High-resolution crystallographic studies of the hydration of the coenzyme cob(II)alamin have provided hydrogen-bond parameters of unprecedented accuracy for a biomacromolecule.
Keywords: cob(II)alamin, neutron crystallography, hydration, hydrogen bonding, high resolution, D19, TOPAZ
Abstract
The hydration of the coenzyme cob(II)alamin has been studied using high-resolution monochromatic neutron crystallographic data collected at room temperature to a resolution of 0.92 Å on the original D19 diffractometer with a prototype 4° × 64° detector at the high-flux reactor neutron source run by the Institute Laue–Langevin. The resulting structure provides hydrogen-bonding parameters for the hydration of biomacromolecules to unprecedented accuracy. These experimental parameters will be used to define more accurate force fields for biomacromolecular structure refinement. The presence of a hydrophobic bowl motif surrounded by flexible side chains with terminal functional groups may be significant for the efficient scavenging of ligands. The feasibility of extending the resolution of this structure to ultrahigh resolution was investigated by collecting time-of-flight neutron crystallographic data during commissioning of the TOPAZ diffractometer with a prototype array of 14 modular 2° × 21° detectors at the Spallation Neutron Source run by Oak Ridge National Laboratory.
1. Introduction
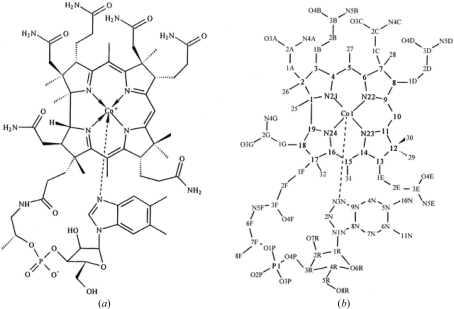
Cobalamin (Cbl) is a large heterocyclic molecule in which a central cobalt ion is equatorially coordinated by four donor N atoms from a corrin ring and axially coordinated at its lower (α) position by an N atom from 5,6-dimethylbenzimidazole (DMB) as shown in Fig. 1 ▶. In the Co(III) state the upper (β) axial position can be occupied by a large number of different ligands and several liganded Cbl X-ray crystal structures have been reported (Hannibal et al., 2010 ▶; Gruber et al., 1999 ▶; Randaccio et al., 2006 ▶). Cbls cannot be synthesized by higher organisms. In dietary vitamin B12 (CNCbl) a cyanide group is the β-axial ligand, but this form is enzymatically hydrolyzed to cob(II)alamin [Cbl(II)] and then transformed into one of two biologically active forms, AdoCbl and MeCbl, in which the β axial ligand is either a 5′-deoxyadenosyl (Ado) group or a methyl (Me) group, respectively (Chu et al., 1993 ▶; Froese et al., 2009 ▶; Kim et al., 2008 ▶).
Figure 1.
(a) Structural formula of Cbl(II). (b) Atom numbering and side-chain labeling used for description.
In mammals, several proteins are involved in the uptake, transport and storage of dietary Cbls (Banerjee et al., 2009 ▶). X-ray crystallographic structures of complexes of Cbl with the intrinsic factor (IF; Mathews et al., 2007 ▶) and trans-Cbl (TC; Wuerges et al., 2006 ▶) transport proteins have been reported. AdoCbl and MeCbl are the largest and most complex cofactors in biology and are required by two mammalian enzymes: MeCbl is the cofactor of methionine synthase (MES), whose mechanism involves generating alkyl cations by heterolytic cleavage of the Co—C bond, while AdoCbl is the cofactor of methylmalonyl coenzyme A mutase (MCAM), whose mechanism involves generating an Ado radical by homolytic dissociation of the Co—C bond. X-ray crystallography has shown that although DMB is coordinated to the α-axial position in the ‘base-on’ configuration in the free coenzyme and in complexes with dehydratase enzymes and the transport proteins IF and TC, it is found in the ‘base-off’ configuration in complexes with the enzymes MES and MCAM, displaced from the cobalt ion by a histidine amino-acid side chain of the protein (Banerjee et al., 2009 ▶).
Understanding how an enzyme facilitates controlled and reversible cleavage of the Co—C bond and therefore transition between Co(III) and Co(I,II) is an outstanding problem in biochemistry. Although early ideas from classical coordination chemistry suggested the importance of the Co—N bond to the DMB α-ligand in modulating cleavage, more recent X-ray crystallographic studies and electronic structure calculations of enzyme–Cbl complexes (Jensen & Ryde, 2009 ▶) and similarities between the structures of MeCbl and its homolysis product Cbl(II) (Kräutler al., 1989 ▶) suggest that enzymes play a dominant role in pulling the adenosyl radical away from the corrin ring. However, the exact architecture of the corrin ring, with its seven amide side chains (three acetamides and four propionamides) projecting above and below the plane, and coordination with DMB are likely to contribute to stabilizing the low-spin Co(I), Co(II) and Co(III) states involved in catalysis and recombination (Jensen & Ryde, 2009 ▶).
