Abstract
Expression profiles of 5058 human gene transcripts represented by an array of 7451 clones from the first IMAGE Consortium cDNA library from infant brain have been collected by semiquantitative hybridization of the array with complex probes derived by reverse transcription of mRNA from brain and five other human tissues. Twenty-one percent of the clones corresponded to transcripts that could be classified in general categories of low, moderate, or high abundance. These expression profiles were integrated with cDNA clone and sequence clustering and gene mapping information from an upgraded version of the Genexpress Index. For seven gene transcripts found to be transcribed preferentially or specifically in brain, the expression profiles were confirmed by Northern blot analyses of mRNA from eight adult and four fetal tissues, and 15 distinct regions of brain. In four instances, further documentation of the sites of expression was obtained by in situ hybridization of rat-brain tissue sections. A systematic effort was undertaken to further integrate available cytogenetic, genetic, physical, and genic map informations through radiation-hybrid mapping to provide a unique validated map location for each of these genes in relation to the disease map. The resulting Genexpress IMAGE Knowledge Base is illustrated by five examples presented in the printed article with additional data available on a dedicated Web site at the address http://idefix.upr420.vjf.cnrs.fr/EXPR/welcome.html.
The genomes of individuals remain virtually the same throughout their lifetimes, with the notable exceptions of specific rearrangements such as those of the immunoglobulin genes in B lymphocytes and T-cell-receptor genes in T lymphocytes, and a limited number of mutations arising as errors of DNA replication during cell division or DNA damage induced by external agents (radiation, viruses) and that remain uncorrected by the DNA proofreading systems.
In contrast, each cell of the hundreds of cell types that make up the human body expresses different sets of genes at different levels as the result of differentiation, development, environmental influences, disease, or treatment. Each physiological and pathological situation can thus be characterized by a specific set of gene transcripts (transcriptome) and protein products (proteome). The term transcriptome was coined by one of us (C. Auffray) in 1996 to characterize entire sets of transcripts; it has been used by others in the context of simpler unicellular model organisms such as yeast (Velculescu et al. 1997; Dujon, 1998). Gene expression is regulated at the level of transcription to a large extent. Therefore assessment of the variation of transcriptomes provides a global initial appraisal of the dynamic aspects of these regulations. Description of the corresponding proteomes to complement the expression profiles is under way with the development of methods for large-scale systematic analyses of proteins (see, e.g., Humphery-Smith et al. 1997; VanBogelen et al. 1997; Anderson and Anderson 1998; Wilkins et al. 1998).
The ultimate outcome of the Human Genome Project will be to provide biologists with the basic knowledge necessary to decipher the structure and function of all human genes and their products in relation to physiology and disease. As part of this effort, a Consortium for Integrated Molecular Analyses of Genomes and their Expression (IMAGE, Lennon et al. 1996) has established a common resource of publicly available cDNA libraries from which the genome community has collected a wealth of sequence, map, and expression data. Thus, from the circa one million human partial cDNA sequences registered in GenBank, >80% have been derived from IMAGE Consortium cDNA collections (Auffray et al. 1995; Hillier et al. 1996).
Several groups have developed clustering approaches to establish links between the cDNA clones and sequences derived from transcripts of the same gene as the result of alternate initiation and termination of transcription and alternate splicing (Adams et al. 1995; Houlgatte et al. 1995; Schuler et al. 1997; Burke et al. 1998), indicating that most of the estimated 60,000–80,000 human genes (Antequera and Bird 1994; Fields et al.1994) are already represented in the IMAGE cDNA collections.
The most recent versions of the human gene maps that have been assembled by an international consortium of laboratories using radiation-hybrid panels and physical resources are based mostly on expressed sequence tag site (ESTS) markers derived from these IMAGE Consortium resources, and provide localizations for some 30,000 distinct genes (Hudson et al. 1995; Gyapay et al. 1996; Schuler et al. 1996; Stewart et al. 1997; Deloukas et al. 1998).
A wide variety of cytogenetic, genetic, and physical mapping data is available on a genome, chromosome, or local scale. Because the methods and resources used to build these maps vary in their basic principles and resolution power, their integration and the assessment of the precision of the position of a given gene pose both fundamental and practical problems and difficulties that remain largely unresolved. Even in regions in which extensive genomic sequences are available, this remains an important task to achieve to facilitate the identification of the genes involved in inherited diseases and physiological traits within the frame of positional cloning approaches. The scaling up of human genome sequencing that is underway makes it an even more important issue to address.
