Abstract
Bivalve molluscs are newly discovered models of successful aging, and this invertebrate group includes Arctica islandica, with the longest metazoan life span. Despite an increasing biogerontological focus on bivalves, their life history traits in relation to maximum age are not as comprehensively understood as those in vertebrate model aging organisms. We explore the allometric scaling of longevity and the relationship between development schedules (time to maturity and growth rate) and longevity in the Bivalvia. Using a traditional nonphylogenetic approach and the phylogenetically independent contrasts method, the relationship among these life history parameters is analyzed. It is demonstrated that in bivalves, maximum shell size, development, and growth rates all associate with longevity. Our findings support the observations of life history patterns in mammals and fish. This is the first investigation into the relationship among longevity, size, and development schedules throughout this group, and the results strengthened by the control for phylogenetic independence.
Keywords: Bivalves, Longevity, Phylogenetically independent contrast analysis
BIVALVE molluscs (clams, oysters, and mussels) are newly discovered models of natural aging (1), and the taxonomic class includes species with the longest metazoan life span— exceeding 400 years (2). The class Bivalvia also holds the record within the animal kingdom for the greatest number of species, which attain ages in excess of 150 years (1). For example, the Geoduck clam (Panopea abrupta), the freshwater pearl mussel (Margaritifera margaritifera), and the ocean quahog (Arctica islandica) are all exceptionally long lived attaining maximum ages of 163, 190, and 405 years, respectively (2–4).
In recognition of these animals attaining exceptional ages, there has been an increasing biogerontological focus on the group (3,5,6), and they have also been the subject of several aging reviews (1,7,8). In addition to its use in determining age, the hard calcareous shell of many bivalves also contains an ontogenetic record of growth in the form of annually resolved growth lines and increments (9). The advantage of this is that bivalves sampled from a range of environmental settings can provide reliable and accurate information on their individual ontogenetic life history, including their chronological age (10). Despite an increasing focus on bivalves from a biogerontological perspective, the relationship of their other life history traits to maximum age is not as comprehensively understood as those in vertebrate groups, such as mammals and birds. It would therefore be interesting to understand how life history traits and development schedules in bivalves are related to longevity and compare these with the relationships observed in humans and with traditional vertebrate model aging organisms.
Factors correlating with the maximum longevity in animal groups other than molluscs have received considerable attention, particularly with regard to life history theory (11,12). In mammals and birds, adult body size (most commonly represented as body mass) correlates positively with longevity, larger animals living, on average, longer than smaller ones (13–16). The most plausible explanation for the widely established relationship between body size and longevity appears to be the role of ecological factors. For example, larger animals are less prone to predation and thus have lower mortality rates, which in turn leads to a greater longevity and, according to evolutionary theory, the evolution of a slower aging process (17,18). Developmental schedules, such as time to maturity and postnatal growth rate, have also been associated with longevity in birds and mammals (16,19). Specifically, age at maturity is positively correlated with maximum species longevity (20) and negatively correlated with demographic aging rate (19), whereas postnatal growth rate are negatively correlated with maximum longevity in mammals (16) and with demographic aging rates in terrestrial vertebrates (19,21). The present study explores whether the same relationships hold within the Bivalvia. In the only study to investigate these life history traits in bivalves (22), concentrating on the freshwater mussels, concluded that overall, longevity was negatively related to the growth rate, which explained a high percentage of variation in longevity. By contrast, size and relative shell mass explained little variation in longevity.
Through the application of the phylogenetically independent contrast (PIC) method of Felsenstein (23), which statistically removes bias due to the effects of shared evolutionary history, de Magalhães and colleagues (16) demonstrated that time to maturity is associated with adult life span in mammals, with evidence suggesting an even stronger association than previously thought. The length of time a mammal will live after maturity, that is, its adult life span, is proportional to the amount of time it took to reach maturity. Ricklefs (19) noted the same relationship between time to maturity and actuarial aging more generally across all terrestrial vertebrates.
We investigate the allometric scaling of longevity and whether there is a relationship between development schedules and longevity in bivalves. Using a traditional nonphylogenetic approach (ie, simple regression of species values at the tip of the phylogeny), the relationship between maximum asymptotic size and maximum longevity was initially analyzed (see Table 1). Although traditional nonphylogenetic approaches have been historically used in comparative biology; it has been clearly demonstrated during the last 20 years that interspecific comparisons can be potentially compromised by statistical nonindependence of species values (23–26), leading to unacceptably high Type I error rates (incorrectly accepting alternate hypotheses) and inaccurate estimations of correlations or slopes (27–29), although under what circumstances such concerns are warranted is not clear (30).
Table 1.
