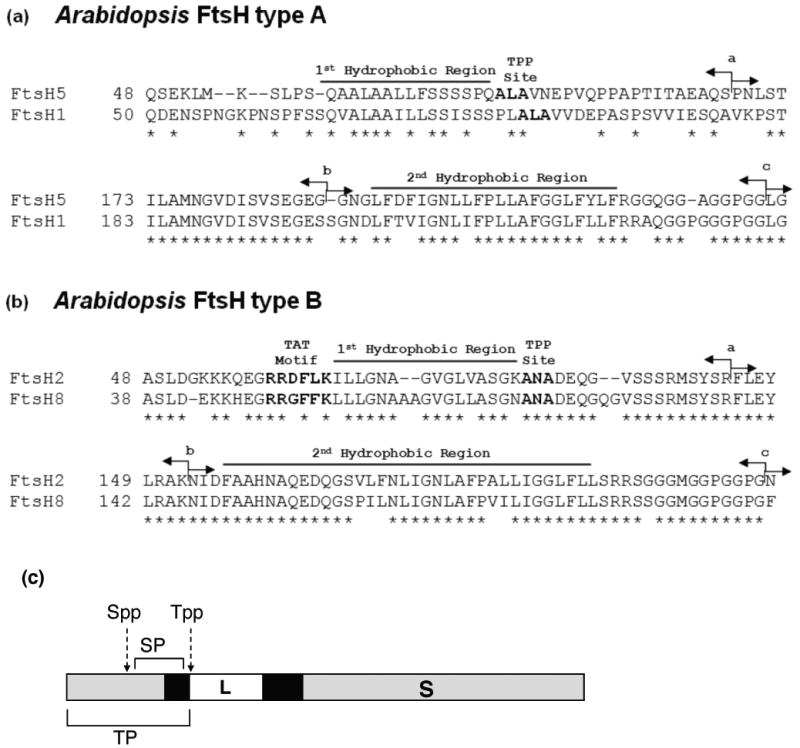
FtsH type A and B differ by the presence/absence of the Twin Arginine Motif. The amino acid sequences of the N-terminal region of four Arabidopsis FtsH proteases are aligned. (a) Precursors to type A, FtsH5 and FtsH1, have a chloroplast-targeting domain followed by a hydrophobic domain and the tripeptide A-L-A, a cleavage motif for the thylakoid signal peptidase (Tpp). The latter two features are characteristic of a hydrophobic signal peptide. (b) Precursors to type B, FtsH2 and FtsH8, possess a chloroplast targeting domain, a classical Tat motif (R-R-X-F-L-K), a hydrophobic domain, and an A-N-A Tpp cleavage site. The latter three features are characteristic of a Tat pathway signal peptide. (c) Diagram of a generic (i.e. type A or type B) FtsH precursor protein. In the present work, the chloroplast targeting domain and the signal peptide combined are called the ‘transit peptide’ (TP). The lumenal domain (L), transmembrane anchor (T), and stromal domain (S) are marked. In (a) and (b) of the figure, the amino-acid sequences start after the predicted Spp cleavage site. The arrows in sequence panels (a) and (b) are swapping points for fusions between FtsH5 and FtsH2 or truncation points (see below).
pFtsH2 and pFtsH5 are localized in vitro by a two step pathway
In vitro translated precursor to FtsH2 (pFtsH2; 74-kDa) and precursor to FtsH5 (pFtsH5; 75-kDa) were incubated with intact pea chloroplasts. The precursors were imported into the organelle and processed to a mature form of 65-kDa for mFtsH2 and 67-kDa for mFtsH5 (Figure 2a and 2b, lanes 2). Protease treatment of recovered chloroplasts confirmed that the mature forms were inside of the chloroplasts (lanes 3). Fractionation of the recovered chloroplasts showed that both mature proteins were present in the thylakoid fraction (T, lanes 5) rather than the soluble stromal fraction (S, lanes 4).
Figure 2.
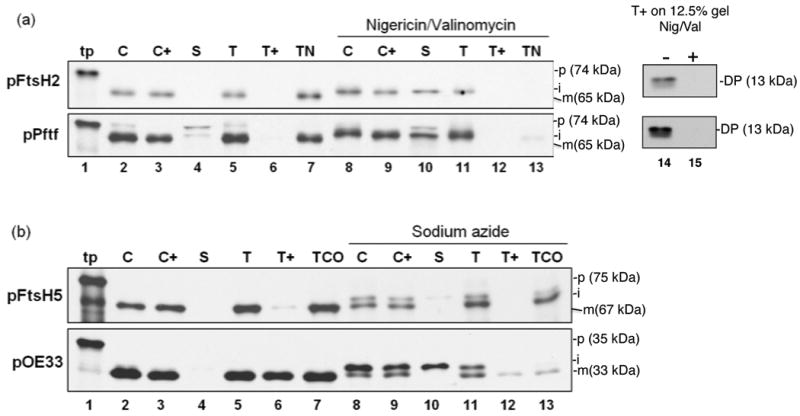
pFtsH2 and pFtsH5 are imported into isolated chloroplasts, processed to an intermediate form, and integrated into thylakoids. Radiolabeled in vitro translated pFtsH2, pPftf, (a) pFtsH5 or pOE33 (b) (lanes 1, tp) were incubated with intact isolated pea chloroplasts in a reaction containing 5mM ATP and ∼ 100 μE/m2 −s light at 25 °C for 20 min, in the absence or presence of nigericin and valinomycin (for pFtsH2 and pPftf) or sodium azide (for pFtsH5 and pOE33), as depicted above the panels. Intact chloroplasts were recovered from the reaction (C, lanes 2 and 8) and treated with thermolysin (C+, lanes 3 and 9). Untreated intact chloroplasts were fractionated into stroma (S, lanes 4 and 10) and thylakoids. Thylakoids aliquots were washed with import buffer (T, lanes 5 and 11), treated with thermolysin (T+, lanes 6 and 12) or treated with 100 mM NaOH or 200 mM Na2CO3 (TN or TCO, respectively, lanes 7 and 13). Samples were analyzed by SDS-PAGE on 7.5% gels and fluorography, except for thermolysin treated thylakoids, which were analyzed on 12.5% gels (a, lanes 14 and 15). Translation products (tp, lanes 1) represent 2% and all other samples represent 5% of the amount present in each assay. The precursor, intermediate, and mature forms of the proteins are designated p, I, and m, respectively, on the right side of the panels. Mr values in parentheses next to the various species were estimated from their migration compared to standard marker proteins.
