The spectroscopic and stability properties and X-ray crystal structure of the H87C mutant of the 2Fe–2S ligand mitoNEET are reported. Strikingly, the single point mutation leads to changes in its absorbance and CD spectra and an increase of around sixfold in the stability of the 2Fe–2S clusters over the pH range 5–7. However, the crystal structure of the H87C mutant displays heterogeneity in a few key residues, including the introduced cysteine ligand. Nonetheless, the cluster is highly stabilized from release.
Keywords: mitochondrial outer membrane, 2Fe–2S clusters, CISD1, diabetes, pioglitazone
Abstract
MitoNEET is the only identified Fe–S protein localized to the outer mitochondrial membrane and a 1.5 Å resolution X-ray analysis has revealed a unique structure [Paddock et al. (2007 ▶), Proc. Natl Acad. Sci. USA, 104, 14342–14347]. The 2Fe–2S cluster is bound with a 3Cys–1His coordination which defines a new class of 2Fe–2S proteins. The hallmark feature of this class is the single noncysteine ligand His87, which when replaced by Cys decreases the redox potential (E m) by ∼300 mV and increases the stability of the cluster by around sixfold. Unexpectedly, the pH dependence of the lifetime of the 2Fe–2S cluster remains the same as in the wild-type protein. Here, the crystal structure of H87C mitoNEET was determined to 1.7 Å resolution (R factor = 18%) to investigate the structural basis of the changes in the properties of the 2Fe–2S cluster. In comparison to the wild type, structural changes are localized to the immediate vicinity of the cluster-binding region. Despite the increased stability, Cys87 displays two distinct conformations, with distances of 2.3 and 3.2 Å between the Sγ and the outer Fe of the 2Fe–2S cluster. In addition, Lys55 exhibits multiple conformations in the H87C mutant protein. The structure and distinct characteristics of the H87C mutant provide a framework for further studies investigating the effects of mutation on the properties of the 2Fe–2S cluster in this new class of proteins.
1. Introduction
MitoNEET is the first outer mitochondrial membrane (OMM) protein that has been identified to contain a 2Fe–2S cluster (Wiley, Paddock et al., 2007 ▶). Interestingly, 32 amino acids of its N-terminus comprise an atypical mitochondrial targeting sequence (Wiley, Murphy et al., 2007 ▶). The ability to localize green fluorescent protein to the OMM with this 32-amino-acid sequence confirmed this function. The C-terminus (33–108) of mitoNEET is cytoplasmically exposed; despite initial annotation as a zinc-finger protein, this domain contains a novel redox-active 2Fe–2S cluster (Paddock et al., 2007 ▶). X-ray structures (1.4–1.8 Å resolution) of mitoNEET (Paddock et al., 2007 ▶; Hou et al., 2007 ▶; Lin et al., 2007 ▶; Conlan, Paddock et al., 2009 ▶) revealed that the protomers of the homodimer intertwine to form the newly named ‘NEET’ fold (Paddock et al., 2007 ▶). This folded dimer consists of two domains: a β-cap domain at the C-terminus of the protein and a 2Fe–2S cluster-binding domain where each protomer binds one 2Fe–2S cluster.
MitoNEET was initially identified as a mitochondrial target for the antidiabetes drug pioglitazone (Colca et al., 2004 ▶). In recent years, mitochondrial dysfunction has been implicated in type 2 diabetes and the malfunction is thought to result from a diminished oxidative capacity of the mitochondrial electron-transport chains (Lowell & Shulman, 2005 ▶). Interestingly, knockout mitoNEET and mitoNEET-homolog mice studies showed diminished mitochondrial oxidative capacity and degenerative disease states (Wiley, Murphy et al., 2007 ▶, Chen, Kao, Chen et al., 2009 ▶). These findings imply a role for mitoNEET in the oxidation–reduction states of mitochondria, thereby underscoring the importance of understanding its redox activity.
The 2Fe–2S cluster of mitoNEET is coordinated by three cysteines (Cys72, Cys74 and Cys83) and one histidine (His87). Recently, we showed that the endoplasmic reticulum associated family member Miner1 shares the same unique 2Fe–2S ligation and protein fold (Conlan, Axelrod et al., 2009 ▶). The distinctive fold and binding pocket makes mitoNEET representative of a new structural class of Fe–S-containing proteins (Meyer, 2008 ▶; Lill & Mühlenhoff, 2008 ▶). Distinct from the classic 4Cys ferrodoxin and 2Cys,2His Reiske 2Fe–2S-bound clusters, mitoNEET represents a novel and interesting hybrid 2Fe–2S cluster protein.
In an effort to understand the unique properties of the NEET family of proteins, we now report the X-ray structure to 1.7 Å resolution of the H87C mutant as well as a more detailed characterization of its 2Fe–2S clusters. The H87C protein is reversibly reducible and the stability of the 2Fe–2S clusters is increased sixfold over a range of pH values. Unexpectedly, Cys87 shows two distinct conformations with 2.3 and 3.2 Å cluster bond lengths. In addition, the nearby residue Lys55, which hydrogen bonds to His87 in the wild-type protein, now displays concomitant changes, with distances of 2.5 and 3.6 Å to Cys87. The increased stability despite this structural heterogeneity demonstrates the likely importance of the His87–Lys55 interaction in the wild-type protein in tuning the appropriate redox potential and lability/stability of cluster transfer for the biological function of this protein.