Collecting expression profiles of human genes in the hundreds of cell types that make up the human body would further bridge the gap between the structure of the genome and biology. The basic methods for cDNA array hybridization have been available since the inception of the genetic engineering revolution >20 years ago. Advances in robotics, imaging technologies, and informatics that are the hallmark of the genome era made it possible to collect expression profiles by semiquantitative hybridization on thousands of cDNA targets spotted at high density on membranes (Nguyen et al. 1995; Takahashi et al. 1995; Zhao et al. 1995; Piétu et al. 1996). However, it is only recently that, with the emergence of novel platforms using microarray formats of increased densities and fluorescent probes, the potential of this approach has received more attention and acceptance in the community (Schena et al. 1995, 1996; De Risi et al. 1996, 1997; Lockhart et al. 1996; Lashkari et al. 1997; Zhang et al. 1997; Wodicka et al. 1997; Cho et al. 1998; de Saizeu et al. 1998; Gray et al.1998) with other applications such as genotyping (Winzeler et al. 1998). Whether based on arrays of cDNA clones or arrays of oligonucleotides synthesized in situ on glass, these techniques take advantage of the physical and/or information resources of the IMAGE Consortium.
In this context, the Genexpress team developed the IMAGE concept (Auffray et al. 1995) and cofounded the IMAGE Consortium (Lennon et al. 1996). After the initial characterization of one-third of the first IMAGE Consortium cDNA library from infant brain (Soares et al. 1994), we developed the Genexpress Index (Houlgatte et al. 1995), a resource for gene discovery and the genic map of the human genome based on clustering of annotated cDNA clones, sequences, and ESTS genic markers assigned to specific human chromosomes. We also established a method for semiquantitative analysis of the expression levels of thousands of gene transcripts and identified by differential hybridization a set of novel genes transcribed preferentially in human muscles (Piétu et al. 1996).
Here we further characterize 5058 human genes represented in the infant brain library, and describe a novel, prototype resource for functional and computational genomics of the human brain transcriptome, the Genexpress IMAGE Knowledge Base, integrating curated sequence, map, and expression annotations.
RESULTS AND DISCUSSION
To obtain a preliminary documentation of the expression profiles of 5058 human gene transcripts, hybridization (Piétu et al. 1996) of an array of 7451 clones from the infant brain cDNA library (Soares et al. 1994) was performed with complex cDNA probes derived from brain, muscle, heart, testis, placenta and thymus mRNA.
Because the hybridization signal intensity of individual clones is correlated in most cases with the number of copies of the corresponding gene transcript, it is possible to classify transcripts into general categories of abundance. Because it has not been demonstrated how well the observed absolute signals correlate with the actual number of molecules per cell, result of these first-pass hybridizations should be regarded as semiquantitative.
As a reflection of the high complexity of the brain transcriptome, only 759 of the clones (10%) can be ascribed to the categories of moderate-to-high abundance, having hybridization intensity values with the two brain complex cDNA probes >1.96 (95% confidence interval to differ from the normal distribution of low signals, see Piétu et al. 1996).
Similarly, hybridizations with the complex probes from other tissues reflected the complexity of their transcriptomes, with 1.8% of the clones classified in these categories with the two muscle probes and 6.3% for the two testis probes. In addition, 103 of the clones (1.4%) hybridized at significant levels with one or more complex probes derived from the five other tissues tested but not with the brain probes.
To assess if hybridization with several probes could be because of the presence of repetitive sequences, we hybridized the cDNA array with a mixture of Alu and LINE probes, and 731 clones (10%) were found positive. Of those, 285 did not hybridize significantly with any of the complex probes, indicating that they contain members of the repetitive sequence families that are not represented often in the transcriptomes. A similar number of the clones containing repetitive sequences (283) represented 80% of the clones hybridizing with four to six complex probes. These results should therefore be interpreted with caution as an indication of ubiquitous expression of the corresponding genes, as a large fraction of them may actually be expressed at lower levels in some or all of the tissues analyzed.