In addition to its role as a coenzyme, Cbl has also been implicated in the inhibition of nitric oxide-induced physiologies and pathologies (Hassanin et al., 2010 ▶). NO is a short-lived radical that is produced by tumor and host cells and is thought to function as an important modulator of tumor progression and angiogenesis. It has been suggested that in vivo Cbl(II) directly scavenges NO to form NOCbl, which has renewed interest in the exact structure of Cbl(II) and its β-ligand binding. Recent X-ray crystal structures of NOCbls have featured remarkably long Co—N(DMB) α-axial bond lengths, suggesting that this bond may well be correlated with direct β-ligand scavenging (Hassanin et al., 2010 ▶). Cbls have also been implicated in the dechlorination by anaerobic organisms of the toxic compounds perchloroethylene and trichloroethylene (Randaccio et al., 2006 ▶).
Cbls possess a number of functional groups found in biomacromolecules and can be crystallized in forms that contain several ordered solvent molecules and which diffract X-rays to high resolution. They have therefore been suggested as particularly useful models for studying the interaction of water with proteins and nucleic acids (Bouquiere et al., 1994 ▶; Randaccio et al., 2006 ▶; Savage et al., 1987 ▶; Savage, 1986 ▶). Neutrons offer important advantages for determining solvent structures hydrating biomacromolecules because the relatively strong scattering power of hydrogen (H) and deuterium (D) can allow the location of all atoms in a water molecule even at medium resolution (>1.5 Å; Blakeley et al., 2008 ▶); D atoms (neutron scattering length 6.67 × 10−15 m) appear as strong peaks in neutron scattering density maps, thereby revealing the location of isotopically exchanged H atoms, while H atoms (−3.74 × 10−15 m) appear as negative troughs.
The water structure in crystals of vitamin B12 has been studied in detail previously using high-resolution monochromatic neutron data collected at 279 K to a resolution of 0.95 Å on the former instrument D8 with a point detector at the high-flux reactor neutron source run by the Institute Laue–Langevin (ILL; Savage et al., 1987 ▶). This study revealed intricate networks of partially disordered solvent running throughout the crystal (Savage, 1986 ▶). A subsequent high-resolution study on the original D19 at 15 K extended the resolution to 0.90 Å (Bouquiere et al., 1994 ▶). We have previously studied the water structure in Cbl(II) at room temperature using quasi-Laue neutron data collected using the original LADI diffractometer also at ILL (Langan et al., 1999 ▶). LADI has recently been significantly upgraded (to LADI-III) in order to improve its performance (Blakeley et al., 2010 ▶). Limitations set by the wavelength band used on the original LADI (which had a peak at 2.8 Å and a FWHH of 19%) restricted the resolution of the data to d > 1.42 Å. The final nominal resolution (at which the completeness fell below 70%) of the data set collected in 36 h was around 1.8 Å. Even at this medium resolution we were able to obtain coordinates for all of the atoms involved in seven ordered water and two acetone solvent molecules. From these coordinates, hydrogen-bonding parameters were determined that contributed to an improved general understanding and modeling of the hydration of biomacromolecules.
In addition to the data set collected on LADI, another more complete neutron data set was collected from Cbl(II) at room temperature on the original D19 with its small prototype 4° × 64° area detector. We were able to collect data to a higher resolution with a data-collection time of 28 d. The resolution of the data was still limited to d > 0.92 Å because of the use of a wavelength of 1.54 Å and the geometry of the original D19. Recently, a new single-crystal diffractometer called TOPAZ has been built at the Spallation Neutron Source (SNS) run by Oak Ridge National Laboratory. At the SNS neutrons are produced by bombarding a mercury target with pulses of high-energy protons. Neutrons produced by proton pulses are ‘time-stamped’ and travel as a function of their energy so that neutrons of different energies are detected at different arrival times. By recording the time-of-flight (TOF) information, the corresponding energy and wavelength of each neutron can be calculated. TOF techniques therefore allow wavelength-resolved Laue patterns to be collected using all of the available useful neutrons. In particular, the tunable wavelength range on TOPAZ in combination with an array of position-sensitive detectors will offer the possibility of collecting data to ultrahigh resolution from biological molecules with unit-cell parameters of up to about 70 Å in length.
In this study, we report the structure of Cbl(II) determined from the monochromatic neutron data set collected on the original D19 at the H11 thermal beam at the ILL to 0.92 Å resolution. We also report preliminary studies of the feasibility of extending the resolution of this structure even further by collecting TOF neutron data during a short commissioning experiment on TOPAZ. Particular attention is given to a description of the data-collection and integration process on TOPAZ since this is a new instrument and it has not been reported in full before. The results from D19 provide detailed hydrogen-bonding parameters for the hydration of different functional groups in a biological molecule at room temperature to unprecedented resolution and accuracy. The features of the Cbl(II) structure may promote the capture and scavenging of small molecules such as NO. The results from TOPAZ demonstrate that neutron data can be collected from biological molecules rapidly to high resolution but that there is also the potential to collect to ultrahigh resolution with longer data-collection times.