2. Materials and methods
2.1. Protein cloning and mutagenesis
For crystallization, the portion of the CISD1 cDNA encoding the cytoplasmic domain of the protein (residues 33–108) and containing the point mutation H87C was originally subcloned as an sf-GFP fusion protein in a modified pET28a(+) vector (Novagen) that contained the superfolder GFP cDNA (Pédelacq et al., 2006 ▶) as described previously (Conlan, Paddock et al., 2009 ▶). This construct contained a thrombin cleavage site between the sfGFP and mitoNEET genes, facilitating purification. For all other studies, the portion of CISD1 cDNA (purchased from Open Biosystems) encoding the cytoplasmic domain was subcloned into bacterial expression vector pET28a(+) (Novagen) containing an N-terminal thrombin-cleavable His tag. This clone served as a template to create the H87C point mutation by PCR using the forward primer 5′-CTGTGATGGGGCTTGCACAAAACATAACG-3′ and the reverse primer 5′-CGTTATGTTTTGTGCAAGCCCCATCACAG-3′.
2.2. Protein expression and purification
When using the sfGFP fusion-protein construct, BL21 (DE3) RIL cells (Stratagene) transformed with the plasmid were grown and protein expression was induced as described previously (Wiley, Paddock et al., 2007 ▶; Conlan, Paddock et al., 2009 ▶). The cells were harvested by centrifugation and lysed by sonication. The fused protein was partially purified by use of ammonium sulfate cuts as described previously (Paddock et al., 2007 ▶). Subsequently, the fused protein was cleaved using thrombin. MitoNEET was then further purified on an S-100 size-exclusion column (GE Healthcare) equilibrated with 50 mM Tris–HCl pH 8.0 and 100 mM NaCl (Conlan, Axelrod et al., 2009 ▶). The mitoNEET fractions from size-exclusion chromatography were pooled and concentrated to 10 mg ml−1. The protein purity was assessed to be >99% using SDS–PAGE and an A 280/A 458 optical ratio of <2.3. The protein concentrations were determined by UV-absorbance spectroscopy using ∊280 = 9.13 mM −1 cm−1.
A second plasmid was constructed in which CISD1 (33–108) H87C was subcloned into pET28a containing an N-terminal cleavable His tag. This plasmid was transformed into BL21 (DE3) RIL competent cells and grown as described previously (Wiley, Paddock et al., 2007 ▶; Conlan, Paddock et al., 2009 ▶). The cells were harvested and resuspended in binding buffer (20 mM Tris–HCl pH 7.9, 5 mM imidazole, 500 mM NaCl). The protein was purified by Ni–NTA chromatography as described previously (Conlan, Axelrod et al., 2009 ▶) followed by size-exclusion chromatography (S-200, GE Healthcare) in 50 mM Tris–HCl pH 8.0, 100 mM NaCl. Cation-exchange chromatography (HiTrap, GE Healthcare) was performed for the H87C protein used in crystallization (Paddock et al., 2007 ▶).
2.3. Optical spectroscopy
All UV–visible absorption spectra were measured from the near-UV to the near-IR (250–700 nm) on a Cary50 spectrometer (Varian Inc., Palo Alto, California, USA) equipped with a temperature-controlled cell (T = 308 K; 10–20 µM protein in 25 mM Tris–HCl pH 8.0 and 100 mM NaCl). All CD spectra were recorded on an Aviv-2 Circular Dichroism Spectrometer at 298 K in a 0.2 cm (UV spectra) or 1.0 cm (visible spectra) path-length quartz cuvette. Each sample was at 0.3 mg ml−1 final protein concentration in 25 mM Tris–HCl pH 8.0 and 100 mM NaCl.
2.4. Stability measurements
The stability of the 2Fe–2S clusters was determined by monitoring their characteristic absorbance at 460 nm as a function of time at 308 K with ∼20 µM protein in 100 mM citrate or 100 mM Tris–HCl at varying pH values.
2.5. Crystallization
Crystals were obtained as described previously for wild-type mitoNEET from the same construct (Conlan, Paddock et al., 2009 ▶). The final crystallization conditions were 100 mM Tris–HCl pH 8.0, 100 mM NaCl, 15–20% polyethylene glycol 3000 and 5 mg ml−1 protein in the well equilibrated against 100 mM Tris–HCl pH 8.0 and 30–33% polyethylene glycol 3000 in the reservoir. Crystals were frozen (77 K) after a 1 min soak in 100 mM Tris–HCl pH 8.0 and 40% polyethylene glycol 3000 and were maintained frozen (77 K) in liquid nitrogen until data collection. X-ray diffraction data were collected at the Stanford Synchotron Radiation Laboratory.
2.6. X-ray data collection and structural determination
Frozen crystals were screened using the Stanford Automated Mounter (Cohen et al., 2002 ▶) operated by Blu-Ice (McPhillips et al., 2002 ▶). The data were recorded on BL9-2 using a Rayonix MX-325 CCD detector and on BL7-1 using an ADSC Q315R CCD detector. The data used to determine the structure of H87C mitoNEET were collected on the same beamlines using Blu-Ice (McPhillips et al., 2002 ▶) to a resolution of 1.7 Å, autoindexed and integrated with MOSFLM (Winn et al., 2011 ▶) and scaled with SCALA (Winn et al., 2011 ▶).