Another 10% of the clones are associated with hybridization intensity values close to the 1.96 threshold and represent transcripts of low abundance in the different tissues tested, whereas the remainder of the clones have hybridization intensity values that cannot be distinguished from background with high confidence with the methodology used.
The results of these expression-profiling experiments form the basis of the expression module of the Genexpress IMAGE Knowledge Base. They are presented in a table on the web site (http://idefix.upr420.vjf.cnrs.fr/EXPR/welcome.html/).
The corresponding comprehensive clone and sequence clustering and integrated gene mapping information of Genexpress Index2 (Mariage-Samson et al., in prep.), an upgraded version of the Genexpress Index (Houlgatte et al. 1995), are also included.
Entry into and navigation through the resulting Genexpress IMAGE Knowledge Base of the brain transcriptome can be envisioned in a variety of different ways. This depends on whether initial emphasis is placed on questions such as where, when, and at what level is a gene expressed, or on the relatedness of the sequence of the gene and its products (transcripts and proteins) to structures of known function, or on map information: Where is the gene located precisely, and is there a human pathology associated with the corresponding genomic region?
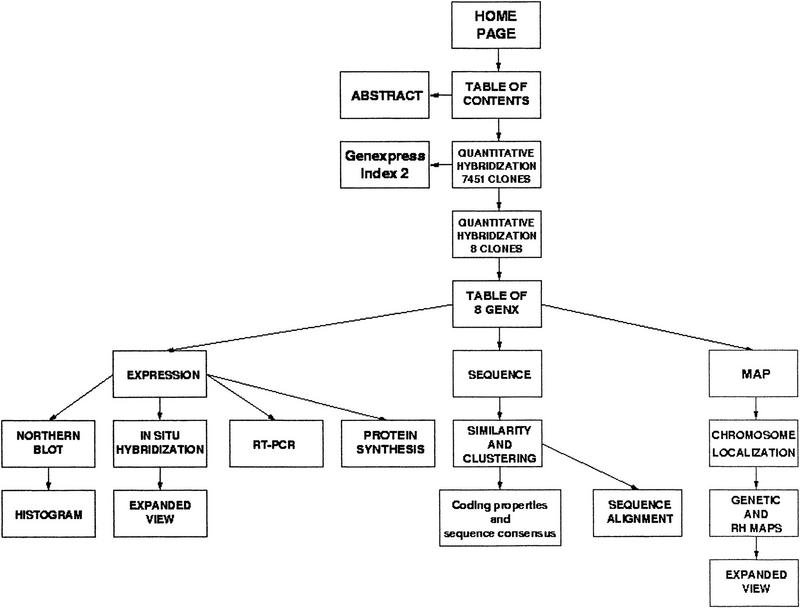
Four of the genes detected as expressed preferentially or specifically in brain based on the semiquantitative differential hybridization results (Table 1) and further characterized by in situ hybridization on rat tissues sections are taken as examples to discuss the problems encountered and the solutions implemented. One other gene is discussed more briefly to illustrate another possible navigation route taking first advantage of sequence information. Only the figures relevant to the first example are presented in printed form. The others (Table 2) are discussed more briefly, and the relevant figures are available on the web site, the content of which is presented in schematic form in Figure 1.
Table 1.