To verify the proper integration into the membrane, aliquots of the thylakoid fraction were extracted with 100 mM NaOH, 200 mM Na2CO3, or treated with thermolysin. The FtsH2 protein was resistant to extraction with NaOH (Figure 2 a, TN, lane 7) and thermolysin treatment produced a partial degradation product of ∼ 13-kDa (Figure 2a, lane 14), which is the expected size of the lumenal tail plus one transmembrane domain. That the protease resistance of the FtsH2 fragment is due to protection by the membrane bilayer was verified by treatment of thylakoids with thermolysin plus 1% Triton X-100, which resulted in complete degradation (Figure S2). These characteristics are in agreement with those observed for the previously characterized pepper chromoplast FtsH protein Pftf (Summer et al., 2000) (Figure 2a). For the following experiments, the presence of the characteristic protease protected fragment or resistance to NaOH was taken as an indicator of proper integration of FtsH2.
In vitro translation of FtsH5 either with a homemade wheat germ system or a TNT coupled Transcription and Translation Kit (Promega) produced both the 75-kDa full length precursor protein and an additional C–terminally truncated protein of ∼ 25-kDa (Figure S3). Production of C-terminally truncated proteins (early quitters) from large mRNAs is not unusual. This truncated precursor appeared to be imported into the chloroplasts and processed to ∼17-kDa, which is very close to the migration of the expected protease-protected product. For this reason, resistance to extraction with Na2CO3 was used to assess integration of FtsH5 into membrane. Imported and thylakoid localized FtsH5 was fully resistant to extraction with the alkaline 200 mM Na2CO3 (Figure 2 b, TCO, lane 7), although not resistant to extraction with 100 mM NaOH (data not shown). Resistance to 200 mM Na2CO3 is a generally accepted criterion for a membrane anchored proteins, although some membrane proteins are resistant to the harsher 100 mM NaOH treatment. It's not clear why FtsH2, but not FtsH5, is resistant to NaOH, as both have strongly hydrophobic transmembrane anchors as determine by Membrane Protein Explorer. Nevertheless, resistance to 200 mM Na2CO3 was taken as an indicator of proper integration of FtsH5.
A two-step processing mechanism for localization of FtsH2 and FtsH5 is suggested by the prediction of sequentially cleaved targeting peptides, i.e. cleavage by the stromal peptidase produces an intermediate sized stromal form; cleavage by the thylakoid signal peptidase produces a smaller integrated mature protein. This was tested by import in the presence of inhibitors characteristic for the Tat and SecA/SecYE pathways. For pFtsH2, the chloroplast import reaction was conducted in the presence of ionophores nigericin and valinomycin, which dissipates the proton motive force, the driving force for the Tat pathway. This prevented FtsH2 membrane integration (Figure 2a, lanes 13 and 15) and resulted in accumulation of an intermediate sized form (lane 10) that was found both in the stromal fraction and associated with but not integrated into the membranes. Nigericin/valinomycin has a limited and variable effect on the Sec pathway (Yuan and Cline, 1994) and had virtually no effect on pFtsH5 integration (data not shown and Figure 3, lane 6). However, azide, which inhibits SecA of the Sec pathway (Yuan and Cline, 1994), partially inhibited integration of FtsH5 (Figure 2b, lane 13), resulting in accumulation of an intermediate-sized form largely associated with the membrane fraction (lane 11) and which was largely extracted by Na2CO3. Interestingly, the truncated FtsH5 protein accumulated an intermediate-sized form in the stroma and membranes of azide-treated chloroplasts, and the membrane-associated form was extracted by Na2CO3 (Fig S3). Thylakoid transport of the control SecA/SecYE substrate pOE33 was also inhibited by sodium azide and accumulated the intermediate iOE33 (Figure 2b).
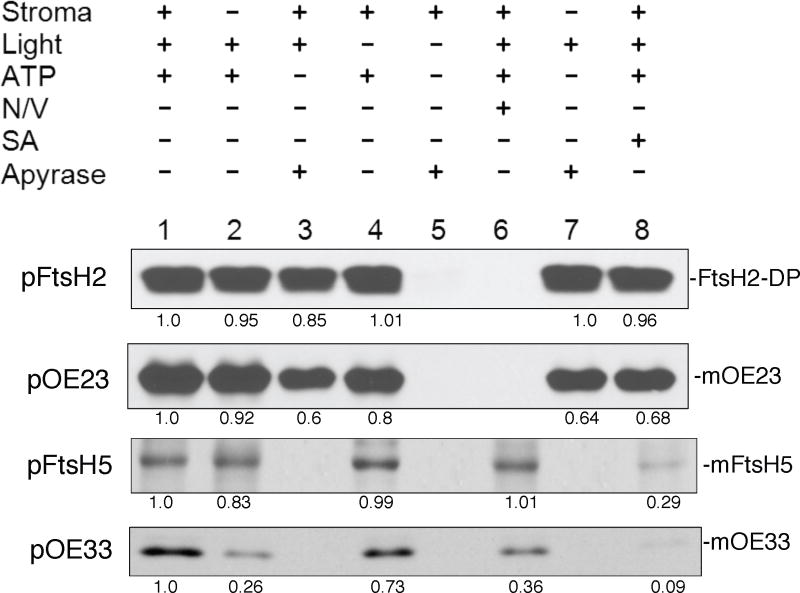
Figure 3.
pFtsH2 integration into isolated thylakoids requires the thylakoidal proton motive force whereas FtsH5 integration requires NTPs. The requirements for integration into isolated pea thylakoids was examined by incubating radiolabeled pFtsH2, pFtsH5, pOE23 or pOE33 for 30 min at 25 °C under the conditions depicted above the panels. These include the absence or presence of stromal extract, light, 5 mM ATP, nigericin and valinomycin (N/V), sodium azide (SA), and apyrase. The recovered thylakoids were treated with thermolysin (for pFtsH2, pOE23 and pOE33) or extracted with 200mM Na2CO3 (for pFtsH5). The samples were analyzed by SDS-PAGE and fluorography on 12.5% gels. Bands in the fluorograms were quantified using Image J software and the amounts in each band relative to the band in lane 1 of each panel are depicted below the panels.