The structure of H87C was determined by molecular replacement using MOLREP (Winn et al., 2011 ▶) with the wild-type mitoNEET structure (PDB entry 2qh7) as the starting model (Pähler et al., 1990 ▶; Terwilliger & Berendzen, 1999 ▶). Data reduction and primary phasing at a resolution of 1.7 Å were accomplished as described previously (Paddock et al., 2007 ▶). Further structural refinement was achieved using REFMAC (Winn et al., 2011 ▶) and WinCoot (Emsley & Cowtan, 2004 ▶). The figures presented here were generated using PyMOL (DeLano & Lam, 2005 ▶).
3. Results
3.1. Differences in the optical properties indicate changes in the 2Fe–2S cluster of the H87C mutant protein
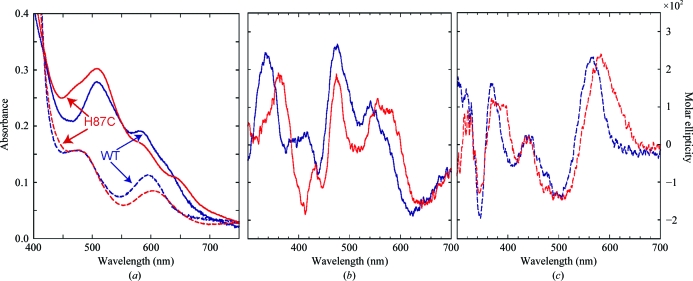
Engineering new ligands into Fe–S proteins can often be difficult owing to cluster instability or can be disruptive to the native fold of the protein (Kimura et al., 2005 ▶; Kounosu et al., 2004 ▶). The titratable His87 located near the 2Fe–2S cluster of human mitoNEET is implicated in affecting the assembly and stability of the cluster (Wiley, Paddock et al., 2007 ▶). To determine the extent to which this amino acid influences the properties of the cluster (e.g. redox potential/stability), we characterized the properties of mutant mitoNEET in which His87 was replaced by Cys (H87C). We collected both optical absorbance and circular-dichroism (CD) spectra. The UV–Vis absorbance spectra of wild-type mitoNEET and the H87C protein show the presence of 2Fe–2S clusters, but differ significantly from each other, indicating a change in the interaction between the 2Fe–2S cluster and the protein (Fig. 1 ▶ a). In contrast, the UV–Vis spectra obtained when another nearby amino acid Asp84 is replaced by Asn is essentially identical to that of the wild type (Wiley, Paddock et al., 2007 ▶), suggesting that changes to nearby residues do not necessarily affect the optical properties of the protein. The absorption spectrum of the H87C mutant (red trace) has a shoulder near 425 nm on its major peak near 460 nm; the 425 nm peak is indicative of a ferredoxin 4Cys-coordinated 2Fe–2S center (Jung et al., 1999 ▶). In addition, the 530 nm peak is diminished and shifted. These changes are consistent with an alteration in the coordinating ligands in the H87C mutant and the potential for heterogeneity in the coordinating geometries. As with wild-type mitoNEET, the H87C mutant protein is reversibly reducible (Fig. 1 ▶ a, dashed line).
Figure 1.
The optical properties of H87C differed from those of wild-type mitoNEET under both oxidizing and reducing conditions. (a) The absorbance spectrum (solid line) of wild-type mitoNEET is shown in blue and that of the point mutant is shown in red. While the overall shapes of the peaks are similar, the 330 and 460 nm peak intensities differ between the wild type and the H87C mutant. In addition, the 460 nm peak exhibits a shift and the smaller 530 nm peak is diminished and shifted. The absorbances of isolated wild-type and H87C mitoNEET are reduced (dotted line) upon addition of sodium dithionite. (b) The CD spectra of oxidized wild-type mitoNEET (blue) and H87C (red) protein are presented. The visible CD spectrum of the mutant also differs from that of the wild type. It has shifted peaks and an additional trough under oxidizing conditions (b). Once reduced (c), the visible spectrum of the mutant has shifted and broadened peaks and troughs compared with those of wild-type mitoNEET.
To further investigate the changes near the 2Fe–2S centers, we obtained circular-dichroism (CD) spectra. The UV CD spectra, which monitor the backbone conformation, of wild-type and H87C mitoNEET are largely similar (data not shown). This is true for both the oxidized and the reduced states of the 2Fe–2S clusters, thus indicating that the overall fold remains intact in the mutant.
The near-UV to visible wavelength range (300–700 nm) of the CD spectrum assesses changes to the stereochemistry of the 2Fe–2S clusters. Clear differences were noted between the spectra of the wild type and the H87C mutant in both oxidized and reduced forms (Figs. 1 ▶ b and 1 ▶ c). The wild-type and H87C mutant mitoNEET CD spectra are different from one another but are also distinct from those for both Reiske and ferrodoxin 2Fe–2S cluster proteins (Kimura et al., 2005 ▶; Jung et al., 1999 ▶).
3.2. The overall fold is intact in the mutant H87C protein
The H87C mutant protein crystallized in the same orthorhombic space group P212121 as the wild type, with unit-cell parameters a = 49.587, b = 49.311, c = 59.222 Å, α = β = γ = 90° (Table 1 ▶). The Matthews coefficient (V M) of the crystal was 2.0 Å3 Da−1, with an estimated solvent content of 37%. We determined the crystal structure of H87C by molecular replacement using the wild-type structure (PDB entry 2qh7; Paddock et al., 2007 ▶) as a starting model. X-ray diffraction intensities were collected on Stanford Synchrotron Radiation Laboratory (SSRL) BL7-1 and BL9-2 to an upper resolution of 1.7 Å (Table 1 ▶). The structural model refined with an R factor of 17.8% (R free = 22.6%).