Symbols and Abbreviations Used in This Study
| Parameter | Definitions |
| K | Parameter of the von Bertalanffy growth function, of dimension per year, expressing the rate at which the asymptotic length is approached |
| L∞ | Asymptotic length in millimeters; parameter of the VBGF expressing the mean length that the species would reach if indefinite growth |
| Tsex | Age at first maturity (years) |
| Tmax | Maximum age or life span reached in a population (years) |
| Tad | Age at first maturity substituted from the maximum age (Tmax − Tsex) |
| VBGF | von Bertalanffy growth function, used to describe the growth in length of fish and marine invertebrates |
The overall aim of our study is to investigate whether bivalves demonstrate similar life history strategies to those of terrestrial vertebrates where body size and developmental schedules are associated with longevity and therefore possibly with aging rates. In addition, species that do not fit the allometry of life span may hold clues about the evolutionary forces shaping longevity and aging (16). In this article, we specifically investigate using previously published data three hypotheses related to bivalve longevity (a) that bivalve size correlates positively with maximum longevity, (b) that postmaturational longevity is proportional to time to maturation, and (c) that the earlier a bivalve attains its maximum asymptotic size, the shorter will be its life span.
MATERIALS AND METHODS
The symbols and abbreviations of the growth parameters and life history traits to be investigated in this study are listed and defined in Table 1.
Data collection: Allometric Scaling of Longevity
Initially, we set out to investigate the relationship between body mass and maximum life span (Tmax) in bivalves in a similar manner to that which has been carried out by de Magalhães and colleagues (16) for a range of other animal groups. Unfortunately, body mass and maximum age data are unavailable for molluscs in sufficient quantities to allow meaningful analyses to be undertaken. However, data were available on maximum bivalve size, that is, the maximum asymptotic size (L∞; shell length or shell height) derived from the Von Bertalanffy growth equation (31,32). We have therefore used this dimension rather than mass in our analyses.
The values for Tmax that we have used refer to the published reported maximum life span of the oldest animal recorded in a population or the longevity estimate obtained from an age frequency analysis. Many species of bivalves contain a growth record in the form of annually deposited internal shell increments, and therefore, estimates of the age of individual bivalves are accurate and reliable (9). Using these two parameters (L∞ and Tmax), the allometric relationship between body size and longevity was investigated. To what extent Tmax is an accurate estimation of the rate of aging depends critically on the number of individuals sampled in a population, and the rate of mortality increase with age, so its utility has been a source of debate (15,16). The consensus is that to the extent that it measures real differences in achieved longevity, differences in Tmax are proportional to genetic limitations on longevity among species. Thus, it has been argued to be related to a species’ rate of physiological aging (33–35). There are additional estimates of aging rate, which could be used in place or alongside Tmax, such as the mean adult longevity or adult mortality rate doubling time, which is a demographic measurement of aging. However, the greater availability of estimates of Tmax in the literature governed our decision to use this parameter as the estimator of aging rate.
Data Collection: Developmental Schedules and Longevity
In the second part of our study, we investigated the relationship between developmental rate and longevity. Any estimate of species longevity incorporates developmental time, that is, time from birth to maturation, potentially adding bias to the analysis as discussed by de Magalhães and colleagues (16). Therefore, for this analysis, we have used the maximum adult life span (Tad), which is defined as Tmax minus age at sexual maturity (Tsex). Bivalves have varying reproductive strategies, including brooding, for example, some oyster species, although the majority employ external fertilization; therefore, Tsex can be defined simply as the age at which reproductive maturity is attained. Using these two parameters (Tad and Tsex), the relationship between development rate and longevity was investigated.
For proxy measures of developmental schedules, Tsex was used as a measure of a bivalve’s growth rate. Previous investigations of terrestrial vertebrates have used postnatal growth rate as a measure of developmental schedule (16,19); however, such data are unavailable for bivalves. Instead, we used the van Bertalanffy growth coefficient K (per year), an appropriate alternative that is widely available in the literature. This parameter expresses the rate at which L∞ is approached. Tsex is later demonstrated (see results later) to be directly proportional to adult life span. Therefore, the relationship between K and total longevity is investigated, not simply adult life span, due to the nature of available data.
We obtained from the literature growth and life history parameters for a total of 111 species and populations of bivalves from natural marine or estuarine or freshwater environments. The phylogenetic range of the bivalves included in each of the regression analyses and the PICs are presented in Table 2. In instances where the literature provided estimates for both sexes of a species, the arithmetic mean of the two was used. To exclude potentially problematic growth parameter estimates, data from species sampled from within heavily fished or other anthropogenically impacted areas were not included in our analyses. It is reasonably assumed that in such areas, estimates of Tmax are not reliable or representative of the actual achievable longevity for that species.
Table 2.