The results demonstrate that pFtsH2 and pFtsH5 possess bipartite transit peptides with a chloroplast-targeting domain that is removed after import into the stroma and a signal peptide that is removed after integration into thylakoids. Combined with the protease and alkaline extraction data in Figure 2 and Figure S3, this supports a model in which both FtsH2 and FtsH5 have a single transmembrane anchor and a large carboxyl-proximal stroma-exposed catalytic domain.
pFtsH2 requires the proton gradient whereas pFtsH5 requires NTPs for integration
To further examine the requirements specific for thylakoid integration, pFtsH2 and pFtsH5 were directly assayed for integration into isolated thylakoids and compared with the Tat substrate pOE23 and the SecA/SecY substrate pOE33 (Figure 3). In the presence of stromal extract, light to generate a proton gradient, and ATP, both proteins were integrated into the membrane (lanes 1) as assessed by the partial protection against thermolysin post-treatment (for FtsH2) or resistance to extraction with 200 mM Na2CO3 (for FtsH5). FtsH2 integration was dependent on the proton motive force as it was inhibited by the ionophores nigericin and valinomycin in the light (lane 6) and in the dark was ATP-dependent, via reverse action of the ATP synthase (Figure 3, lanes 4 and 5). In the presence of light FtsH2 integration occurred without stromal extract (Figure 3, lanes 2 and 7) and without NTPs, which were scrubbed from the reaction with apyrase (lanes 3, and 7), and in presence of sodium azide (lane 8). Identical requirements were exhibited for transport of the Tat substrate pOE23. pFtsH5 integration was dependent on NTPs (lanes 3 and 7) and reduced in samples treated with sodium azide (lane 8). The integration was unaffected by ionophores (lane 6). pFtsH5 requirements are similar to the requirements for transport of the SecA/SecY substrate pOE33, with two notable differences; FtsH5 integration was virtually unaffected by the absence of stromal extract, the main source of SecA (lane 2), whereas pOE33 transport was significantly reduced, and FtsH5 integration was reduced by sodium azide whereas pOE33 transport was eliminated (lane 8). This might indicate a smaller dependence of FtsH5 than pOE33 on SecA, or possibly that an unidentified NTPase serves as a translocation motor for FtsH5 integration.
Antibody to Hcf106 inhibits pFtsH2 integration; antibody to SecY inhibits pFtsH5 integration
The involvement of Tat and Sec translocation machinery in FtsH integration was tested by antibody inhibition assays, whereby thylakoids were pretreated with antibodies to components of the three thylakoid pathways (Mori et al., 1999a; Mori and Cline, 2001). As shown in Figure 4, treatment of thylakoids with antibody to the Tat component Hcf106 prevented integration of FtsH2 (lanes 2), whereas treatment of thylakoids with antibody to chloroplast SecY (lane 3) inhibited the integration of FtsH5 (lanes 3). Integration of either protein was unaffected by antibodies to the chloroplast SRP pathway component Alb3 (lanes 4). The effects on antibody treatments on the control proteins pOE23 (Tat), pOE33 (SecA/SecYE), and pLHCP (SRP/Alb3) confirmed the efficacy and specificity of the treatments. As an additional test for pathway specificity, the integration pathways of FtsH2 and FtsH5 was queried by an in organello competition assay, wherein a pre-import incubation accumulates thylakoid saturating concentrations of either a Tat substrate (iOE23) or a Sec substrate (iOE33) before challenge with radiolabeled precursor proteins. As expected, the Tat substrate competed for FtsH2 localization, whereas the SecA/SecYE substrate competed for FtsH5 localization (Figure S4).
Figure 4.
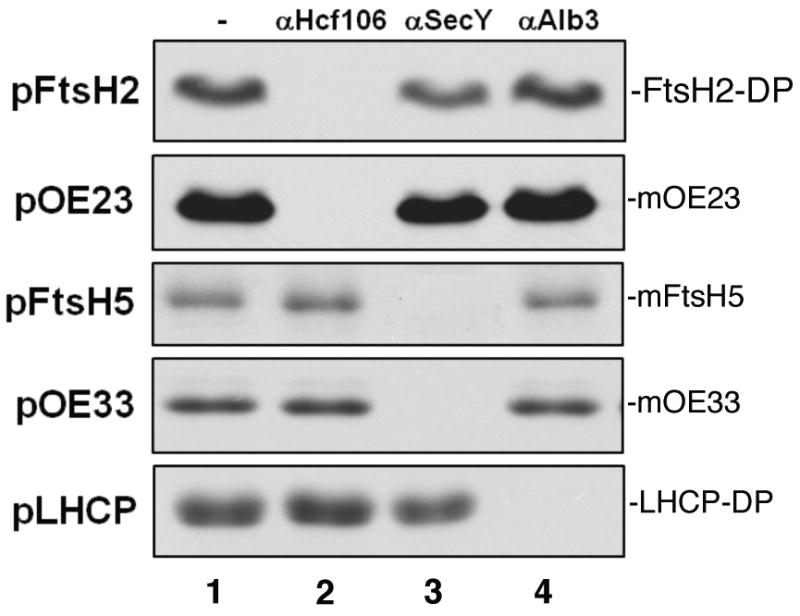
Pre-treating thylakoids with Hcf106 antibody prevents integration of pFtsH2, whereas pre-treating with SecY antibody prevents pFtsH5 integration. For antibody inhibition, pea thylakoids were incubated without (-) or with anti-Hcf106 IgG (1 mg/ml), anti-SecY serum, or anti-Alb3 IgG (1 mg/ml) for 1 h on ice. Thylakoids were washed with import buffer, supplemented with stromal extract, and incubated with radiolabeled pFtsH2, pFtsH5, pOE23, pOE33 or pLHCP and 5 mM ATP in the light at 25 °C for 30 min. The samples were then treated with thermolysin (for pFtsH2, pOE23, pOE33 and pLHCP) or 200 mM Na2CO3 (for pFtsH5). Samples were analyzed by SDS-PAGE and fluorography on 12.5% gels. FtsH2-DP is the thermolysin degradation product of the integrated protein and LHCP-DP is the characteristics thermolysin degradation product of integrated LHCP.
The amino-proximal regions of pFtsH2 and pFtsH5 are sufficient for thylakoid integration
Previous work in many laboratories has shown that targeting specificity to SecA/SecYE or Tat pathways is governed by the signal peptide. However, a previous attempt by us to characterize targeting determinants in vivo suggested that the mature domain of FtsH5 was important for thylakoid targeting (unpublished results). Specifically GFP was fused in frame to the amino terminal sequences of FtsH2 and FtsH5 from the amino terminus to the second hydrophobic domain carboxyl flanking sequence. Although pFtsH2-GFP was correctly localized to thylakoids in tobacco transgenic plants, pFtsH5-GFP accumulated in the stromal compartment. In order to clarify whether or not all of the targeting specificity resides in the amino terminal domains of pFtsH2 and pFtsH5, truncated versions of pFtsH2 (pFtsH2Δ) and pFtsH5 (pFtsH5Δ), containing the amino terminal sequences but lacking the stroma-facing enzymatic domain, were prepared by in vitro transcription and translation. As shown in Figure 5, both truncated precursors, pFtsH2Δ and pFtsH5Δ, were able to correctly localize in an in vitro chloroplast import assay. pFtsH2Δ was imported into chloroplasts (left panel, C, C+) and inserted into thylakoids, as confirmed by protection against protease treatment (T, T+) and NaOH extraction (TN). pFtsH5Δ was imported into chloroplasts (right panel, C, C+) and inserted into thylakoids, as confirmed by protection against protease treatment (T, T+) and Na2CO3 extraction (TCO). These results demonstrate that all necessary requirements for import of pFtsH2 and pFtsH5 into isolated chloroplasts and subsequent integration into thylakoids reside in the N-terminal domain.