Table 1. Summary of crystal parameters and data-collection and refinement statistics for H87C mitoNEET.
Values in parentheses are for the highest resolution shell.
| Space group | P212121 |
| Unit-cell parameters (Å, °) | a = 49.587, b = 49.311, c = 59.222, α = 90.00, β = 90.00, γ = 90.00 |
| Data collection | |
| Wavelength (Å) | 0.9795 |
| Resolution range (Å) | 38.0–1.70 |
| No. of observations | 114199 |
| No. of unique reflections | 16489 |
| Completeness (%) | 99.7 (99.8) |
| Mean I/σ(I) | 16.5 (2.8) |
| Rmerge† on I (%) | 6.2 (72.6) |
| Model and refinement statistics | |
| Cutoff criterion | |F| > 0 |
| Resolution range (Å) | 37.895–1.70 |
| No. of reflections (total) | 15652‡ |
| No. of reflections (test) | 793 |
| Completeness (%) | 99.511 |
| Rcryst§ | 0.1779 |
| Rfree¶ | 0.2256 |
| Stereochemical parameters | |
| Restraints (r.m.s. observed) | |
| Bond angles (°) | 1.7218 |
| Bond lengths (Å) | 0.0117 |
| Average isotropic B value (Å2) | 4.0451 |
| ESU†† based on Rfree (Å) | 0.0778 |
R
merge = 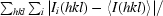
 , where I
i(hkl) is the scaled intensity of the ith measurement and 〈I(hkl)〉 is the mean intensity for that reflection.
, where I
i(hkl) is the scaled intensity of the ith measurement and 〈I(hkl)〉 is the mean intensity for that reflection.
Typically, the number of unique reflections used in refinement is slightly lower than the total number that were integrated and scaled. Reflections are excluded owing to systematic absences, negative intensities and rounding errors in the resolution limits and unit-cell parameters.
R
cryst = 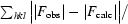
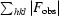 , where F
calc and F
obs are the calculated and observed structure-factor amplitudes, respectively.
, where F
calc and F
obs are the calculated and observed structure-factor amplitudes, respectively.
R free is the same as R cryst, but calculated using 5% of the total reflections that were chosen at random and omitted from refinement.
Estimated overall coordinate error.
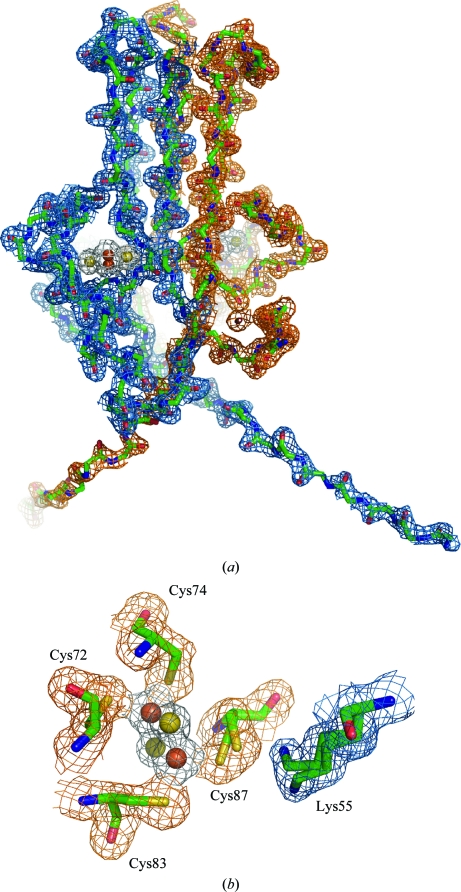
The H87C structure shares the same NEET fold as the wild type, with two intertwined protomers creating two domains: a β-cap and a cluster-binding domain (Fig. 2 ▶ a). The N-termini, which protrude from the bottom of the cluster-binding domain, are almost entirely resolved, similar to the wild-type structrure obtained using the same superfolder construct (Conlan, Paddock et al., 2009 ▶).
Figure 2.
MitoNEET H87C retains the CDGSH fold with multiple confirmations of the new coordinating Cys87. The H87C mutant protein retains the NEET-family fold (a), while the electron density of the iron–sulfur cluster domain (b) clearly shows the two confirmations of Cys87 (orange) and Lys55 (blue) from the partner protomer. (a) A backbone tracing of homodimeric H87C (green) with the observed 2F o − F c electron-density map (gray) contoured at 1.0σ. A 2Fe–2S cluster is present in each protomer and is rendered as yellow (sulfur) and red (iron) spheres. (b) An expanded view of one 2Fe–2S cluster (rotated ∼90° counterclockwise along the dyad axis) showing the cluster and iron ligands (O colored red, N blue, C green and S yellow) and the corresponding observed 2F o − F c electron density (gray) map contoured at 0.5σ and 1.5σ, respectively. The amino-acid ligands are indicated. The electron-density map was obtained from crystals of the soluble portion of the H87C mutant (resolved for residues 33–108) that diffracted to 1.7 Å resolution.