2. Experimental methods
The preparation of Cbl(II) and its crystallization with hexadeuteroacetone in D2O have been described previously (Langan et al., 1999 ▶). A crystal of dimensions 4.5 × 1.4 × 1.3 mm was mounted in a sealed quartz capillary with its mother liquor. Data were collected on the original D19 using a vertically mounted cylindrical electronic position-sensitive detector which subtended 4° × 64° at the sample position at room temperature with a wavelength of 1.54 Å in normal-beam geometry with a goniometer step size in ω of 0.1° and over a period of about 28 d [space group P212121; unit-cell parameters a = 15.9795 (9), b = 21.8889 (13), c = 26.8095 (18) Å; V = 9377.26 Å3; 11 986 measured reflections, 7970 unique reflections and 5619 unique reflections with F o > 4σ; R σ = 0.1009; R merge = 0.042 for 6625 unique reflections with positive intensities]. Reflections were integrated with the three-dimensional procedure of Wilkinson et al. (1988 ▶), which uses an empirical library of peak shapes from the stronger reflections to improve the integration for the weak peaks. D19 has recently been upgraded with a new horizontally mounted 30° × 120° cylindrical detector with corresponding gains of up to 50-fold in data-collection efficiency from biological samples (Nishiyama et al., 2008 ▶; Kovalevsky et al., 2010 ▶), so that high-resolution data sets on macromolecules can be measured in days rather than weeks.
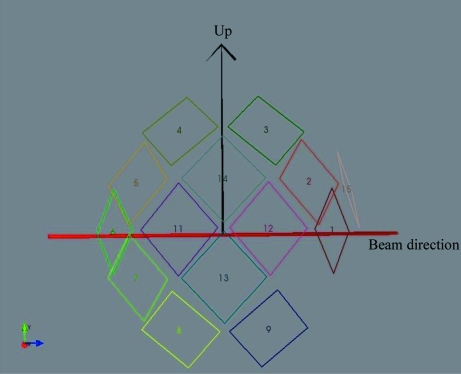
Data were then collected during the commissioning of TOPAZ at room temperature in TOF Laue mode (Hoffmann et al., 2009 ▶) using neutrons with wavelengths in the range 0.65–3.4 Å. For commissioning purposes, the instrument was equipped with a prototype array of 14 modular area detectors at distances of 39.5–42.5 cm from the sample. Each detector module had a square active area of 150.2 × 150.2 mm (corresponding to an angular coverage of about 21° × 21°) that was divided into 256 × 256 pixels (0.578 mm per pixel). The detector modules were positioned on the surface of a spherical detector array with a total vertical angular coverage of ±32° from the horizontal plane and 18.0–144.0° in the horizontal (2θ) plane, as shown in Fig. 2 ▶. In its final configuration TOPAZ will have 48 detectors and therefore more than three times the detector coverage used in this short commissioning experiment. Since this experiment, significant gains have also been made in the signal-to-noise ratio of data collected on TOPAZ by reducing the instrument scattering background.
Figure 2.
The geometrical arrangement of the initial 14 detector modules in the detector array during the commissioning of TOPAZ.
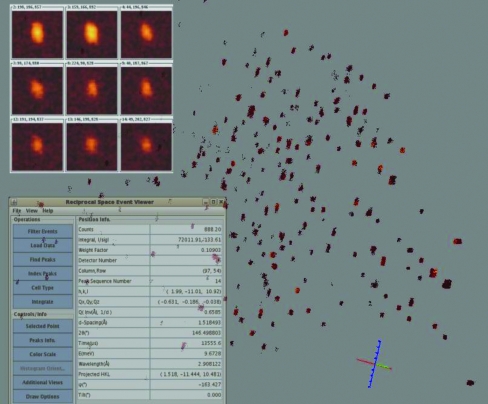
Raw data were recorded in neutron event mode with a time resolution of 1 µs, which is equivalent to a neutron wavelength resolution of 2.15 × 10−4 Å at 1.0 Å for a sample at 18.0 m from the neutron source. The detected neutron events were recorded as a function of TOF, detector number, detector pixel address (ID) and scattering angle. Positions of detector pixel IDs were mapped initially from their engineering values and then calibrated using a standard sapphire crystal with known unit-cell parameters. The calibrated detector information was stored in Nexus format (Konnecke, 2006 ▶), in which the Z axis is along the neutron beam direction, the Y axis is up and the X axis follows the right-handed rule. During data collection, the neutron event data were saved and broadcast to a listening server in a local EventViewer program in ISAW (Mikkelson et al., 2009 ▶) for live viewing of the measured diffraction data in three-dimensional reciprocal q-space, as shown in Fig. 3 ▶. The EventViewer program is capable of indexing peaks, generating an orientation matrix with user input and carrying out data integration in real time.
Figure 3.