Figure 5.
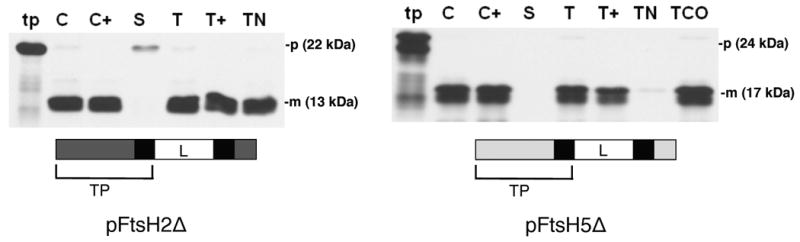
The N-terminal regions of pFtsH2 and pFtsH5 are able to insert into thylakoid membranes. Radiolabeled pFtsH2Δ or pFtsH5Δ (as diagrammed below the panels) was incubated with intact pea chloroplasts in a reaction containing 5mM ATP and light at 25 °C for 20 min. Intact chloroplasts were recovered (C) and treated with thermolysin (C+). Untreated intact chloroplasts were fractionated into stroma (S) and thylakoids. Thylakoid aliquots were washed with import buffer (T), treated with thermolysin (T+) or extracted with 100 mM NaOH (TN). For pFtsH5Δ, a thylakoid aliquot was extracted with 200 mM Na2CO3 (TCO). Samples were analyzed by SDS-PAGE on 12.5% gels and fluorography.
The pFtsH2 mature domain is incompatible for integration by the Sec-pathway
As an initial test for the underlying molecular basis for dual integration pathways for homologous proteins of the same multimeric complex, we prepared chimeric precursor proteins, in which the transit peptide (including the signal peptide) of one precursor protein was fused to the mature domain of the other, producing TP2Mat5 and TP5Mat2 (see Figure 1a and 1b, points a). As shown in Figure 6a the chimeric precursor TP2Mat5 (76-kDa) was imported into chloroplasts, localized to thylakoids, and processed to a mature form (67-kDa, lane 2). Because in vitro translation of TP2Mat5 only produces the full length precursor protein, protease treatment of thylakoids was used to verify membrane integration. The protease treated membranes produced a protected fragment of ∼17–kDa (lane 11), confirming integration. Essentially the same results were obtained in a parallel assay but with the chloroplasts pre-treated with sodium azide (lanes 8, 9, 10 and 13). By contrast, when the chloroplasts were treated with nigericin and valinomycin (N/V), the imported protein accumulated as intermediate sized precursor (∼72-kDa) in the stromal fraction (lane 6) and thylakoids, which was completely degraded by protease treatment (lane 12). These results indicate that the pFtsH2 transit peptide and signal peptide are able to direct import and integration of the FtsH5 mature domain by the Tat pathway.
Figure 6.
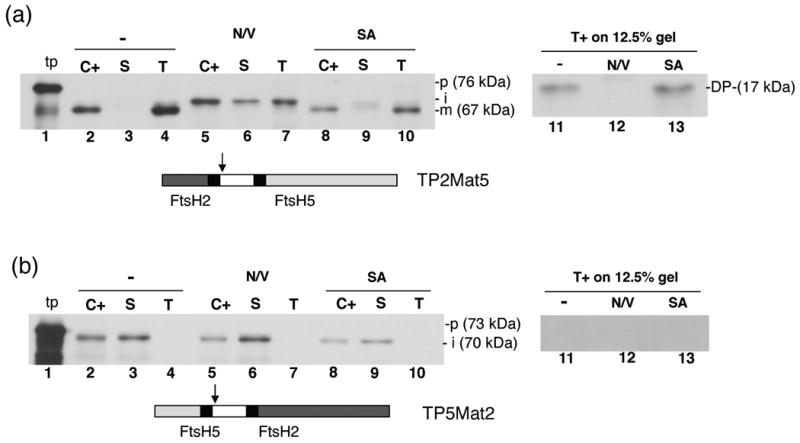
The FtsH2 mature domain is incompatible for transport by the Sec-pathway. Radiolabeled TP2Mat5 (a) or TP5Mat2 (b) as diagrammed below the panels was incubated with intact pea chloroplasts in a reaction containing 5 mM ATP and light at 25 °C for 20 min, in the presence or absence of nigericin and valinomycin (N/V) or sodium azide (SA). Intact chloroplasts were recovered from reaction and treated with thermolysin (C+). Untreated intact chloroplasts were fractionated into stroma (S) and thylakoids. Thylakoid aliquots were washed with import buffer (T) or treated with thermolysin (T+). Samples were analyzed by SDS-PAGE and fluorography on 7.5% gels except for the T+ samples, which were analyzed on a 12.5% gel. Arrows on diagrams denote swapping points between FtsH5 and FtsH2 sequences.
The chimeric precursor TP5Mat2 (73-kDa) was imported into chloroplasts and processed to a 70-kDa form (Fig 6, lane 2), which is slightly larger than the 65-kDa band expected after a thylakoid processing step. Interestingly, the processed form remained exclusively associated with the stromal fraction (lane 3). The same result was obtained with the nigericin/valinomycin treated chloroplasts (lanes 5, 6 and 7), and sodium azide treated chloroplasts (lanes 8, 9 and 10). This result indicates that the pFtsH5 transit peptide was not able to direct integration of the FtsH2 mature region into thylakoids, suggesting that the FtsH2 mature region has a conformational or sequence-based incompatibility with the Sec machinery.