3.3. Multiple conformations of the Cys87 2Fe–2S ligand
Given the change in the optical and CD spectra discussed above, we performed a more detailed analysis of the cluster geometry and compared it with that of wild-type mitoNEET (Table 2 ▶). For comparison, the mitoNEET geometric parameters obtained by averaging the four deposited structures (PDB entries 2qh7, 2qd0, 2r13 and 3ew0; Paddock et al., 2007 ▶; Hou et al., 2007 ▶; Lin et al., 2007 ▶; Conlan, Paddock et al., 2009 ▶) are shown in Table 2 ▶. The most significant difference between the H87C cluster geometry and that of wild-type mitoNEET is the interaction between the His87/Cys87 side chain and the outer Fe.
Table 2. Summary of cluster geometry for wild-type and H87C mitoNEET.
| Parameter | Wild type | H87C | H87C − wild type |
|---|---|---|---|
| Bond lengths (Å) | |||
| Fe1—Fe2 | 2.72 | 2.73 | 0.01 |
| S1—S2 | 3.48 | 3.53 | 0.05 |
| Fe1—S1 | 2.19 | 2.20 | 0.01 |
| Fe1—S2 | 2.20 | 2.23 | 0.03 |
| Fe2—S1 | 2.24 | 2.24 | 0.00 |
| Fe2—S2 | 2.23 | 2.23 | 0.00 |
| Cys72—Sγ—Fe1 | 2.34 | 2.35 | 0.01 |
| Cys74—Sγ—Fe1 | 2.25 | 2.19 | −0.06 |
| Cys83—Sγ—Fe2 | 2.31 | 2.33 | 0.02 |
| His87—Nδ—Fe2 | 2.08 | ||
| Cys87—Sγ—Fe2 | |||
| Conformation 1 | 2.37 | 0.29 | |
| Conformation 2 | 3.18 | 1.10 | |
| Torsion angle (°) | |||
| Fe1—S1—S2—Fe2 | −177 | −176 | 1 |
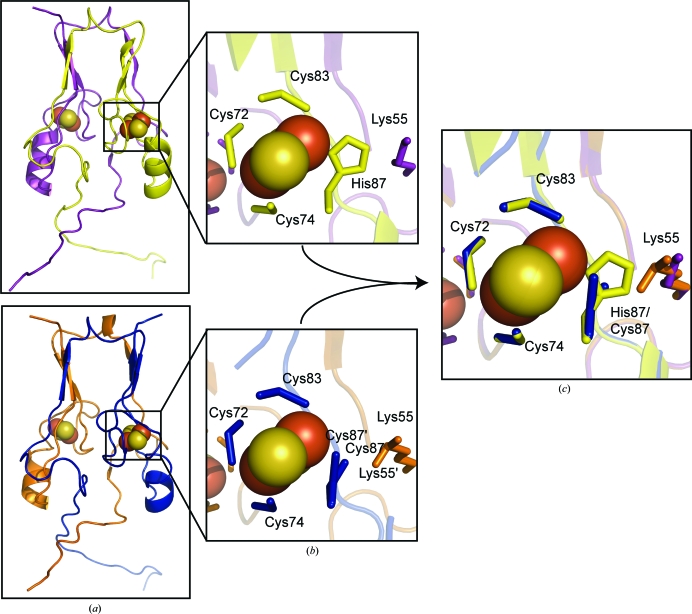
The sequence spanning Cys72–Cys87 comprises the segment containing the ligands of the 2Fe–2S clusters (Fig. 2 ▶ b). We find that the position of the Sγ atoms of Cys72, Cys74 and Cys83 are essentially the same as those observed in the wild-type protein (Fig. 3 ▶ c, Table 2 ▶). However, the side chain of Sγ of the mutant Cys87 displays two distinct positions in the crystal structure. Cys87 shows two distinct conformations with bond lengths of 2.3 and 3.2 Å, both of which are longer than the 2.08 Å distance from His87 to the outer Fe in the wild-type structures (Table 2 ▶). This heterogeneity is consistent with the complexity seen in the CD spectra and discussed above. An interesting additional structural change in the cluster-binding region is a rearrangement in the position of Lys55, which interacts with His87/Cys87 (Figs. 3 ▶ b and 3 ▶ c).
Figure 3.
H87C shares the same overall fold as mitoNEET, with specific changes in the 2Fe–2S cluster region. (a) The point mutant H87C (bottom) shares the homodimeric structure as wild-type mitoNEET (top). The overall structural features and fold of the mutant protein remain the same as those of the wild-type protein, indicating that any changes in properties are the result of the ligand change at the 2Fe–2S cluster. The cluster regions (b) differ owing to the substitution of the ligand. The distance of the coordinating Sγ of Cys87 in the mutant is different from the Nδ of His87 in the wild-type protein. In addition, the substitution causes a reorientation of Lys55 from the neighboring protomer. (c) shows a superposition of the 2Fe–2S region of wild type and H87C to better illustrate the specific changes in the 2Fe–2S cluster region.