Background, neutron events (20 out of 259 million) for one sample position as displayed by the three-dimensional reciprocal-space EventViewer software. Upper left insert, nine of the strong peaks found in Nexus image frames using the peak-search program in ISAW. Lower left insert, display parameters.
An initial orientation matrix was determined by indexing the diffraction data after the first few minutes of data collection. The matrix was then imported into the CrystalPlan program (Zikovsky et al., 2011 ▶) for data-collection strategy optimization. Event data were collected at 11 crystal goniometer settings over the course of 2 d (42 h of neutron beam time) and translated into Nexus frames for data integration. During translation, TOF data were binned into neutron image frames logarithmically with a constant Δt/t resolution of 0.004 (0.4%). Integrated diffraction intensities were obtained using the program ISAW. Data reduction and absorption correction were carried out with the local ANVRED2 program (Schultz et al., 1984 ▶) launched from within the ISAW program suite (Mikkelson et al., 2005 ▶). The Becker–Coppens procedure (Becker & Coppens, 1974 ▶) implemented in GSAS (Larson & Von Dreele, 2004 ▶) was used for the neutron wavelength-dependent extinction correction. The resulting reflection data were converted to SHELX format for structural analysis [space group P212121; unit-cell parameters a = 16.005 (2), b = 21.931 (3), c = 26.911 (4) Å, V = 9446 (2) Å3; 10 706 observed reflections with I > 2σ(I); R merge = 0.1008 for 7323 unique reflections].
The structure was refined first against the D19 data using SHELX (Sheldrick, 2008 ▶; C68H/D114CoN13O23P; R = 0.1232 for all 7970 data and R = 0.0923 for 5904 reflections with F o > 4σ) and included seven water (D2O) and two acetone [(CD3)2CO] solvent molecules. All atoms except those of the acetone molecules were refined with anisotropic atomic displacement parameters. The occupancies of the D atoms bound to the amide groups (0.77), the acidic C10 corrin atom (0.69) and the hydroxyl groups (0.71) were allowed to refine. The occupancies of the water and acetone solvent molecules were also allowed to refine. Significant (>3σ) density was present in the final 2F o − F c map in the large solvent channel which could not be easily interpreted as further ordered water or acetone molecules. There were also clear indications that the acetone molecule sitting near the β-axial position over the cobalt cation is disordered. We did not attempt to model these features as our primary interest was in determining detailed hydrogen-bonding parameters for ordered water molecules. The final structure is shown in Fig. 4 ▶ and a CIF file is available as supplementary material1. Hydrogen-bond geometries determined from these parameters are given in Table 1 ▶. The structure was also refined against the TOPAZ data. We investigated first using a resolution cutoff of d > 0.9 Å and with all atoms having isotropic atomic displacement parameters (R = 0.1108 for 7323 data). The resulting structure was essentially identical to that obtained with the D19 data. When the resolution range was extended to d < 0.9 Å the R factor increased dramatically without significantly improving the accuracy of the structure. This effect suggests that although there is some diffraction at ultrahigh resolution beyond 0.9 Å, the nominal diffracting power of the crystal is about 0.9–1.0 Å.
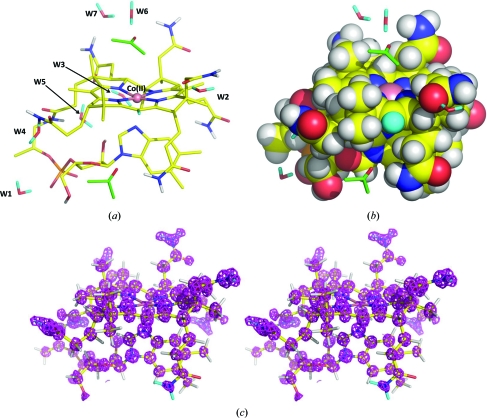
Figure 4.
(a) Skeletal representation of the neutron structure of Cbl(II) with the ordered solvent molecules labeled W1–W7. (b) Space-filling representation illustrating the presence of a hydrophobic bowl above the corrin plane. (c) Stereogram of positive neutron scattering density in a 2F o − F c map displayed at a level of 5σ.
Table 1. Hydrogen-bond parameters for donor (D), acceptor (A) and deuterium (H) atoms.