Hybridization Signal Intensity of the Seven Gene Transcripts Selected for Detailed Characterization
| Genexpress clone (1NIB) name | Image clone ID | GENX Index1 | GENX Index2 | Alu | Brain1 | Brain2 | Heart | Muscle1 | Muscle2 |
|---|---|---|---|---|---|---|---|---|---|
| c-0ob03 | 34591 | 465 | 465 | −0.19 | 4.13 | 5.6 | 1.51 | −1.27 | 1.39 |
| c-11h05 | 49279 | 3960 | 3960 | −0.34 | 4.06 | 3.33 | 0.38 | −2.29 | 1.59 |
| c-0ge07 | 26501 | 5201 | 4044 | 0.43 | 4.12 | 3.56 | −0.44 | −0.91 | 0.25 |
| c-03a10 | 32335 | 4858 | 4858 | −0.78 | 2.31 | 2.03 | −0.65 | −1.08 | −0.18 |
| c-0oa03 | 34458 | 3111 | 200991 | 1.91 | 4.35 | 4.4 | 1.65 | −1.59 | −0.02 |
| c-08e08 | 57147 | 769 | 769 | 1.6 | 5.81 | 7.5 | −0.09 | −1.02 | 2.44 |
| c-01d10 | 31734 | 3809 | 3809 | 0.11 | 4.64 | 4.54 | 3.98 | −1.7 | 4.37 |
| Genexpress clone (1NIB) name | Image clone ID | GENX Index1 | GENX Index2 | Placenta1 | Placenta2 | Thymus1 | Thymus2 | Testis1 | Testis2 |
| c-0ob03 | 34591 | 465 | 465 | −0.77 | 0.76 | 0.12 | 0.6 | −0.22 | 1.58 |
| c-11h05 | 49279 | 3960 | 3960 | 0.02 | −0.66 | −0.04 | 0.01 | 0.3 | 0.31 |
| c-0ge07 | 26501 | 5201 | 4044 | −0.57 | −0.86 | 0.06 | −0.72 | −0.08 | −0.77 |
| c-03a10 | 32335 | 4858 | 4858 | −1.24 | −0.64 | −2.3 | −0.56 | −1.55 | −1.3 |
| c-0oa03 | 34458 | 3111 | 200991 | −0.08 | 1.01 | 0.23 | 0.99 | 0.9 | 1 |
| c-08e08 | 57147 | 769 | 769 | −0.52 | 1.07 | 0.53 | 0.54 | 0.51 | 1.16 |
| c-01d10 | 31734 | 3809 | 3809 | −0.75 | 0.69 | −0.94 | −0.74 | −0.5 | 0.03 |
Clones were selected based on hybridization intensity Ri > 1.96 for the brain probe and a maximum of two other probes. Hybridization intensity values used for selection are in dark shade.
Table 2.
Characterization of Seven Gene Transcripts Selected as Preferentially Expressed in Brain
| GENXa | Sequence similarityb | GenBank, SWISS-PROT* accession no.c | Genexpress clone (1NIB) name | IMAGE clone ID | GenBank/ EMBL accession no. | Insert size (kb) | Transcript size (kb) | Chromosomal localization | Orphan neurologic pathologiesd |
|---|---|---|---|---|---|---|---|---|---|
| 465 | REL Rattus norvegicus calcium- independent α-latrotoxin receptor | U72487 U78105O09026* O35818* | c-0obo3 | 34591 | Z42577F02089 | 1.4 | 7.5 | 4q12 | |
| 769 | ID clone 23682 mRNA sequence ID clone 24440 mRNA sequence ID mRNA for KIAA0513 protein | U79288 AF055005AB011085 |
c-08e08 | 57147 | Z42089F01625 | 1.7 | 7.5 | 16q24.3 | |
| 3809 | ID protein Armadillo multigene family (Hatzfeld and Natchtsheim 1996) | X81889 Q99569* |
c-01d10 | 31734 | Z42486Z38681 | 1.5 | 4.6 | 2q22.3–q24.2 | |
| 3960 | ID visinin-like protein1 (Polymeropoulos et al. 1995) | U14747 P42323* | c-11h05 | 49279 | F06242F02525 | 1.5 | 2 | 2 | |
| 4044 | REL mouse and cattle cyclin-dependent kinase regulatory subunit P35 | S73375 S82819 |
c-0ge07 | 26501 | Z42350Z38574 | 1.8 | 4.2 | 17p11.2–q12 | DFNB3 DDPAC |
| 4858 | ID human DNA sequence Chr22 | Z99716Z82192 | c-03a10 | 32335 | Z41917Z38211 | 1.8 | 6.0/2.6 | 22q13.1–q13.3 | |
| 200991 | ID clone 23876 neuronal olfactomedin-related ER localized protein mRNA | U79299 AF035301Q99784* | c-0oa03 | 34458 | F05835F02082 | 1.4 | 2.6 | 9q34.1–34.3 | ALS4 |
GENX number refers to the Genexpress Index 2 cluster of cDNA clones, sequences, and ESTS markers.
Classification of each GENX cluster is based on data base similarities. Sequences were classified as known (ID) if identical to a human sequence and related (REL) if having a partial similarity to a sequence in another species.