3.4. Increased stability of the 2Fe–2S clusters in H87C while maintaining pH sensitivity
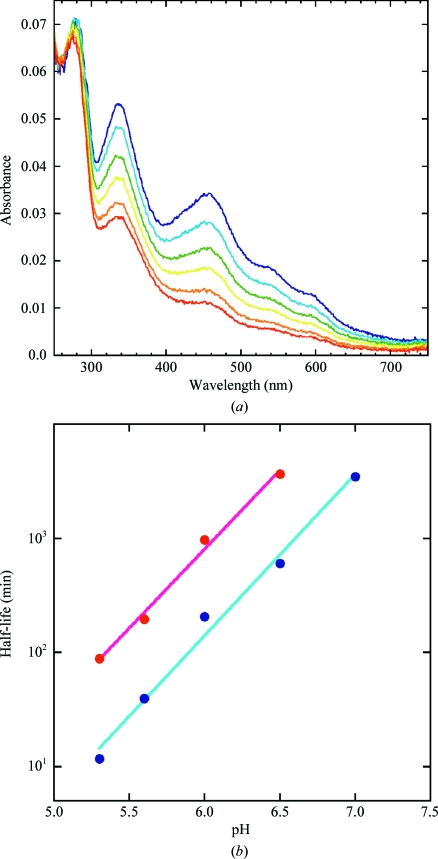
Typically, histidine can be a strong ideal ligand for an Fe–S cluster, as is found in Rieske-type 2Fe–2S clusters. However, the stability of the 2Fe–2S clusters of the H87C mutant was previously found to be an order of magnitude greater at pH 6. These observations led to the suggestion that the strong pH-dependence of the stability of wild-type mitoNEET was associated with protonation of His87 (Wiley, Paddock et al., 2007 ▶). For this reason, we pursued a more detailed comparative study of the stabilities of the 2Fe–2S clusters for both the wild type and H87C as a function of pH. The cluster integrity was monitored by optical methods as described by Wiley, Paddock et al. (2007 ▶). The lifetime was determined by following this decay as a function of time. The H87C-mutant mitoNEET displayed a consistent sixfold-increased stability over the entire pH range from pH 5.0 to 7.0 (Fig. 4 ▶). Thus, contrary to expectations, we conclude that His87 is not responsible for the pH-sensitivity of cluster release in the isolated protein, as this phenomenon is maintained despite the increase in cluster stability.
Figure 4.
The H87C 2Fe–2S cluster is stabilized from release compared with that of wild-type mitoNEET. (a) The optical spectrum of H87C mitoNEET was collected at multiple time points at pH 5 (0, 11, 33, 66, 144 and 986 min). The decay of the 460 nm peak in the absorbance spectrum is monitored as a function of time and corresponds to the loss of the 2Fe–2S cluster. (b) The cluster is much more stable in the H87C mutant (red) compared with the wild-type protein (blue). From this decay a half-life can be obtained at multiple pH values. Across this pH range, the 2Fe–2S cluster of H87C (red) is around sixfold more stable than that of wild-type mitoNEET (blue).
4. Discussion
The single coordinating His87 is one of the unique structural features of the NEET protein family (Paddock et al., 2007 ▶; Hou et al., 2007 ▶; Lin et al., 2007 ▶). Substitution of His87 of wild-type mitoNEET by Cys (the H87C mutant) resulted in a protein that retained the same fold but contained a 2Fe–2S cluster with dramatically different properties. The ability to substitute Cys for His is somewhat surprising, since the side chains are neither strictly isosteric nor of similar size or shape. In contrast to what is reported here, in other 2Fe–2S proteins substitution of coordinating histidines often leads to less stable proteins in the oxidized and/or reduced states (Kounosu et al., 2004 ▶; Kimura et al., 2005 ▶).
4.1. Dramatic increase in the stability of the 2Fe–2S cluster in H87C mitoNEET despite structural heterogeneity of Cys87
Protonation of His87 in the wild-type protein has been implicated to trigger release of the 2Fe–2S cluster and to be responsible for the pH-dependent stability (Wiley, Paddock et al., 2007 ▶). Resonance Raman studies showed pH-dependent changes that were attributed to the interaction between His87 and the cluster (Tirrell et al., 2009 ▶). Despite predictions that alteration of His87 would remove the pH-sensitivity, this is not observed. That is, direct protonation of His87 is not responsible for the observed pH-dependence of the stability, as the H87C mutant and the wild type show essentially the same pH-dependence of their lifetimes. The observed Fe—Sγ bond lengths are on average greater than the ideal canonical distance of 2.30 Å found between Cys ligands and the 2Fe–2S centers in most ferredoxins and Rieske cluster-containing proteins (Yeh et al., 2002 ▶; Hunsicker-Wang et al., 2003 ▶). The longer Fe—S—Cys bond lengths are expected to result in a less-than-ideal interaction with the 2Fe–2S centers. Given this mixture of bond lengths between Cys87 and the outer Fe, the increased stability (Fig. 4 ▶) is even more surprising.
An explanation for the increased stability is suggested by the crystal structure. The mutation results in structural changes that are localized to the immediate region of position 87, near the outer Fe of the 2Fe–2S cluster. Since Cys87 becomes a ligand to the cluster in the H87C mutant, it is presumed to be anionic. Thus, a local electrostatic change occurs. In addition, there are changes to the nearby Lys55 which induce additional changes in the local electrostatic environment. This provides a qualitative explanation of the effect of the mutation on the change in stability.