The Phylogenetic Coverage of the Bivalves Included in the Regression Analyses and the Analyses of Phylogenetic Independent Contrasts
| Parameters | N | No. of Species | No. of Families | No. of Orders | No. of Bivalve Subclasses | % Heterodonta |
| L∞ vs. Tmax | 56 | 56 | 25 | 7 | 4 | 70 |
| Tad vs. Tsex | 50 | 50 | 20 | 6 | 3 | 70 |
| Tmax vs. K | 35 | 35 | 20 | 6 | 4 | 82 |
| Contrasts | ||||||
| Contrasts of L∞ vs. Tmax | 55 | 56 | 25 | 7 | 4 | n/a |
| Contrasts of Tad vs. Tsex | 49 | 49 | 20 | 6 | 3 | n/a |
| Contrasts of Tmax vs. K | 34 | 35 | 20 | 6 | 4 | n/a |
Statistical Analyses
A traditional nonphylogenetic approach (ie, simple regression of species values ignoring phylogeny) was initially used to compare the relationship between five different parameters (L∞, Tmax, Tad, Tsex, and K). To meet the assumptions required for least squares linear regression, data were ln-transformed prior to statistical analysis. Although traditional nonphylogenetic approaches have been widely used in comparative biology, computer simulations (27,28) have demonstrated that such approaches can lead to unacceptably high Type I error rates (incorrectly accepting alternate hypotheses) and inaccurate estimations of correlations or slopes (29). To assess these potential problems, significant correlations from the traditional analyses were further investigated using the PIC method of Felsenstein (23).
Standardized independent contrasts were computed following the methods of Garland and Janis (29) and Garland and Adolph (36). To assure contrasts were appropriately standardized, the absolute values of the standardized contrasts were correlated with their standard deviations; if no correlation was apparent, the branch lengths adequately standardized the independent contrasts as described (37). The relationship between the standardized independent contrasts is investigated through ordinary least squares regression analysis, with regression lines constrained to pass through the origin (24,27,37–39). During the statistical analysis of PICs, associated p values can generally be determined by reference to conventional statistical tables (36). Therefore, the significance level was set at .05.
Phylogenetic Information
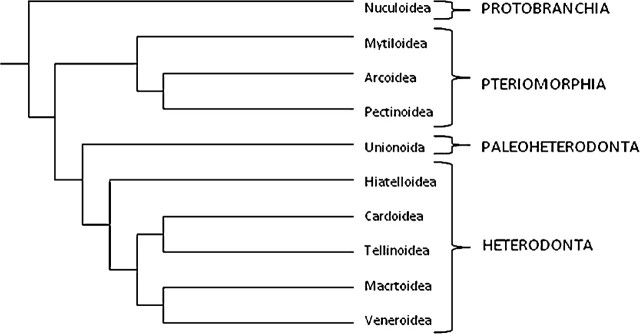
Although the precise topological structure of the bivalve phylogenetic tree is not completely resolved, the general relationships between the taxa are (for bivalve phylogenies, see (40–45)). This enabled a phylogenetic tree to be created (Figure 1), which contained accurate relationships of all the species used in the study. Where possible, the branch length data from Taylor and colleagues (45) were used; where no branch length data were available, an arbitrary branch length was used, this length was the mean of all branch lengths contained in the Taylor and colleagues (45) phylogeny, with branch lengths in the units substitutions per site. Although this latest molecular phylogeny was largely constructed to describe the phylogeny of the Heterodonta, Taylor and colleagues (45) utilized species from the order Palaeoheterodonta (Trigoniidae, Margaritiferidae, and Unionidae) as outgroups, so there is broad coverage of the class Bivalvia.
Figure 1.
Phylogenetic relationships of the class Bivalvia documenting the relationships between the four main subclasses and the main speciose orders and superfamilies (drawn from (45)).
Provided accurate phylogenetic topologies and branch lengths are used, the statistical power of independent contrasts is identical to that of traditional nonphylogenetic approaches (36). Although it is not ideal to utilize arbitrary branch lengths, computer simulations have demonstrated that independent contrasts are reasonably robust with respect to violation of assumptions (including errors in branch lengths: (28,46,47) providing arbitrary branch lengths are appropriately checked for statistical adequacy (37)).
RESULTS
Allometric Scaling of Longevity With Size
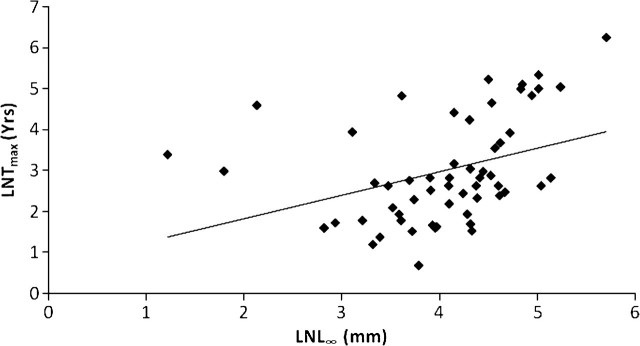
Data from 56 species of bivalves reveal a statistically significant positive impact of L∞ on Tmax (Figure 2, Table 2), although only 13.1% of the variation in maximum longevity is accounted for by asymptotic size. PIC analysis of the same 56 species confirmed this relation and even enhanced the statistical significance (Table 3). This result suggests that the evolution of body size in bivalves is associated with that of longevity.