| Donor | Acceptor | D—H (Å) | H⋯A (Å) | D⋯A (Å) | D—H⋯A (°) |
|---|---|---|---|---|---|
| Water–water hydrogen bonds | |||||
| O7W | O6W | 1.007 (16) | 1.94 (3) | 2.95 (3) | 179 (3) |
| O1W | O2W | 0.988 (13) | 1.81 (3) | 2.80 (3) | 176 (2) |
| O4W | O2W | 1.008 (16) | 1.98 (3) | 2.95 (3) | 162 (2) |
| O5W | O1W | 0.995 (17) | 1.85 (3) | 2.84 (3) | 173 (3) |
| Mean | 1.000 (10) | 1.90 (8) | 2.88 (8) | 173 (7) | |
| Amine–water hydrogen bonds | |||||
| N5E | O4W | 1.010 (13) | 1.88(2) | 2.89(2) | 177.2 (18) |
| N4A | O6W | 1.028 (12) | 2.098 (15) | 3.121 (11) | 173.2 (13) |
| N5B | O5W | 1.014 (12) | 2.46 (3) | 3.25 (3) | 134.7 (16) |
| N4C | O3W | 1.023 (11) | 2.041 (18) | 2.902 (18) | 140.3 (11) |
| N4G | O7W | 1.024 (12) | 1.80 (2) | 2.82 (2) | 173.0 (15) |
| Mean | 1.020 (7) | 2.06 (20) | 3.00 (18) | 160 (20) | |
| Water–phosphate hydrogen bonds | |||||
| O1W | O2P | 0.999 (13) | 1.701 (17) | 2.692 (19) | 170.2 (19) |
| O2W | O3P | 0.998 (14) | 1.678 (19) | 2.66 (2) | 166.4 (18) |
| O3W | O2P | 0.978 (14) | 1.667 (17) | 2.63 (2) | 168.3 (18) |
| Mean | 0.997 (11) | 1.682 (17) | 2.66 (3) | 168.3 (19) | |
| Water–ketone hydrogen bonds | |||||
| O2W | O3C | 1.004 (14) | 1.65 (2) | 2.65 (2) | 176 (2) |
| O3W | O3G | 0.982 (13) | 1.832 (17) | 2.813 (19) | 176.3 (17) |
| O5W | O3G | 0.997 (17) | 1.98 (3) | 2.97 (3) | 168 (3) |
| Mean | 0.994 (11) | 1.82 (16) | 2.81 (16) | 173 (5) | |
| Other hydrogen bonds | |||||
| N4A | O4D | 1.014 (12) | 2.02 (2) | 3.02 (2) | 169.7 (17) |
| N5B | O3P | 1.025 (12) | 1.917 (19) | 2.937 (17) | 173.4 (19) |
| N5B | O1P | 1.025 (12) | 2.60 (2) | 3.204 (15) | 117.7 (15) |
| N4C | O3P | 1.029 (11) | 1.981 (19) | 3.003 (16) | 171.6 (15) |
| N5D | O64G | 1.029 (15) | 2.07 (5) | 3.10 (5) | 172 (3) |
| N5E | O3A | 1.011 (13) | 1.992 (19) | 2.861 (14) | 142.6 (17) |
| N4G | O4E | 1.020 (11) | 2.033 (19) | 2.987 (17) | 154.6 (16) |
| O7R | O1W | 0.971 (14) | 1.808 (15) | 2.773 (17) | 172.4 (13) |
| O8R | O3W | 1.00 (3) | 1.94 (3) | 2.91 (3) | 163 (3) |
| O4W | O4F | 1.004 (17) | 1.75 (2) | 2.75 (3) | 170 (3) |
| O7W | O4B | 0.992 (16) | 1.673 (19) | 2.66 (3) | 171 (3) |
| O6W | O54G | 0.98 | 1.84 (4) | 2.81 (4) | 171.5 (12) |
3. Results
The experimental standard deviation (e.s.d.) range for bond lengths in Cbl(II) involving all non-H atoms in the high-resolution (0.92 Å) D19 neutron structure reported here is 0.007–0.020 Å. This is similar to the range of values found in the previous high-resolution (0.95 Å) neutron study of vitamin B12 (0.007–0.021 Å; Savage et al., 1987 ▶) and indicates a relatively high level of precision for a biomacromolecule. The range of values obtained for the neutron structure of Cbl(II) determined with the TOPAZ data was slightly higher (0.008–0.030 Å), indicating a slightly less accurate structure. However, this is only to be expected given the drastically shorter data-collection time of 2 d on TOPAZ. The purpose of collecting the TOPAZ data was not to improve the accuracy of the data and therefore the structure already determined at d > 0.9 Å, but rather to explore the possibility of collecting further diffraction data at ultrahigh resolutions of d < 0.9 Å. It is clear that both the current D19 with its newly upgraded detector and the future as-designed TOPAZ with its larger detector array will be capable of collecting highly accurate data from small to medium-sized biological macromolecules at unprecedented rates.
The overall geometry of Cbl(II) has been described previously and its corrin moiety is very similar to that of vitamin B12 (Kräutler al., 1989 ▶; Fig. 4 ▶). However, significant differences exist between the conformations adopted by some of the side chains. In particular, the conformation of the nucleotide F loop is different and the phosphate groups therefore hydrogen bond to different water moieties. W1–W5 are found in the hydrophilic region at the interface of stacked planes of Cbl(II) molecules and are involved in hydrogen bonding to the phosphate groups (Langan et al., 1999 ▶; Fig. 4 ▶ a). Another difference is the existence of a large open solvent channel in Cbl(II). W6 and W7 and the two acetone molecules are found in this channel, but it is mostly occupied by disordered solvent (Langan et al., 1999 ▶) which we did not attempt to model. In vitamin B12 the DMB ligand occupies this channel and is surrounded by a complex pattern of partially occupied water hydrogen-bonding networks that extend throughout the crystal (Savage, 1986 ▶). The water hydrogen-bonding parameters derived from this study are therefore based on a different water structure to that found in vitamin B12 (Bouquiere et al., 1994 ▶; Randaccio et al., 2006 ▶; Savage, 1986 ▶; Savage et al., 1987 ▶).