4.2. Dramatic shift in the 2Fe–2S cluster properties in H87C-mutant mitoNEET
In addition to cluster stability, another significant difference upon mutation of His87 is the large decrease of ∼300 mV in the redox potential (E m) of the H87C mutant protein (Zuris et al., 2010 ▶). This shift in E m implies a large change in the environment near the 2Fe–2S cluster. The crystal structure of the mutant unequivocally shows that Cys87 becomes the new ligand for the 2Fe–2S cluster and no other surrounding residue moves into ligating distance. In addition, the mutant and wild-type proteins are superimposable except near the Cys87 mutation and the nearby Lys55 side chain. The lack of any major structural changes is also seen in the matching UV CD spectra of the wild-type and the mutant proteins. Thus, global conformation changes are not a major cause of the observed change in E m.
Given that the redox potential becomes more like that of a 4Cys-coordinated 2Fe–2S cluster, it is reasonable to attribute a large part of the change to the introduction of the anionic Cys. The magnitude of the change would require that the wild-type His ligand be neutral; the Cys replacement results in the introduction of a negative charge near the cluster only if His87 is neutral. Thus, we conclude that the nonligating N∊ of His87 is protonated in the reduced state. A similar conclusion was drawn from pulsed EPR studies owing to the axial nature of the hyperfine tensor (Dicus et al., 2010 ▶).
4.3. The stability of the 2Fe–2S cluster is by design
Since the stability can readily be increased by the single amino-acid substitution of His87 by Cys, the wild-type stability (the ability to release the 2Fe–2S cluster) appears to be by evolutionary design. This idea is further supported by the high degree of sequence conservation from archaea to mammals of the 16-residue loop composing the 2Fe–2S cluster-binding region of human mitoNEET (Wiley, Murphy et al., 2007 ▶). In particular, His87 of mitoNEET is strictly conserved. Thus, having a single His coordinating ligand must confer some evolutionary advantage. Perhaps our first clue as to what advantage it may offer comes from knockout studies of the mitoNEET paralog Miner1, which resulted in impaired oxidative capacity (Wiley, Murphy et al., 2007 ▶; Chen, Kao, Chen et al., 2009 ▶). Studies on mice knockouts of Miner1, which has the same fold and redox potential as mitoNEET (Conlan, Axelrod et al., 2009 ▶), showed the mice to have a decreased lifespan and a generally lower quality of health (Chen, Kao, Chen et al., 2009 ▶; Chen, Kao, Kirby et al., 2009 ▶). Furthermore, loss of the cluster-binding domain of Miner1 is causative of the genetic human disease Wolfram syndrome 2 (Amr et al., 2007 ▶). The physical effects of this disease show similarities to those of the knockout mice. More recently, Miner1 was found to interact with Bcl-2 at the ER and to antagonize the pro-autophagic response of beclin 1 (Chang et al., 2010 ▶). The authors showed that the presence of the 2Fe–2S cluster ligands was important for this interaction. Thus, the ability to assemble and disassemble the 2Fe–2S clusters of Miner1 may be important in regulating cellular autophagy.
Our results show that the single His ligand confers two special properties to mitoNEET: (i) the ease of cluster removal in the isolated protein and (ii) the redox potential are dramatically changed upon ligand substitution. The 3Cys–1His coordination is likely to have been finely tuned such that it has biophysical properties that are suitable for its as yet unknown biological function. Although 4Cys coordination creates a more stable cluster, this protein is not found in nature. Having a 4Cys-coordinated 2Fe–2S cluster results in a mitoNEET with a hindered ability to transfer electrons to partner(s) (energetically more difficult to reduce) and to transfer 2Fe–2S clusters to the cytosol or partner(s) (the 2Fe–2S center is bound more tightly). Thus, we expect that the His ligand is optimized by evolutionary design for possible functions in electron transfer and/or Fe/Fe–S cluster transfer within the cellular environment.
Supplementary Material
PDB reference: mitoNEET, 3lpq
Acknowledgments
This work was supported by NIH grants GM41637 (to M. Okamura and MLP), GM54038 and DK54441 (to PAJ). Students were supported by HEME 5T32DK007233-34 (JAZ) and CMG training grant 2T32GM007240-29 (ARC). RN thanks the Zevi Hermann Shapira Foundation for supporting the collaborative USA–Israeli efforts. We thank Christopher L. Rife at the Joint Center for Structural Genomics (JCSG) for providing an automated programming script for coordinate validation. Parts of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program and the National Institute of General Medical Sciences.
References
- Amr, S., Heisey, C., Zhang, M., Xia, X.-J., Shows, K. H., Ajlouni, K., Pandya, A., Satin, L. S., El-Shanti, H. & Shiang, R. (2007). Am. J. Hum. Genet. 81, 673–683. [DOI] [PMC free article] [PubMed]
- Chang, N. C., Nguyen, M., Germain, M. & Shore, G. C. (2010). EMBO J. 29, 606–618. [DOI] [PMC free article] [PubMed]
- Chen, Y.-F., Kao, C.-H., Chen, Y.-T., Wang, C.-H., Wu, C.-Y., Tsai, C.-Y., Liu, F.-C., Yang, C.-W., Wei, Y.-H., Hsu, M.-T., Tsai, S.-F. & Tsai, T.-F. (2009). Genes Dev. 23, 1183–1194. [DOI] [PMC free article] [PubMed]
- Chen, Y.-F., Kao, C.-H., Kirby, R. & Tsai, T.-F. (2009). Autophagy, 5, 1043–1045. [DOI] [PubMed]
- Cohen, A. E., Ellis, P. J., Miller, M. D., Deacon, A. M. & Phizackerley, R. P. (2002). J. Appl. Cryst. 35, 720–726. [DOI] [PMC free article] [PubMed]
- Colca, J. R., McDonald, W. G., Waldon, D. J., Leone, J. W., Lull, J. M., Bannow, C. A., Lund, E. T. & Mathews, W. R. (2004). Am. J. Physiol. Endocrinol. Metab. 286, E252–E260. [DOI] [PubMed]
- Conlan, A. R., Axelrod, H. L., Cohen, A. E., Abresch, E. C., Zuris, J., Yee, D., Nechushtai, R., Jennings, P. A. & Paddock, M. L. (2009). J. Mol. Biol. 392, 143–153. [DOI] [PMC free article] [PubMed]
- Conlan, A. R., Paddock, M. L., Axelrod, H. L., Cohen, A. E., Abresch, E. C., Wiley, S., Roy, M., Nechushtai, R. & Jennings, P. A. (2009). Acta Cryst. F65, 654–659. [DOI] [PMC free article] [PubMed]
- DeLano, W. & Lam, J. (2005). Abstr. Pap. Am. Chem. Soc. 230, u1371–u1372.