Figure 2.
The relationship between the natural log of the asymptotic length (L∞; centimeters), derived from the von Bertalanffy growth formula, and the natural log of the maximum age (Tmax; years) attained by bivalve species. Nonphylogenetic regression analysis: n = 56, F = 8.72 and p = .004, r2 = .137.
Table 3.
Results of the Regression Analysis for Each Set of ln-Transformed Data, Documenting the df, r2 Value, F Statistic, and Associated p Value
| Parameters | df | r2 | F statistic | Associated p Value |
| L∞ vs. Tmax | 56 | .131 | 8.721 | .004* |
| PIC L∞ vs. Tmax | 55 | .138 | 11.278 | .001* |
| Tad vs. Tsex | 50 | .484 | 30.912 | <.001* |
| PIC Tad vs. Tsex | 49 | .302 | 18.995 | <.001* |
| Tmax vs. K | 35 | .643 | 86.343 | <.001* |
| PIC Tmax vs. K | 34 | .619 | 80.718 | <.001* |
Indicates the result is statistically significant at the 95% level. PICs = phylogenetically independent contrasts.
Developmental Schedules and Longevity
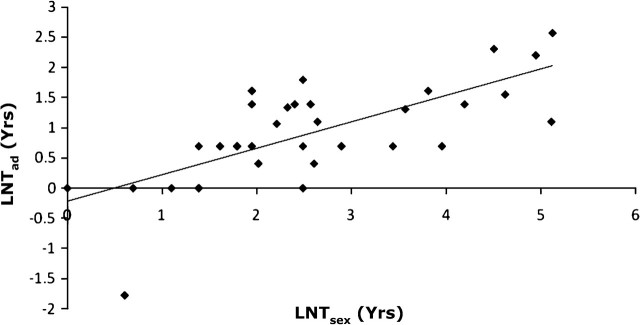
Data from 35 bivalve species were used to investigate the relationship between adult life span and age at maturity (in years; Table 2). In both traditional (Figure 3) and PIC analysis (Table 3), age at sexual maturity was significantly related to adult life span, explaining 48.4% and 30.2% of the variation, respectively. These results strongly suggest that development time, measured as time to maturity, not only influences adult life span but also may coevolve with adult life span. The amount of time a bivalve will live after maturity, that is, its adult life span, is proportional to the amount of time it took to attain maturity.
Figure 3.
The relationship between the natural log of the age at sexual maturity (Tsex) and the natural log of the adult life span (Tad; years) attained by bivalve species. Nonphylogenetic regression analysis: n = 50, F = 30.912 and p ≤ .001, r2 = .484.
Growth Rate and Longevity
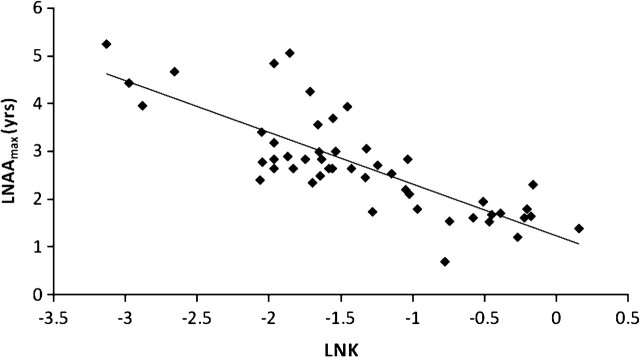
We then investigated the relationship between growth rate (K per year) and longevity (Figure 4). Analysis of 50 species demonstrated a robust significant negative relationship between K and Tmax using either nonphylogenetic (Figure 4) or PIC analyses (Table 3), explaining 64.3% and 61.9% of the variance, respectively. These results indicate that the faster a bivalve grows, the shorter its life span.
Figure 4.
The relationship between the natural log of the maximum age (years) attained by bivalve species (Tmax) and the natural log of the growth coefficient K as derived from the von Bertalanffy growth formula for bivalve species. Nonphylogenetic regression analysis: n = 35, F = 86.343 and p ≤ .001, r2 = .643.
DISCUSSION
This study is the first to investigate in invertebrates the much documented relationships between longevity and size and development schedules that occur in terrestrial vertebrates (16,19). More importantly, it investigates these relationships in the exceptionally long-lived molluscan class Bivalvia.
Allometric Scaling of Body Size and Longevity
In multiple vertebrate taxa, there is a well-established relationship between body size and longevity (16,48), for which de Magalhães and colleagues (16) suggest that evolutionary ecological factors appear the most plausible explanation. Simply, larger species are believed to be less prone to predation and therefore experience lower mortality rates, as seen in fish by Pauly (49). Greater survivorship in turn leads to longer life spans and, according to evolutionary theory, the evolution of a slower aging process (17,18).