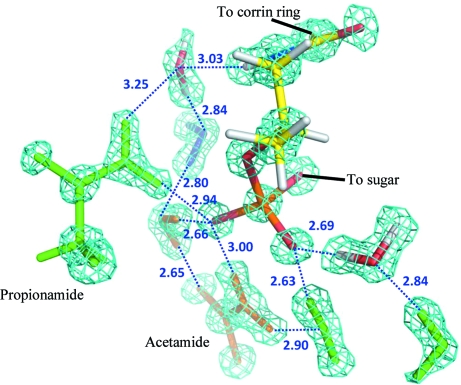
The accuracy of the hydrogen-bonding parameters involving water is significantly improved in the high-resolution neutron structure presented here compared with the previously reported neutron structure of Cbl(II) to medium resolution (1.8 Å; Langan et al., 1999 ▶), e.g. the mean values of the e.s.d. of donor–acceptor distances and angles for hydrogen bonds between water molecules are 0.03 Å and 2.5°, respectively, in this study, compared with 0.07 Å and 5.2° in the previous study. Interestingly, this improved accuracy reveals a more linear geometry, with the mean hydrogen-bond angle being 173 (7)° in this study compared with 152° in the previous study. The former value is still significantly smaller than 180° and is very similar to the values of 170° and 175° found in a statistical study of the distributions of hydrogen-bond angles for N—H⋯O and O—H⋯O, respectively, in protein–ligand complexes (Sarkel & Desiraju, 2004 ▶). Furthermore, the improved accuracy allows the identification of some differences in the mean solvent hydrogen-bonding parameters for different functional groups which were not evident in the previous study. In particular, the mean donor–acceptor hydrogen-bond lengths decrease in the order amine–water > water–water > water–ketone > water–phosphate [3.00 (18) > 2.88 (8) > 2.81 (16) > 2.66 (3) Å; Fig. 5 ▶]. This trend agrees with an increase in hydrogen-bond strength in the series N—H⋯O—H > O—H⋯O—H > O—H⋯O=C > O—H⋯−O—P (Grabowski, 2006 ▶; Rozas, 2007 ▶).
Figure 5.
A detail of a 2F o − F c map displayed at a level of 5σ showing how the improved accuracy of this structure allows the identification of some differences in the mean solvent hydrogen-bonding parameters for different functional groups.
The relatively small hydrogen-bond lengths for water molecules hydrating the phosphate group reflect a larger electrostatic contribution from the charge on the phosphate. These hydrogen bonds fall into a group of hydrogen bonds called negatively charge-assisted hydrogen bonds (Perrin & Nielson, 1997 ▶). There is a significant difference between the P1—O3P and P1—O2P bond lengths [1.53 (1) and 1.47 (1) Å, respectively] which indicates that most of the charge is associated with O3P, as correctly represented in Fig. 1 ▶. O3P and O2P participate in three and two hydrogen bonds, respectively. In addition to having the longest mean lengths, hydrogen bonds donated by amine groups to water molecules have the largest range of angles [134.7 (16)–177.2 (18)°]. Although the largest range of hydrogen-bond angles involve the hydrogen bonds involving water molecules, there is one hydrogen bond (N5B—O3P) which is clearly an outlier as it has both the smallest angle (117.7°) and the longest H⋯A distance (2.6 Å).
The locations of H and D atoms covalently bound to Cbl(II) are well defined by negative troughs and positive peaks in 2F o − F c, σA maps and F o − F c OMIT density maps, respectively. These atoms were refined without constraints on their positions or atomic displacement parameters (ADP). Table 2 ▶ shows the mean ADPs for H/D atoms and the heavy atoms that they are covalently bonded to. The values of the ratio of the mean ADPs suggests that when refining a biomacromolecular structure at lower resolutions with H atoms included using a ‘riding model’ (Sheldrick, 2008 ▶) the ADPs of H atoms should be tied to about 1.5 or 1.1 times the ADP of a bound C or N atom, respectively. It is surprising to find that the ratios of the ADPs for H to C in CH, CH2 and CH3 groups are similar given the different possible motions of these groups, although we acknowledge that the sampling array is quite limited.