- Dicus, M. M., Conlan, A., Nechushtai, R., Jennings, P. A., Paddock, M. L., Britt, R. D. & Stoll, S. (2010). J. Am. Chem. Soc. 132, 2037–2049. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Hou, X., Liu, R., Ross, S., Smart, E. J., Zhu, H. & Gong, W. (2007). J. Biol. Chem. 282, 33242–33246. [DOI] [PubMed]
- Hunsicker-Wang, L. M., Heine, A., Chen, Y., Luna, E. P., Todaro, T., Zhang, Y. M., Williams, P. A., McRee, D. E., Hirst, J., Stout, C. D. & Fee, J. A. (2003). Biochemistry, 42, 7303–7317. [DOI] [PubMed]
- Jung, Y.-S., Gao-Sheridan, H. S., Christiansen, J., Dean, D. R. & Burgess, B. K. (1999). J. Biol. Chem. 274, 32402–32410. [DOI] [PubMed]
- Kimura, S., Kikuchi, A., Senda, T., Shiro, Y. & Fukuda, M. (2005). Biochem. J. 388, 869–878. [DOI] [PMC free article] [PubMed]
- Kounosu, A., Li, Z., Cosper, N. J., Shokes, J. E., Scott, R. A., Imai, T., Urushiyama, A. & Iwasaki, T. (2004). J. Biol. Chem. 279, 12519–12528. [DOI] [PubMed]
- Lill, R. & Mühlenhoff, U. (2008). Annu. Rev. Biochem. 77, 669–700. [DOI] [PubMed]
- Lin, J., Zhou, T., Ye, K. & Wang, J. (2007). Proc. Natl Acad. Sci. USA, 104, 14640–14645. [DOI] [PMC free article] [PubMed]
- Lowell, B. B. & Shulman, G. I. (2005). Science, 307, 384–387. [DOI] [PubMed]
- McPhillips, T. M., McPhillips, S. E., Chiu, H.-J., Cohen, A. E., Deacon, A. M., Ellis, P. J., Garman, E., Gonzalez, A., Sauter, N. K., Phizackerley, R. P., Soltis, S. M. & Kuhn, P. (2002). J. Synchrotron Rad. 9, 401–406. [DOI] [PubMed]
- Meyer, J. (2008). J. Biol. Inorg. Chem. 13, 157–170. [DOI] [PubMed]
- Paddock, M. L., Wiley, S. E., Axelrod, H. L., Cohen, A. E., Roy, M., Abresch, E. C., Capraro, D., Murphy, A. N., Nechushtai, R., Dixon, J. E. & Jennings, P. A. (2007). Proc. Natl Acad. Sci. USA, 104, 14342–14347. [DOI] [PMC free article] [PubMed]
- Pähler, A., Smith, J. L. & Hendrickson, W. A. (1990). Acta Cryst. A46, 537–540. [DOI] [PubMed]
- Pédelacq, J. D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. (2006). Nature Biotechnol. 24, 79–88. [DOI] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Tirrell, T. F., Paddock, M. L., Conlan, A. R., Smoll, E. J., Nechushtai, R., Jennings, P. A. & Kim, J. E. (2009). Biochemistry, 48, 4747–4752. [DOI] [PMC free article] [PubMed]
- Wiley, S. E., Murphy, A. N., Ross, S. A., van der Geer, P. & Dixon, J. E. (2007). Proc. Natl Acad. Sci. USA, 104, 5318–5323. [DOI] [PMC free article] [PubMed]
- Wiley, S. E., Paddock, M. L., Abresch, E. C., Gross, L., van der Geer, P., Nechushtai, R., Murphy, A. N., Jennings, P. A. & Dixon, J. E. (2007). J. Biol. Chem. 282, 23745–23749. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yeh, A. P., Ambroggio, X. I., Andrade, S. L., Einsle, O., Chatelet, C., Meyer, J. & Rees, D. C. (2002). J. Biol. Chem. 277, 34499–34507. [DOI] [PubMed]
- Zuris, J. A., Halim, D. A., Conlan, A. R., Abresch, E. C., Nechushtai, R., Paddock, M. L. & Jennings, P. A. (2010). J. Am. Chem. Soc. 132, 13120–13122. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: mitoNEET, 3lpq