When a significant sequence similarity was found, the GenBank or SWISS-PROT (with an asterisk) accession no. is reported.
(DFNB3) Neurosensory recessive deafness 3; (DDPAC) frontotemporal dementia with parkinsonism with variable phenotypes; ALS4) familial ALS.
Figure 1.
Schematic representation of the Web site content. Available at http://idefix.upr420.vjf.cnrs.fr/EXPR/welcome.html.
From Expression to Sequence to Map: GENX-769
Expression
The results of semiquantitative hybridizations (Table 1) indicated that the GENX-769 gene was expressed at a significantly higher level in brain and to a lesser extent in muscle, when compared to testis, placenta, and thymus. RT–PCR analysis of 14 human tissues (Nagase et al. 1998) confirmed and extended these results (see web site).
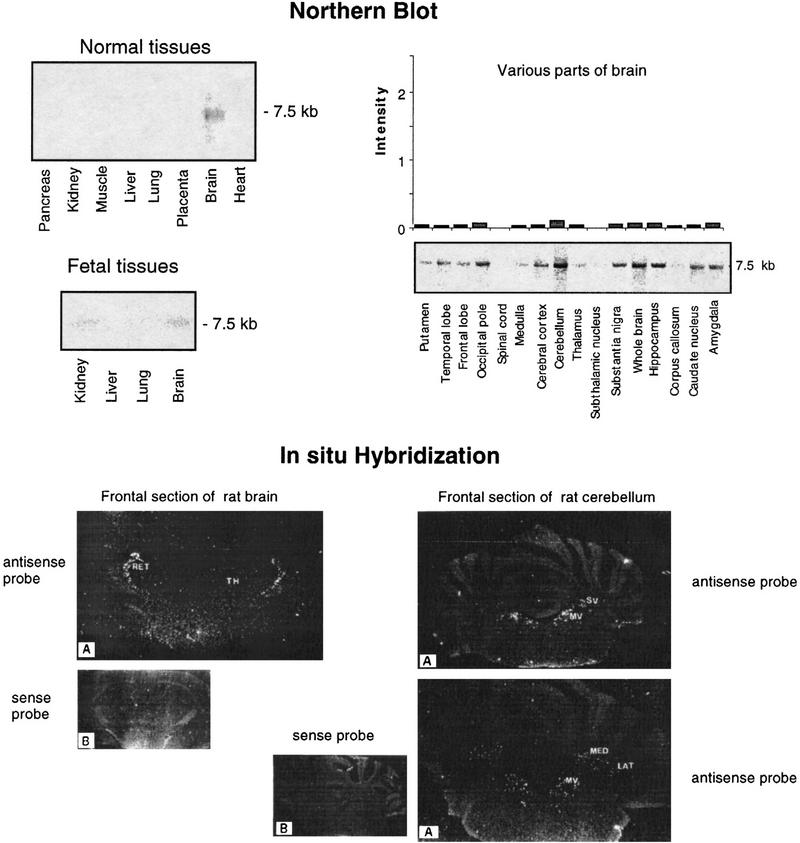
Northern blot analysis of eight adult tissues and four fetal tissues revealed a faint signal in brain only, corresponding to a 7.5-kb mRNA. The signal was relatively weaker in fetal brain as compared to adult brain. Further Northern analysis of mRNAs from 15 brain regions indicated similar low expression in most regions, with a slightly stronger signal in cerebellum (Fig. 2, top).
Figure 2.
Expression profiles of the GENX-769 mRNA. (Top) Northern blot analysis of the GENX-769 transcript in 8 normal tissues, 15 regions of brain, and 4 fetal tissues. The insert of cDNA clone c-08e08 (IMAGE-57147) was used as a probe. The size of the transcript is indicated in kb. Each lane contained 2 μg of poly(A)+ mRNA from the various human sources. For expression in 15 regions of brain, intensity signals observed on Northern blots was quantitated using ImageQuant software (Molecular Dynamics). To normalize the signal in relation with the quantity of RNA in each lane and compare variations from one tissue to another, each value was divided by those obtained by hybridization of the Northern blots with ubiquitous control actin and ubiquitin cDNA probes to control the amount of mRNA. The signal intensity is represented in arbitrary units. (Bottom) In situ hybridization of GENX-769. Rat brain coronal sections through the diencephalon hybridized with antisense (A) and sense (B) probes. Only the reticular nuclei (RET) in the thalamus (TH) are labeled. A very weak signal is observed in the hippocampus (Ammon’s horn and dentate gyrus). On sections through the cerebellum, labeled cells are localized in the medial vestibular (MV) and superior vestibular (SV) nuclei, and in the medial (ME) and lateral (LAT) cerebellar nuclei.