Table 2. Mean (U eq) atomic displacement parameters for H/D atoms in different groups.
| Group | No. | C or N | H | Ratio |
|---|---|---|---|---|
| CH | 14 | 0.054 (8) | 0.079 (18) | 1.5 |
| CH2 | 13 | 0.067 (15) | 0.094 (19) | 1.4 |
| CH3 | 11 | 0.078 (27) | 0.116 (34) | 1.5 |
| ND2 | 6 | 0.102 (40) | 0.112 (60) | 1.1 |
Interestingly, the methyl H atoms that project up above the corrin plane, together with the aliphatic H atoms from the three acetamide side chains, form a hydrophobic perimeter wall around the upper (β) axial ligand position, as illustrated in Fig. 4 ▶(b). The upper side of Cbl(II) therefore has the shape of a bowl with a hydrophobic bottom and inside, and an opening that faces the large solvent channel. At the opening of this barrel lies the partially disordered acetone molecule, tethered above the cobalt cation to the functional amide group of the A acetamide side chain by a bridging W6 water molecule (Fig. 4 ▶ a). Only the A and C acetamide side chains (located to the right of the cobalt ion in Fig. 4 ▶ a) point upwards to the upper side of the corrin ring. These A and C side chains are found in a variety of different conformations in X-ray crystal structures of different Cbls and their amine functional groups are found both pointing towards or away from the inside of the hydrophobic bowl. It is therefore likely that they are highly flexible. It seems likely that the functional groups at the end of these flexible side chains play a role in concentrating physiological ligands around the hydrophobic bowl and perhaps even in guiding them towards the cobalt cation at the bottom of the bowl (Fig. 4 ▶ b).
4. Discussion
Crystallographic refinement of biomacromolecular structures minimizes a target function that involves diffraction data and typically a force field. Obtaining highly accurate hydrogen-bonding parameters for water hydrating biological structures is important because it provides experimental data that can be used to parameterize the force field. The structure reported and the resulting water hydrogen-bonding parameters are of unprecedented resolution for a neutron study of a biomacromolecule. Nonbonded interactions in the force field are typically treated as a simple repulsive term (referred to as the ‘Repel’ force field; Adams et al., 1997 ▶). This repulsive force depends on the atomic radii of the interacting atoms and it is able to model the larger hydrogen-bonding distances observed in this study for N⋯O interactions compared with O⋯O interactions. However, it does not model the significant differences observed in the O⋯O distances when different functional groups are involved and thus does not take into account the different strengths of hydrogen bonds. The hydrogen-bonding parameters reported here can therefore be used to improve the Repel force field by taking into account the involvement of different functional groups. We expect that this will increase the accuracy of biomacromolecular structure refinement. Furthermore, the hydrogen-bonding parameters reported here provide a basis for testing the accuracy of newly emerging force fields and approaches in biomacromolecular refinement that include electrostatic interactions (Fenn et al., 2011 ▶).
D19 with its newly upgraded detector and the future as-designed TOPAZ with its larger detector array will be capable of collecting highly accurate data from small to medium-sized biological macromolecules at unprecedented rates. Furthermore, in our exploration of the possibility of extending the resolution of the Cbl(II) structure beyond 0.9 Å, we observed some diffraction at ultrahigh resolution beyond 0.9 Å, although the nominal diffracting power of the crystal prevented its use in structure refinement. This suggests that TOPAZ may be an ideal instrument for developing new research based on ultrahigh-resolution neutron studies of biological macromolecules.
Supplementary Material
Crystal structure: contains datablocks global, B12r. DOI: 10.1107/S090744491101496X/tm5035sup1.cif
Acknowledgments
We acknowledge the ILL and SNS for provision of facilities. TOPAZ is funded by the Office of Basic Energy Sciences of the Department of Energy. PL, MM, AK and ZF were partly funded by the Office of Biological and Environmental Research of the Department of Energy. PL and MM were partly supported by an NIH–NIGMS grant (R01GM071939-01) to develop computational tools for neutron protein crystallography.
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: TM5035). Services for accessing this material are described at the back of the journal.
References
- Adams, P. D., Pannu, N. S., Read, R. J. & Brünger, A. T. (1997). Proc. Natl Acad. Sci. USA, 94, 5018–5023. [DOI] [PMC free article] [PubMed]
- Banerjee, R., Gherasim, C. & Padovani, D. (2009). Curr. Opin. Chem. Biol. 13, 484–491. [DOI] [PMC free article] [PubMed]
- Becker, P. J. & Coppens, P. (1974). Acta Cryst. A30, 129–147.
- Blakeley, M. P., Langan, P., Niimura, N. & Podjarny, A. (2008). Curr. Opin. Struct. Biol. 18, 593–600. [DOI] [PMC free article] [PubMed]
- Blakeley, M. P., Teixeira, S. C. M., Petit-Haertlein, I., Hazemann, I., Mitschler, A., Haertlein, M., Howard, E. & Podjarny, A. D. (2010). Acta Cryst. D66, 1198–1205. [DOI] [PubMed]
- Bouquiere, J. P., Finney, J. L. & Savage, H. F. J. (1994). Acta Cryst. B50, 566–578.