The FtsH2 lumenal tail, transmembrane anchor, and stromal exposed regions are all required for incompatibility
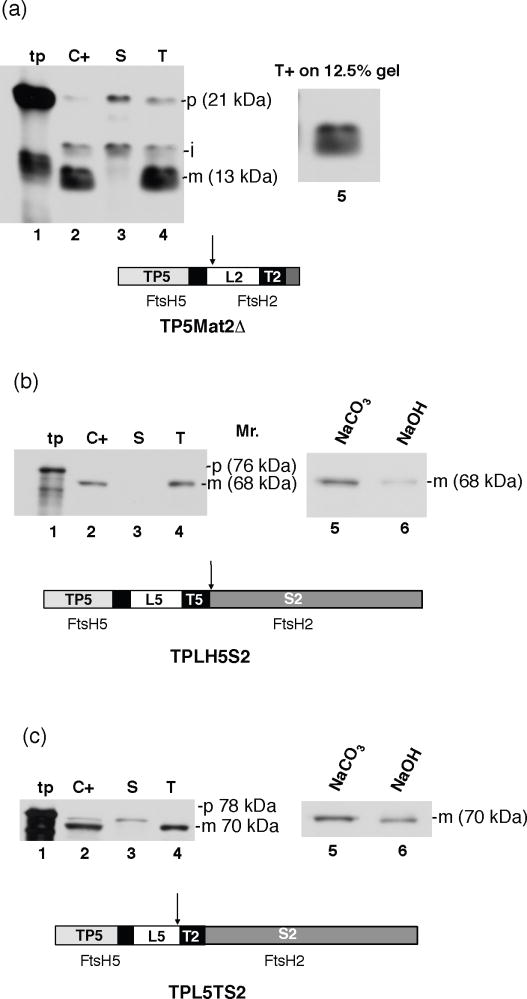
There are three different domains of the mature FtsH2 that could potentially cause incompatibility, the lumenal tail, the transmembrane anchor, and the large catalytic stromal domain. Three different fusion precursor proteins were constructed to test these domains for incompatibility: TP5Mat2Δ, which has the same swapping point of TP5Mat2 but lacks the stroma-exposed domain to test the FtsH2 lumenal (L) and transmembrane (T) domains (Figure 7a); TPLT5S2, in which the FtsH5 sequence through the transmembrane domain is fused to the FtsH2 stromal domain (Figure 7b); and TPL5TS2 in which the FtsH5 transit peptide and lumenal tail are fused to the FtsH2 transmembrane domain and stromal domain (Figure 7c). As shown in Figure 7, all three fusion proteins were imported into chloroplasts (lanes 2), proteolytically processed, and localized to thylakoids (lanes 4). Each protein was properly integrated into thylakoids as determined by either partial protease protection (Figure 7a, lane 5) or by resistance to 100 mM NaOH (Figure 7 c, lane 6) or resistance to 200 mM Na2CO3 (Figure 7 b, lane 5), depending on the source of the transmembrane anchor in the fusion protein. This result indicates that no single FtsH2 domain or combination of two FtsH2 domains prevent integration by the Sec machinery. Perhaps there is a folded structure consisting of the all domains that prevents access by the Sec machinery.
Figure 7.
Both the FtsH2 lumen exposed- and stroma exposed-domains are required for incompatibility with the Sec pathway. Radiolabeled TP5Mat2Δ, TPLT5S2, and TPL5TS2 were incubated with intact isolated pea chloroplasts in a reaction containing 5mM ATP in light at 25 °C for 20 min. Intact chloroplasts were recovered and treated with thermolysin (C+). Untreated intact chloroplasts were fractionated into stroma (S) and thylakoids. Thylakoids aliquots were washed with import buffer (T), treated with thermolysin (T+), or extracted with 200 mM Na2CO3 (Na2CO3) or with 100 mM NaOH (NaOH). Samples were analyzed by SDS-PAGE and fluorography. The transit peptide (TP), lumenal domain (L), and transmembrane domain (T) are labeled 2 or 5 to signify their origin from FtsH2 or FtsH5, respectively.
Discussion
Three classes of bacterially conserved ATP-dependent proteases in chloroplasts are the Clp, Lon, and FtsH proteases (for review see (Adam et al., 2006; Sakamoto, 2006)). Clp and Lon are located in the stroma and FtsH is located in the membrane. All possess the ATP-binding motif but differ in their catalytic domains for proteolysis. The FtsH protease was originally described in E. coli and shown to degrade short-lived proteins (for review (Ito and Akiyama, 2005)). FtsH, as well as orthologous FtsH proteins in different organisms, have two transmembrane domains, followed by an ATPase domain, and a zinc-binding motif, which serves as the catalytic site of the protease. These sites face the central pore of hexamers, access to which is controlled by the ATPase domain. The ATPase domain is also thought to translocate the polypeptide though the protease site as well as to dislocate membrane protein substrates out of the bilayer (Ito and Akiyama). Arabidopsis has twelve FtsH genes (FtsH1-12). Eight of the 12 FtsHs (FtsH1, 2, 5, 6, 7, 8, 9, 12) appear to reside in chloroplasts based on GFP fusion localizations (Sakamoto et al., 2003),three appear to be mitochondrial (FtsH3, 4, 10), and FtsH11 is dual targeted to chloroplasts and mitochondria (Urantowka et al., 2005). Chloroplast targeted FtsH1/FtsH5 (type A), FtsH2/FtsH8 (type B), and FtsH7/FtsH9 are closely related pairs based on sequence identity and genetic analysis. It is now clear that type A subunits and type B subunits combine to assemble a single type of multimeric complex (Yu et al., 2004; 2005). Type A and type B are the most abundant plastid FtsHs.
Our original intention was to include FtsH7/FtsH9 pair in this study because FtsH 7 has been detected in a proteomics study of the plastid envelope (Ferro et al., 2003) and appears to possess two transmembrane anchors, rather than the single anchors demonstrated here for FtsH2 and FtsH5. In preliminary work, we determined that pFtsH7 and pFtsH9 were imported into isolated chloroplasts and localized to the membrane fraction in an alkaline extraction resistant form (Rodrigues at al., unpublished). This confirms that FtsH7/9 are indeed plastid membrane proteins. Unfortunately, our attempts to define their routing pathways were unsuccessful.