We have demonstrated that body size in bivalves significantly correlates with longevity, a relationship that has been observed in many other animal taxonomic groups, including mammals (16) and freshwater mussels (22). Although a significant relationship was observed, body size in bivalves explains far less of the variation in longevity than in other taxonomic groups, only 13% compared with 49% in birds (16) and 66% in mammals (16). The mammalian data are probably the least analogous comparison deriving from largely captive populations. This reduced relationship between longevity and size in bivalves may be due to confounding factors such as burrowing ability and shell thickness among the group, factors that strongly influence their vulnerability to predation. For example, Kirby (50) demonstrated that in two species of the oyster genus, Crassostrea, longevity is strongly influenced by shell thickness, suggesting that shell thickness may provide greater resistance to extrinsic hazards (eg, predation or fluctuations in temperature) leading to a longer life.
Recent analyses investigated the relationship between size and longevity in the fresh water mussels and obtained similar results, although a significant positive correlation between size and longevity, size, and relative shell mass explained little variation in longevity (22).
Developmental Schedules and Longevity
Evolutionary theories of aging propose that the rate of aging is caused by the force of natural selection acting to optimize fitness early in life (51). The antagonistic pleiotropy theory of aging suggests the existence of pleiotropic genes that endow benefits early in life at the cost of deleterious effects later to explain the evolution of senescence (52), therefore proposing a trade-off between reproduction and longevity. In a classic article, Williams (52) expounded that reproductive maturation is the most significant milestone in the evolution of senescence and that senescence may theoretically initiate immediately after this stage in development, that is, “the sooner this point is reached, the sooner senescence should begin and the sooner it should have demonstrable effects.” Therefore, according to the theory, the time a bivalve will live after maturity should be proportional to the amount of time it took to reach maturity.
Mammalian maximum adult life span correlates with age at maturity (16,20). In support of these findings, Ricklefs (19) demonstrated that age at maturity is negatively correlated with demographic aging rate in terrestrial vertebrates. Ricklefs (19) states that the rate of aging within a species is primarily related to that of extrinsic mortality, and those young individuals must wait longer to enter the breeding population in species with lower extrinsic mortality rates. We have shown that maximum adult life span in bivalves is also significantly and positively correlated with age at maturity, a correlation that is predicted by the antagonistic pleiotropy theory of aging.
It has been suggested that the optimization theories of aging, including the antagonistic pleiotropy and the disposable soma theories of aging, may not be relevant to animals that demonstrate neither a decrease in fecundity with age or an observable decline in their fitness (53). In longer lived bivalves, there is no evidence of reproductive senility and gonad production continues, regardless of age, and as there is indeterminate growth (although a minimal amount at advanced ages), gonad production is reported to actually increase with age (54,55). Although at the individual level this will be the case, it is almost inconceivable that the mortality rate (age dependant and age independent) would not result in the older cohort of clams leaving less gametes than the younger cohorts (increase in gamete production with age in the population would be offset by loss of individuals from the population), which, according to the two optimization theories of aging, would result in natural selection acting to optimize fitness early in life, that is, senescence will creep into a population, albeit at a slow rate. These longer lived bivalves have also been suggested to be good examples of a group of organisms displaying negligible senescence (56) or possibly negative senescence (57). Our findings here align with, and were predicted by, the antagonistic theory of aging. Regardless of the growth and reproductive characteristics of the taxonomic class, we suggest that antagonistic pleiotropy is still a relevant theory for explaining aging in bivalves because, at the population level, the rate of extrinsic mortality results in a decrease in gamete production per age cohort, despite gamete production increasing with age at the individual level (55) and hence a reduction in the strength of natural selection with age.
The final hypothesis investigated the relationship between shell growth rate, another proxy measure of the developmental schedule, and longevity. Longevity is significantly negatively correlated with growth rate, expressed as the von Bertalanffy growth coefficient (K). In mammalian species, de Magalhães and colleagues (16) demonstrated that a fast postnatal growth rate in mammals is associated with a shorter life span. Furthermore, Ricklefs and Scheuerlein (21) and Ricklefs (19) have demonstrated that postnatal growth rate is negatively correlated with demographic aging rates in terrestrial vertebrates. Although the von Bertalanffy growth coefficient (K) is not the precise equivalent of postnatal growth rate, it is the closest analogous parameter available for bivalves. In the freshwater mussels, Haag and Rypel (22) also demonstrated longevity related negatively to the growth rate, K, which explained more than 75% of variation in longevity. These patterns remained when data were corrected for phylogenetic relationships among species, and furthermore, path analysis supported the conclusion that K was the most important factor influencing longevity both directly and indirectly through its effect on shell mass (22).