In situ hybridization of rat brain and cerebellum sections provided evidence of a very restricted pattern of expression. Labeled cells were specifically found within the reticular thalamic nucleus. Isolated, scattered neurons were also found in the superior and medial vestibular nuclei, which mediates vestibulo-ocular reflexes and in the lateral medial cerebellar nuclei (Fig. 2, bottom). This pattern of expression is reminiscent of a reflex control pathway of the head position correlated with eye movements, in which the vestibular nuclei integrate inputs from the cerebellar nuclei, which are then passed to the cerebral cortex through the reticular thalamic nuclei, and therefore suggests a possible route of investigation to explore in deciphering the function of the protein encoded by the GENX-769 gene.
Sequence
Sequence similarity search in databases detected a match of the GENX-769 consensus with three mRNA sequences (U79288, AF055005, AB011085) encoding an uncharacterized protein (Table 2). AB011085 represents the complete mRNA sequence of 7758 nucleotides with an open reading frame of 412 amino acids. It is related to a genomic sequence of Drosophila (AC003052).
A 57-kD protein is obtained by in vitro-coupled transcription and translation of the AB011085 cDNA clone coding for the KIAA0513 protein (Nagase et al. 1998, see web site).
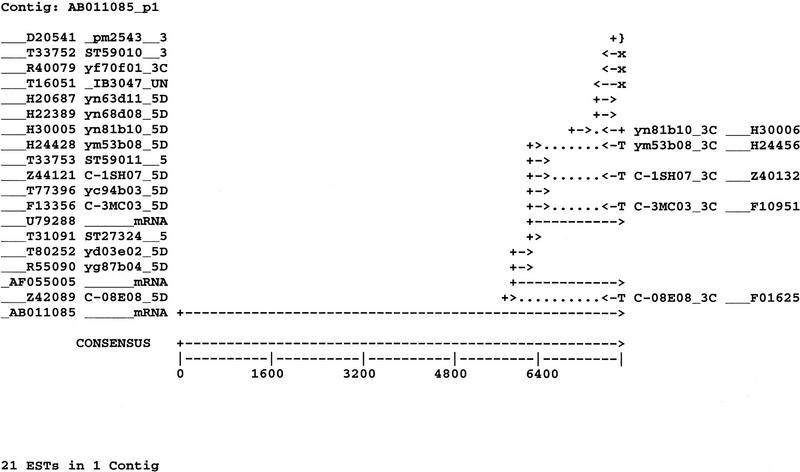
The results of cumulative cDNA clone and sequence clustering of the Genexpress Index2 are displayed in Figure 3 in relation with the 7.7-kb GENX-769 mRNA. Details on the clones, sequences, and their coding properties, as well as full sequence alignments are available in the web site, together with relevant links to the related UniGene and TIGR entries. Among the 21 sequences assembled in the GENX-769 cluster, 17 are present in UniGene cluster Hs.85053 (Build#46), which contains 21 additional sequences. The 21 sequences of the GENX-769 cluster are all represented in three THC and two singleton sequence clusters in the TIGR database (release 3.3), which altogether contains 29 sequences. Two of the THC sequence clusters (THC204729 and THC174938) represent opposite ends of the same transcript, and the third (THC111593) correspond to a possible alternative splicing. The singletons were not clustered because of limited overlaps in the absence of the full-length AB011085.
Figure 3.
The GENX-769 cluster. Sequence accession numbers and clone names are indicated; RNA is indicated by mRNA with its GenBank accession no. (T) Polyadenylation sites if a polyadenylation signal and a poly(T) tail are present at the 5′ end of the sequence; (x) only the polyadenylation signal is present at the 5′ end of the sequence; (}) the polyadenylation site is present at the 3′ end of the sequence.