The routing and integration pathways for type A and the type B subunits were much more amenable to in vitro studies. Our data indicate that both proteins are localized by a two-step pathway in which the precursor is imported into the plastid stroma where the chloroplast targeting domain of the transit peptide is removed, exposing a thylakoid targeting signal peptide that directs thylakoid integration. Following integration into thylakoids, the signal peptide is removed, producing mature proteins with amino-proximal lumenal tails of 118 (FtsH5) and 84 (FtsH2) residues, single transmembrane anchors, and large catalytic stromal domains. The topology of these domains was verified by protease treatment and alkaline extraction (Figures 2; Figures S2, S3). From the stroma, FtsH5 and FtsH2 are targeted to separate membrane integration machineries. The plastid Sec machinery integrates FtsH5, whereas the Tat machinery integrates FtsH2. That FtsH5 is integrated by the Sec pathway was demonstrated by the requirement for ATP, inhibition by the SecA inhibitor azide, competition by the Sec pathway substrate iOE33, and inhibition by pretreatment with antibodies to SecY. By analogy with the operation of the E. coli Sec system (Osborne et al., 2005; Driessen and Nouwen, 2008), we envision that SecA engages the hydrophobic signal peptide, which serves as a start-transfer sequence, and by a processive insertion of unfolded peptide segments through the SecYE channel, SecA delivers the lumenal domain across the thylakoid membrane. The transmembrane anchor, which would function as a stop transfer sequence, arrests further transport and is laterally released to the lipid bilayer.
Evidence that the Tat pathway integrates FtsH2 includes the PMF as sole energy requirement, competition by the Tat substrate iOE23, and inhibition by pretreatment with antibodies to the Tat component Hcf106. Previous work in our lab showed that a Capsicum FtsH protease, Pftf, implicated in chromoplast vesicle fusion, is also integrated by the Tat pathway (Summer et al., 2000). The defining feature of Tat-targeted FtsH proteins is the presence of a twin arginine motif. The consensus Tat motif for thylakoid proteins is R-R-X-Hyd-Hyd, where X is any amino acid and Hyd is a hydrophobic amino acid. FtsH2 and FtsH8 possess Tat motifs that fit the more stringent bacterial Tat motif, R-R-X-F-L-K, with a phenylalanine at position RR+2. The RR +2 F is important for transport efficiency in E. coli (Stanley et al., 2000) and can increase the binding affinity of thylakoid precursors that normally lack phenylalanine at this position (Gerard and Cline, 2007). A brief survey of the plant sequence database indicates that, in general, plants and green algae possess a Tat-targeted FtsH protein. FtsH2 is one of only a handful of membrane proteins integrated by the Tat machinery (Summer et al., 2000; Molik et al., 2001; Hatzixanthis et al., 2003). The manner by which Tat integrates membrane proteins is unclear because Tat transports fully folded protein domains with charged and hydrophilic surfaces (for review, see (Cline and Theg, 2007)). Experiments with artificial substrates showed that even a small exposed hydrophobic domain can terminate Tat transport (Richter et al., 2007), apparently causing disassembly of the transient translocase complex and release of the hydrophobic segment to the lipid bilayer.
Signal peptide swapping experiments with other substrates have shown that the Tat pathway can transport Sec substrate mature domains with moderate efficiency, but that the Sec pathway is nearly unable to transport Tat substrate mature domains (Henry et al., 1997). A similar situation exists for the FtsH proteases. The chimeric precursor TP2Mat5 was readily imported into chloroplasts and integrated into thylakoids by the Tat pathway. By contrast, TP5Mat2 was imported into plastids but incapable of integration. A tightly folded structure and/or the presence of basic residues near signal peptidase cleavage site appear to be two main reasons for incompatibility of Tat substrates on the Sec pathway (Bogsch et al., 1997; Hynds et al., 1998). We attempted to narrow the relevant incompatibility regions of FtsH2 by making chimeric constructs between FtsH5 and FtsH2. However, we found that no individual domain of FtsH2 was sufficient to abort Sec-mediated integration, suggesting that the entire FtsH2 mature domain presents a structure that interferes with either the recognition by or access to the Sec machinery.
Sakamoto (Sakamoto, 2006) notes that the multiplicity of FtsH isoforms is associated with photosynthetic organisms rather than non-photosynthetic organisms. Four FtsH isomers have been identified in the cyanobacterium Synechocystis sp. PCC6803. Of interest is that one of these isomers (sll1463) contains a twin arginine immediately preceding the first hydrophobic region, which is followed by a putative cleavage site (A-P-A). Thus the presence of chloroplast FtsH isomers integrated by the Tat pathway may be due to a gene duplication and pathway segregation that occurred in the photosynthetic prokaryote progenitor of chloroplasts. It is not unusual for a photosynthetic complex to contain subunits targeted by different thylakoid translocation pathways. For example plant chloroplast photosystem 2 contains subunits translocated by all four known thylakoid pathways. However, the situation for thylakoid FtsH proteins, where homologous subunits of the same complex take different integration pathways enroute to their functional form is highly intriguing. The specific factors that dictate the use of mechanistically diverse machinery may yet yield insight into the operations of Tat and Sec pathways for post-translational integration of membrane proteins.
Experimental Procedures
In-silico analysis
Arabidopsis FtsH sequences were obtained from TAIR database (www.arabidopsis.org). Stromal processing sites were predicted using ChloroP (http://www.cbs.dtu.dk/services/ChloroP) and transmembrane domains by TMPred (http://www.ch.embnet.org/software/TMPRED_form.html). Binary alignments were obtained using SIM and LALNVIEW (http://www.expasy.ch/tools/sim-prot.html).
Preparation of precursor proteins
Clones for pLHCP, pOE23 kD and pOE33 kD were previously described (Cline et al., 1993a). Nucleotide sequences encoding the full-length FtsH2 and FtsH5 cDNAs were amplified from clones kindly provided by Dr. W Sakamoto (Okayama University), using primers containing BamHI (forward) and PstI (reverse) restriction sites. Truncated versions for FtsH2 and FtsH5 were amplified with a forward BamHI-containing primer and an internal primer containing a stop codon and PstI site. Swappings between FtsH2 and FtsH5 were prepared by the splicing method (Horton et al., 1989), i.e by first amplifying the appropriate N- and C-terminal regions, purifying the resulting PCR products, and then PCR amplifying the spliced product using the outside forward BamHI and reverse PstI primers. Truncated versions and swapping points are indicated in Figure 1. All PCR products were digested with BamHI and PstI and ligated into BamHI/PstI-digested pGEM-4Z plasmid and transformed into XL1-Blue cells. All the resulting clones were sequenced entirely on both strands. Radiolabeled precursors were produced by transcription with SP6 polymerase (Promega) and translation with a homemade wheat germ translation system in the presence of 3[H]leucine (Cline, 1986). Translation products were diluted with one volume of 60 mM leucine in 2× import buffer (IB; 1× = 50 mM HEPES/KOH, pH 8.0, 0.33 M sorbitol) prior to use.