The two most commonly accepted optimization theories of aging, the antagonistic pleiotropy and the disposable soma theories of aging, suggest that growth rate should be associated with the life span of a species. Kirkwood and Holliday (58), the proponents of the disposable soma theory of aging, argue that “it may be selectively advantageous for higher organisms to adopt an energy-saving strategy of reduced accuracy in somatic cells to accelerate development and reproduction, but the consequence will be eventual deterioration and death.” Therefore, longevity is determined through the trade-offs between resources spent on somatic maintenance and reproduction (59). Williams (52) similarly predicted a trade-off between reproduction (reproductive success, fitness, and vigor) and longevity as part of the antagonistic pleiotropy model stating that “rapid individual development should be correlated with rapid senescence.”
Growth rate had much greater explanatory power (in terms of the r2 value) than either age at maturity or shell size. The explanatory power of growth rate on longevity observed in bivalves was very similar to that in other taxonomic groups, 0.67 in both mammals (not including cetaceans) and birds (16). Haag and Rypel (22) found similar, stating that 75% of the variation in the longevity of freshwater mussels was explained by the growth rate. It is probable that the reduced accuracy in somatic maintenance associated with the rapid growth rate has the most direct influence longevity, whereas the other parameters hold a more indirect complex relationship or there maybe confounding factors. As mentioned previously, the reduced relationship between longevity and size in bivalves may be due to confounding factors, which strongly influence their vulnerability to predation and other extrinsic factors (eg, burrowing and shell thickness).
Conclusions
This is the first demonstration of a relationship between longevity, size, and development schedules in bivalves, newly discovered models of natural aging (1), and the results are strengthened by the control for phylogenetic independence. From our results, it appears that in bivalves, maximum size, development, and growth are associated with longevity, and these findings are in accord with the two main evolutionary theories of aging and support the findings of other researchers working on mammals and fish. However, in comparison with the vertebrate studies to date, there is a reduced relationship between longevity and size in bivalves. These results therefore have implications in the design and interpretation of comparative studies of aging in bivalves. When investigating similar relationships in mammals, de Magalhães and colleagues (16) concluded that “developmental schedules such as the time to maturity and growth rates have a physiological basis that should be taken into consideration in the comparative biology of aging.” Ignoring this requirement may result in incorrectly assuming that the mechanisms involved in development are associated with aging rather than as a consequence of the correlation between developmental schedules and longevity (16,60). To further understand the actual biologic mechanisms that cause species to display different rates of aging, more detailed biochemical and molecular investigations are required (16).
FUNDING
This work was supported by a National Institute on Aging/Biotechnology and Biological Sciences Research Council Partnering Award to Support Collaborative Research on the Biology of Aging (R01).
References
- 1.Abele D, Strahl J, Brey T, Philipp EER. Imperceptible senescence: ageing in the ocean quahog Arctica islandica. Free Radic Res. 2008;42:474–480. doi: 10.1080/10715760802108849. [DOI] [PubMed] [Google Scholar]
- 2.Wanamaker ADJ, Heinemeier J, Scourse JD, et al. Very-long lived molluscs confirm 17th century AD tephra-based radiocarbon reservoir ages for north Icelandic shelf waters. Radiocarbon. 2008;50:399–412. [Google Scholar]
- 3.Ziuganov V, Miguel ES, Neves RJ, et al. Life span variation of the freshwater pearl shell: a model species for testing longevity mechanisms in animals. Ambio. 2000;29:102–105. [Google Scholar]
- 4.Strom A, Francis RC, Mantua NJ, Miles EL, Peterson DL. North pacific climate recorded in growth rings of geoduck clams: a new tool for paleoenvironmental reconstruction. Geophys Res Lett. 2004;31:L06206. [Google Scholar]
- 5.Strahl J, Philipp EER, Brey T, Abele D. Physiological ageing in the ocean quahog Arctica islandica from Iceland. Aquat Biol. 2007;1:77–83. [Google Scholar]