- Chu, R. C., Begley, J. A., Colligan, P. D. & Hall, C. A. (1993). Metabolism, 42, 315–319. [DOI] [PubMed]
- Fenn, T. D., Schnieders, M. J., Mustyakimov, M., Wu, C., Langan, P., Pande, V. S. & Brunger, A. T. (2011). Structure, 19, 523–533. [DOI] [PMC free article] [PubMed]
- Froese, D. S., Zhang, J., Healy, S. & Gravel, R. A. (2009). Mol. Genet. Metab. 98, 338–343. [DOI] [PubMed]
- Grabowski, S. J. (2006). Annu. Rep. Prog. Chem. Sect. C, 102, 131–165.
- Gruber, K., Jogl, G., Klintscher, G. & Kratky, C. (1999). Vitamin B12 and B12-Proteins, edited by B. Kräutler, D. Arigoni & B. T. Gloding, pp. 335–348. Weinheim: Wiley–VCH.
- Hannibal, L., Smith, C. A. & Jacobsen, D. W. (2010). Inorg. Chem. 49, 9921–9927. [DOI] [PMC free article] [PubMed]
- Hassanin, H. A., El-Shahat, M. F., DeBeer, S., Smith, C. A. & Brasch, N. E. (2010). Dalton Trans. 39, 10626–10630. [DOI] [PMC free article] [PubMed]
- Hoffmann, C., Wang, X. P. & Frost, M. (2009). TOPAZ. http://neutrons.ornl.gov/instruments/SNS/TOPAZ.
- Jensen, K. P. & Ryde, U. (2009). Coord. Chem. Rev. 253, 769–778.
- Kim, J., Gherasim, C. & Banerjee, R. (2008). Proc. Natl Acad. Sci. USA, 105, 14551–14554. [DOI] [PMC free article] [PubMed]
- Konnecke, M. (2006). Physica B, 385–386, 1343–1345.
- Kovalevsky, A. Y., Hanson, L., Fisher, S. Z., Mustyakimov, M., Mason, S. A., Forsyth, V. T., Blakeley, M. P., Keen, D. A., Wagner, T., Carrell, H. L., Katz, A. K., Glusker, J. P. & Langan, P. (2010). Structure, 18, 688–699. [DOI] [PMC free article] [PubMed]
- Kräutler, B., Keller, W. & Kratky, C. (1989). J. Am. Chem. Soc. 111, 8936–8938.
- Langan, P., Lehmann, M., Wilkinson, C., Jogl, G. & Kratky, C. (1999). Acta Cryst. D55, 51–59. [DOI] [PubMed]
- Larson, A. C. & Von Dreele, R. B. (2004). GSAS. Report LAUR 86-748. Los Alamos National Laboratory, New Mexico, USA.
- Mathews, F. S., Gordon, M. M., Chen, Z., Rajashankar, K. R., Ealick, S. E., Alpers, D. H. & Sukumar, N. (2007). Proc. Natl Acad. Sci. USA, 104, 17311–17316. [DOI] [PMC free article] [PubMed]
- Mikkelson, D., Mikkelson, R., Schultz, A. J., Hoffmann, C. & Wang, X. P. (2009). ISAW. http://www.pns.anl.gov/computing/isaw/.
- Mikkelson, D. J., Schultz, A. J., Mikkelson, R. & Worlton, T. G. (2005). IUCr Comm. Crystallogr. Comput. Newsl. 5, 2–39.
- Nishiyama, Y., Johnson, G. P., French, A. D., Forsyth, V. T. & Langan, P. (2008). Biomacromolecules, 9, 3133–3140. [DOI] [PubMed]
- Perrin, C. L. & Nielson, J. B. (1997). Annu. Rev. Phys. Chem. 48, 511–544. [DOI] [PubMed]
- Randaccio, L., Geremia, S., Nardin, G. & Wuerges, J. (2006). Coord. Chem. Rev. 250, 1332–1350.
- Rozas, I. (2007). Phys. Chem. Chem. Phys. 9, 2782–2790. [DOI] [PubMed]
- Sarkel, S. & Desiraju, G. R. (2004). Proteins, 54, 247–259. [DOI] [PubMed]
- Savage, H. (1986). Biophys. J. 50, 947–965. [DOI] [PMC free article] [PubMed]
- Savage, H. F. J., Lindley, P. F., Finney, J. L. & Timmins, P. A. (1987). Acta Cryst. B43, 280–295.
- Schultz, A. J., Srinivasan, K., Teller, R. G., Williams, J. M. & Lukehare, C. M. (1984). J. Am. Chem. Soc. 106, 999–1003.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wilkinson, C., Khamis, H. W., Stansfield, R. F. D. & McIntyre, G. J. (1988). J. Appl. Cryst. 21, 471–478.
- Wuerges, J., Garau, G., Geremia, S., Fedosov, S. N., Petersen, T. E. & Randaccio, L. (2006). Proc. Natl Acad. Sci. USA, 103, 4386–4391. [DOI] [PMC free article] [PubMed]
- Zikovsky, J., Peterson, P. F., Wang, X. P., Frost, M. & Hoffmann, C. (2011). J. Appl. Cryst. 44, 418–423.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, B12r. DOI: 10.1107/S090744491101496X/tm5035sup1.cif