Chloroplast import and thylakoid integration
Intact chloroplasts were isolated from 9- to 10-day-old pea seedlings (Pisum sativum cv. Laxton's Progress 9) as described (Cline, 1986). Chlorophyll concentrations were determined according to (Arnon, 1949). Chloroplast lysate was prepared from intact chloroplasts and washed thylakoids and stroma prepared from lysate (Cline et al., 1993b). Import of radiolabeled precursors into pea chloroplasts or transport/integration into washed thylakoids or chloroplast lysate (0.33 mg chlorophyll/ml or equivalent) and 5 mM MgATP (unless otherwise stated) was conducted at 25 °C in 70 to 100 μE/m2-s white light (Cline et al., 1993b) for the time indicated in the figure legends. Where indicated, chloroplasts, lysates, or thylakoids were preincubated with nigericin (0.75 μM final concentration) and valinomycin (1.5 μM final concentration), or sodium azide (10 mM final concentration) on ice for 10 min. Reactions were initiated by the addition of translation products equivalent to one-sixth the assay volume and terminated by transfer to an ice bath. Intact chloroplasts were recovered, with or without protease treatment, by centrifugation through a 35% Percoll cushion. For fractionation, recovered chloroplasts were lysed by resuspension in 20 mM Hepes, KOH, pH 8, and incubation on ice for 5 min. The thylakoid membranes were separated from the stromal fraction by centrifugation for 8 min at 3200 g. Thylakoids were washed with import buffer, extracted with 100 mM NaOH, extracted with 200 mM Na2CO3, or treated with thermolysin as described (Summer et al., 2000). Thermolysin post-treatment of chloroplasts or thylakoids was with 1 μg thermolysin per microgram chlorophyll for at least 40 min at 4 °C followed by washing in import buffer containing 14 mM EDTA. Samples were analyzed by SDS–PAGE and fluorography. On the fluorograms, translation products represent 2% and all other samples represent 5% of the amounts present in each assay. Mr values for bands on gels were estimated from their migration compared to standard curves for marker proteins (EZ Run pre-stained protein ladder, Fisher Scientific)
In organello competition
In organello competition for Tat and Sec pathways was conducted essentially as described (Cline et al., 1993b). Unlabeled inclusion bodies of pOE23 or pOE33 were dissolved in fresh 10 M urea, 10 mM DTT for 3 h at room temperature. Isolated chloroplasts, 5 mM MgATP and 1.5mM DTT were preincubated with solubilized competitors for 7 min in the light at 25 °C. Competitors were aliquotted from stocks, such that the final competitor concentration was 0.75 μM pOE23, 0.75 μM pOE33, or no competitor, and the urea concentration was 0.25 M in all assays. Radiolabeled precursors for pFtsH2, pFtsH5, pOE23 or pOE33 were then added (1/6 volume), and the incubation was continued for an additional 10 min and then transferred to ice. Chloroplasts were repurified by centrifugation through Percoll cushions. Recovered chloroplasts were lysed in 10 mM HEPES/KOH, pH 8.0, and 10 mM MgCl2 buffer, and separated into stroma and thylakoids by centrifugation at 3200 g for 8 min. An aliquot of the thylakoid fraction was treated with 0.2 mg/ml thermolysin 40 min at 4 °C (FtsH2) or extracted with 200 mM Na2CO3 (FtsH5).
Antibody inhibition of thylakoid protein integration
Antibody inhibition of protein transport was conducted as described (Mori et al., 1999b). Briefly, washed thylakoids were suspended in import buffer containing 10 mM MgCl2 plus 3% BSA at 1 mg of chlorophyll/ml. The suspension was adjusted with 20 mM Hepes/KOH, pH 8, to a final chlorophyll concentration of 0.33 mg/ml and antibody concentrations as indicated in figure legends. After 1 h on ice, thylakoids were recovered by centrifugation at 3,200 g for 8 min and washed with import buffer containing 10 mM MgCl2. Aliquots of pretreated thylakoids (equivalent to 25 μg chlorophyll) were supplemented with stromal extract equivalent to 50 μg chlorophyll of intact chloroplasts and Mg-ATP (5 mM final concentration) and then incubated with radiolabeled proteins pFtsH2, pFtsH5, pOE23, pOE33 and pLHCP in a final volume of 75 μl. Reactions were conducted at 25 °C for 30 min in the light and terminated by transfer to ice. Thylakoids, recovered by centrifugation, were treated with 0.2 mg/ml thermolysin for 40 min at 4 °C or were extracted with 200 mM Na2CO3 (FtsH5).
Supplementary Material
Figure S1: Sequence alignment of chloroplast FtsH proteins. (a) FtsH2 and FtsH5 alignment; (b) FtsH5 and FtsH1 alignment (type A); (c) FtsH2 and FtsH8 alignment (type B). Binary alignments were obtained using SIM and LALNVIEW (http://www.expasy.ch/tools/sim-prot.html).
Figure S2: pFtsH2 is completely degraded by thermolysin without protection by the thylakoid bilayer.
Figure S3: In vitro translation of pFtsH5 RNA produces a truncated precursor in addition to the full-length pFtsH5.
Figure S4: pFtsH2 integration is competed in organello by a Tat precursor while FtsH5 is competed by a Sec precursor.
Acknowledgments
We thank Dr. Wataru Sakamoto for the FtsH cDNA clones. We thank Cassie Aldridge, Jose Celedon, Xianyue Ma, and Michael McCaffery for critical review of the manuscript, and Michael McCaffery for excellent technical assistance. This work was supported in part by National Institutes of Health grant R01 GM46951 to KC and Fundação de Amparo à Pesquisa do Estado de São Paulo grant 08/52067-3 to MCSF. MCSF is also a research fellow of CNPq. RAOR is a recipient of CAPES (BEX 0375/09-8) and FAPESP (2007/57806-6) fellowships.