- 6.Abele D, Brey T, Philipp EER. Bivalve models of ageing and the determination of molluscan lifespans. Exp Gerontol. 2009;44:307–315. doi: 10.1016/j.exger.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 7.Bodnar A. Marine invertebrates as models for ageing research. Exp Gerontol. 2009;44:477–484. doi: 10.1016/j.exger.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Philipp EER, Abele D. Masters of longevity: lessons from long-lived bivalves—a mini-review. Gerontology. 2009;56:55–65. doi: 10.1159/000221004. [DOI] [PubMed] [Google Scholar]
- 9.Richardson CA. Molluscs as archives of environmental change. Oceanogr Mar Biol Annu Rev. 2001;39:103–164. [Google Scholar]
- 10.Austad S. The uses of intraspecific variation in ageing research. Exp Gerontol. 1996;31:453–463. doi: 10.1016/0531-5565(95)02068-3. [DOI] [PubMed] [Google Scholar]
- 11.Stearns S. The Evolution of Life Histories. Oxford: Oxford University Press; 1992. [Google Scholar]
- 12.Charnov EL. Life History Invariants: Some Explorations of Symmetry in EvolutionaryEcology. Oxford: Oxford University Press; 1993. [Google Scholar]
- 13.Lindstedt SL, Calder WA., III. Body size and longevity in birds. Condor. 1996;78:91–94. [Google Scholar]
- 14.Schmidt-Nielsen K. Scaling: Why Is Animal Size So Important? Cambridge: Cambridge University Press; 1984. [Google Scholar]
- 15.Promislow DE. On size and survival: progress and pitfalls in the allometry of life span. J Gerontol. 1993;48:B115–B123. doi: 10.1093/geronj/48.4.b115. [DOI] [PubMed] [Google Scholar]
- 16.de Magalhães JP, Costa J, Church GM. An analysis of the relationship between metabolism, developmental schedules, and longevity using phylogenetic independent contrasts. J Gerontol A Biol Sci Med Sci. 2007;62:149–160. doi: 10.1093/gerona/62.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirkwood TB, Austad SN. Why do we age? Nature. 2000;408:233–238. doi: 10.1038/35041682. [DOI] [PubMed] [Google Scholar]
- 18.Austad S. The Comparative Biology of Ageing. New York: Springer; 2001. [Google Scholar]
- 19.Ricklefs RE. Life-history connections to rates of aging in terrestrial vertebrates. Proc Natl Acad Sci U S A. 2000;107(22):10314–10319. doi: 10.1073/pnas.1005862107. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prothero J. Adult life span as a function of age at maturity. Exp Gerontol. 1993;28:529–536. doi: 10.1016/0531-5565(93)90041-b. [DOI] [PubMed] [Google Scholar]
- 21.Ricklefs RE, Scheuerlein A. Comparison of ageing-related mortality among birds and mammals. Exp Gerontol. 2001;36:845–857. doi: 10.1016/s0531-5565(00)00245-x. [DOI] [PubMed] [Google Scholar]
- 22.Haag WR, Rypel AL. Growth and longevity in freshwater mussels: evolutionary and conservation implications. Biol Rev. 2010 doi: 10.1111/j.1469-185X.2010.00146.x. doi:10.1111/j.1469-185X.2010.00146.x. [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- 24.Harvey PH, Pagel MD. The Comparative Method in Evolutionary Biology. Oxford: Oxford University Press; 1991. [Google Scholar]
- 25.Garland TJ, Dickerman AW, Janis CM, Jones JA. Phylogenetic analysis of covariance by computer simulation. Syst Biol. 1993;42:265–292. [Google Scholar]
- 26.Martins EP, Hansen TF, editors. The Statistical Analysis of Interspecific Data: A Review and Evaluation of Comparative Methods. Oxford: Oxford University Press; 1996. [Google Scholar]
- 27.Grafen A. The phylogenetic regression. Proc R Soc Lond B Biol Sci. 1989;326:119–157. doi: 10.1098/rstb.1989.0106. [DOI] [PubMed] [Google Scholar]
- 28.Martins EP, Garland TJ. Phylogenetic analyses of the correlated evolution of continuous characters: a simulation study. Evolution. 1991;45:534–557. doi: 10.1111/j.1558-5646.1991.tb04328.x. [DOI] [PubMed] [Google Scholar]
- 29.Garland T, Janis CM. Does metatarsal/femur ratio predict maximal running speed in cursorial mammals? J Zool. 1993;229:133–151. [Google Scholar]
- 30.Ricklefs RE, Starck JM. The application of phylogenetically independent constrasts: a mixed progress report. Oikos. 1996;77:167–172. [Google Scholar]
- 31.Bertalanffy Lv. A quantitative theory of organic growth (Inquiries on growth laws. II) Hum Biol. 1938;10:181–213. [Google Scholar]
- 32.Beverton RJH, Holt SJ. On the Dynamics of Exploited Fish Populations, Fishery Investigations. Ministry of Agriculture, Fisheries and Food; 1957. [Google Scholar]
- 33.Cutler RG. Evolution of human longevity: a critical overview. Mech Ageing Dev. 1979;9:337–354. doi: 10.1016/0047-6374(79)90110-6. [DOI] [PubMed] [Google Scholar]