References
- Adam Z, Rudella A, van Wijk KJ. Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Current Opinion in Plant Biology. 2006;9:234–240. doi: 10.1016/j.pbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Arnon DI. Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogsch E, Brink S, Robinson C. Pathway specificity for a delta pH-dependent precursor thylakoid lumen protein is governed by a ‘Sec-avoidance’ motif in the transfer peptide and a ‘Sec-incompatible’ mature protein. Embo J. 1997;16:3851–3859. doi: 10.1093/emboj/16.13.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K. Import of proteins into chloroplasts. Membrane integration of a thylakoid precursor protein reconstituted in chloroplast lysates. J Biol Chem. 1986;261:14804–14810. [PubMed] [Google Scholar]
- Cline K, Henry R, Li C, Yuan J. Multiple pathways for protein transport into or across the thylakoid membrane. Embo J. 1993a;12:4105–4114. doi: 10.1002/j.1460-2075.1993.tb06094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Henry R, Li C, Yuan J. Multiple pathways for protein transport into or across the thylakoid membrane. EMBO J. 1993b;12:4105–4114. doi: 10.1002/j.1460-2075.1993.tb06094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Theg SM. The Sec And Tat Protein Translocation Pathways In Chloroplasts. In: Dalbey RE, K C, Tamanoi F, editors. Molecular Machines Involved in Protein Transport across Cellular Membranes. XXV. London: Elsevier; 2007. pp. 463–492. [Google Scholar]
- Cline K, Dabney-Smith C. Plastid protein import and sorting: different paths to the same compartments. Curr Opin Plant Biol. 2008;11:585–592. doi: 10.1016/j.pbi.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Brugiere S, Miras S, Kowalski S, Louwagie M, Garin J, Joyard J, Rolland N. Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol Cell Proteomics. 2003;2:325–345. doi: 10.1074/mcp.M300030-MCP200. [DOI] [PubMed] [Google Scholar]
- Gerard F, Cline K. The thylakoid proton gradient promotes an advanced stage of signal peptide binding deep within the Tat pathway receptor complex. J Biol Chem. 2007;282:5263–5272. doi: 10.1074/jbc.M610337200. [DOI] [PubMed] [Google Scholar]
- Hatzixanthis K, Palmer T, Sargent F. A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol Microbiol. 2003;49:1377–1390. doi: 10.1046/j.1365-2958.2003.03642.x. [DOI] [PubMed] [Google Scholar]
- Henry R, Carrigan M, McCaffrey M, Ma X, Cline K. Targeting determinants and proposed evolutionary basis for the Sec and the Delta pH protein transport systems in chloroplast thylakoid membranes. J Cell Biol. 1997;136:823–832. doi: 10.1083/jcb.136.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Hynds PJ, Robinson D, Robinson C. The sec-independent twin-arginine translocation system can transport both tightly folded and malfolded proteins across the thylakoid membrane. J Biol Chem. 1998;273:34868–34874. doi: 10.1074/jbc.273.52.34868. [DOI] [PubMed] [Google Scholar]
- Ito K, Akiyama Y. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol. 2005;59:211–231. doi: 10.1146/annurev.micro.59.030804.121316. [DOI] [PubMed] [Google Scholar]
- Molik S, Karnauchov I, Weidlich C, Herrmann RG, Klosgen RB. The Rieske Fe/S protein of the cytochrome b6/f complex in chloroplasts: missing link in the evolution of protein transport pathways in chloroplasts? J Biol Chem. 2001;276:42761–42766. doi: 10.1074/jbc.M106690200. [DOI] [PubMed] [Google Scholar]
- Mori H, Summer EJ, Ma X, Cline K. Component specificity for the thylakoidal Sec and Delta pH-dependent protein transport pathways. J Cell Biol. 1999a;146:45–56. doi: 10.1083/jcb.146.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Summer EJ, Ma X, Cline K. Component specificity for the thylakoidal Sec and Delta pH-dependent protein transport pathways. J Cell Biol. 1999b;146:45–56. doi: 10.1083/jcb.146.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Cline K. Post-translational protein translocation into thylakoids by the Sec and DeltapH-dependent pathways. Biochim Biophys Acta. 2001;1541:80–90. doi: 10.1016/s0167-4889(01)00150-1. [DOI] [PubMed] [Google Scholar]
- Niwa H, Tsuchiya D, Makyio H, Yoshida M, Morikawa K. Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8. Structure. 2002;10:1415–1423. doi: 10.1016/s0969-2126(02)00855-9. [DOI] [PubMed] [Google Scholar]
- Osborne AR, Rapoport TA, van den Berg B. Protein translocation by the Sec61/SecY channel. Annu Rev Cell Dev Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- Richter S, Lindenstrauss U, Lucke C, Bayliss R, Bruser T. Functional Tat transport of unstructured, small, hydrophilic proteins. J Biol Chem. 2007;282:33257–33264. doi: 10.1074/jbc.M703303200. [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Zaltsman A, Adam Z, Takahashi Y. Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell. 2003;15:2843–2855. doi: 10.1105/tpc.017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W. Protein degradation machineries in plastids. Annu Rev Plant Biol. 2006;57:599–621. doi: 10.1146/annurev.arplant.57.032905.105401. [DOI] [PubMed] [Google Scholar]
- Stanley NR, Palmer T, Berks BC. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J Biol Chem. 2000;275:11591–11596. doi: 10.1074/jbc.275.16.11591. [DOI] [PubMed] [Google Scholar]
- Summer EJ, Mori H, Settles AM, Cline K. The thylakoid delta pH-dependent pathway machinery facilitates RR-independent N-tail protein integration. J Biol Chem. 2000;275:23483–23490. doi: 10.1074/jbc.M004137200. [DOI] [PubMed] [Google Scholar]
- Urantowka A, Knorpp C, Olczak T, Kolodziejczak M, Janska H. Plant mitochondria contain at least two i-AAA-like complexes. Plant Mol Biol. 2005;59:239–252. doi: 10.1007/s11103-005-8766-3. [DOI] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR. The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. Plant J. 2004;37:864–876. doi: 10.1111/j.1365-313x.2003.02014.x. [DOI] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR. Functional redundancy of AtFtsH metalloproteases in thylakoid membrane complexes. Plant Physiol. 2005;138:1957–1966. doi: 10.1104/pp.105.061234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Cline K. Plastocyanin and the 33-kDa subunit of the oxygen-evolving complex are transported into thylakoids with similar requirements as predicted from pathway specificity. J Biol Chem. 1994;269:18463–18467. [PubMed] [Google Scholar]
- Zaltsman A, Ori N, Adam Z. Two types of FtsH protease subunits are required for chloroplast biogenesis and Photosystem II repair in Arabidopsis. Plant Cell. 2005;17:2782–2790. doi: 10.1105/tpc.105.035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Sequence alignment of chloroplast FtsH proteins. (a) FtsH2 and FtsH5 alignment; (b) FtsH5 and FtsH1 alignment (type A); (c) FtsH2 and FtsH8 alignment (type B). Binary alignments were obtained using SIM and LALNVIEW (http://www.expasy.ch/tools/sim-prot.html).
Figure S2: pFtsH2 is completely degraded by thermolysin without protection by the thylakoid bilayer.
Figure S3: In vitro translation of pFtsH5 RNA produces a truncated precursor in addition to the full-length pFtsH5.
Figure S4: pFtsH2 integration is competed in organello by a Tat precursor while FtsH5 is competed by a Sec precursor.