- 34.Allman J, McLaughlin T, Hakeem A. Brain weight and life-span in primate species. Proc Natl Acad Sci U S A. 1993;90:118–122. doi: 10.1073/pnas.90.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Magalhães JP, Costa J, Toussaint O. HAGR: the Human Ageing Genomic Resources. Nucleic Acids Res. 2005;33:537–543. doi: 10.1093/nar/gki017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garland TJ, Adolph SC. Why not to do two-species comparative studies: limitations on inferring adaptation. Physiol Zool. 1994;67:797–828. [Google Scholar]
- 37.Garland TJ, Harvey PH, Ives AR. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst Biol. 1992;41:18–32. [Google Scholar]
- 38.Garland T. Rate tests for phenotypic evolution using phylogenetically independent contrasts. Am Nat. 1992;140:500–519. doi: 10.1086/285424. [DOI] [PubMed] [Google Scholar]
- 39.Pagel M. A method for the analysis of comparative data. J Theor Biol. 1992;156:431–442. [Google Scholar]
- 40.Adamkewicz SL, Harasewych MG, Blake J, Saudek D, Bult CJ. A molecular phylogeny of the bivalve mollusks. Mol Biol Evol. 1997;14(6):619–629. doi: 10.1093/oxfordjournals.molbev.a025801. [DOI] [PubMed] [Google Scholar]
- 41.Canapa A, Marota I, Rollo F, Olmo E. The small-subunit rRNA gene sequences of venerids and the phylogeny of Bivalvia. J Mol Evol. 1999;48:463–468. doi: 10.1007/pl00006490. [DOI] [PubMed] [Google Scholar]
- 42.Oliver PG, Holmes AM. The Arcoidea (Mollusca: Bivalvia): a review of the current phenetic-based systematic. Zool J Linn Soc Lond. 2007;148:237–251. [Google Scholar]
- 43.Saavedra C, Peña JB. Phylogenetics of American scallops (Bivalvia: Pectinidae) based on partial 16S and 12S ribosomal RNA gene sequences. Mar Biol. 2006;150:111–119. [Google Scholar]
- 44.Owada M. Functional morphology and phylogeny of the rock-boring bivalves Leiosolenus and Lithophaga (Bivalvia: Mytilidae): a third functional clade. Mar Biol. 2007;150:853–860. [Google Scholar]
- 45.Taylor JD, Williams ST, Glover EA, Dyal P. A molecular phylogeny of heterodont bivalves (Mollusca: Bivalvia: Heterodonta): new analyses of 18S rRNA and 28S rRNA genes. Zool Scr. 2007;36:587–606. [Google Scholar]
- 46.Purvis A, Gittleman JL, Luh HK. Truth or consequences: effects of phylogenetic accuracy on two comparative methods. J Theor Biol. 1994;167:293–300. [Google Scholar]
- 47.Diaz-Uriarte R, Garland T., Jr. Effects of branch lengths errors on the performance of phylogenetically independent contrasts. Syst Biol. 1998;47:654–672. doi: 10.1080/106351598260653. [DOI] [PubMed] [Google Scholar]
- 48.Speakman JR. Correlations between physiology and lifespan–two widely ignored problems with comparative studies. Ageing Cell. 2005;4:167–175. doi: 10.1111/j.1474-9726.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- 49.Pauly D. Tropical fishes: patterns and propensities. J Fish Biol. 1998;53:1–17. [Google Scholar]
- 50.Kirby MX. Differences in growth rate and environment between tertiary and quaternary Crassostrea oysters. Paleobiol. 2001;27:84–103. [Google Scholar]
- 51.Chen D, Pan K, Palter JE, Kapahi P. Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell. 2007;6:525–533. doi: 10.1111/j.1474-9726.2007.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 53.Finch CE, Austad SN. History and prospects: symposium on organisms with slow ageing. Exp Gerontol. 2001;36:593–597. doi: 10.1016/s0531-5565(00)00228-x. [DOI] [PubMed] [Google Scholar]
- 54.Peterson CH. Quantitative allometry of gamete production by Mercenaria mercenaria into old age. Mar Ecol Prog Ser. 1986;29:93–97. [Google Scholar]
- 55.Galinou-mitsoudi S, Sinis AI. Reproductive cycle and fecundity of the date mussel lithophaga lithophaga (Bivalvia: Mytilidae) J Molluscan Stud. 1994;60:371–385. [Google Scholar]
- 56.Finch CE. Longevity, Senescence, and the Genome. Chicago: University of Chicago Press; 1990. [Google Scholar]
- 57.Vaupel JW, Baudischa A, Döllinga M, Roach DA, Gampe J. The case for negative senescence. Theor Popul Biol. 2004;65:339–351. doi: 10.1016/j.tpb.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 58.Kirkwood TB, Holliday R. The evolution of ageing and longevity. Proc R Soc Lond B Biol Sci. 1979;205:531–546. doi: 10.1098/rspb.1979.0083. [DOI] [PubMed] [Google Scholar]
- 59.Martin GM, Austad SN, Johnson TE. Genetic analysis of ageing: role of oxidative damage and environmental stresses. Nat Genet. 1996;13:25–34. doi: 10.1038/ng0596-25. [DOI] [PubMed] [Google Scholar]
- 60.de Magalhães JP, Church G.M. Cells discover fire: employing reactive oxygen species in development and consequences for ageing. Exp Gerontol. 2006;41(1):1–10. doi: 10.1016/j.exger.2005.09.002. 2006. [DOI] [PubMed] [Google Scholar]