Similarly, 19 of the 21 sequences of the GENX-769 cluster are constitutive of the STACK cluster “brain 765” in the SANBI database, one in STACK singleton “2415-0-hemat-001-1997-0.1” and one in STACK singleton “18463-0-brain-001-1997-0.1.”
It is worth noting that 15 of the 16 cDNA clones derived from the GENX-769 mRNA and registered in GenBank came from brain libraries and only one from a promyelocyte library (HL60), further underlining the preferential expression of the GENX-769 gene in brain.
Map
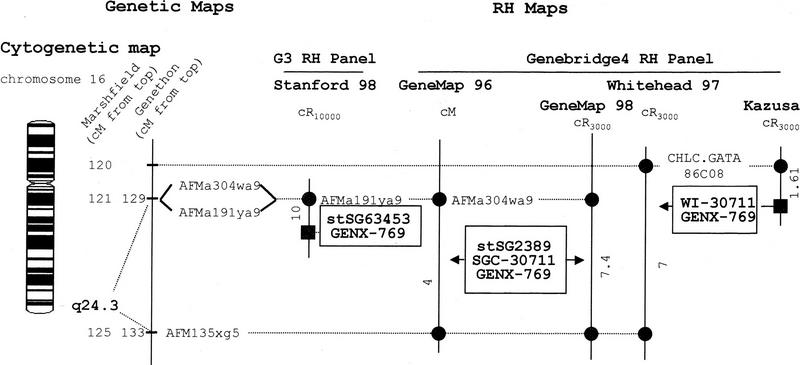
The precise position of the GENX-769 gene on the human genome map was investigated through the exploration of all available data from the various radiation-hybrid and physical maps. Missing data were generated using the radiation hybrid (RH) panel G3. The collected data were integrated with the genetic, cytogenetic, and disease maps in the region.
A total of four ESTS markers were found associated to various sequences present in the GENX-769 cluster. The mapping of three of these markers has been performed using the Genebridge4 (GB4) RH panel and one with the G3 panel. This illustrates the frequent observation that several independent ESTS markers have been used by several groups to map the same gene with the same or different resources.
The integrated mapping data are presented in Figure 4. All the microsatellite markers involved map to the 16q24.3 cytogenetic region according to the GENATLAS data base. The disease map (GENATLAS) does not present any orphan neurologic pathology associated with that region. The localizations of the ESTS markers associated with the GENX-769 gene are presented as intervals delineated by two genetic markers (the AFMa304wa9 and AFM135xg5 markers in the Gene Map ’96 and ’98, the CHLC.GATA86C08 and AFM135xg5 markers in the Whitehead 97 map), or as distance to a framework marker in the GB4-based Kasuza map and in the G3-based Stanford map (Whitehead framework for the GB4-based Kazusa data and Stanford framework for the G3-based map).
Figure 4.
Localization of the GENX-769 gene in the integrated genome maps. The cytogenetic map presented is based on GENATLAS data. The genetic map is displayed with a scale in centiMorgans (cM) from the top of the chromosome according to the Marshfield (CHLC) and the Généthon maps. When the mapping result is expressed as a distance (with a significant lodscore) to a framework marker the GENX-769 marker is represented by a square. When localized in a given interval, the GENX-769 marker is written inside a box and points to this interval with a black arrow for odds >10:1 or lod score >1, a gray arrow for odds <10:1 or lod score <1. The microsatellites or other markers are reported as circles. The distances between markers of the RH maps are indicated in centiRay10000 (cR10000) for the G3 RH panel, in cR3000 for Gene Map ’98, Whitehead ’97, and Kazusa maps and in cM for Gene Map ’96. D numbers or aliases (AFMa304wa9) D16S3061; (AFMa191ya9) D16S3037; (AFM135xg5) D16S520; (CHLC;GATA86C08) 16S2625.
Arbitrary scales have been used to represent units in cM, cR3000, or cR10000 of the various maps. Although tempting, the conversion of the various scales to a common kilobase scale was not chosen as a rule in Figure 4 to respect the original data as much as possible and because it could lead to various distorsions. The factors used for this type of conversion are average factors, calculated for each chromosome by using their total length expressed either in cM, Mb, cR3000, or cR10000 (Gyapay et al. 1996; Slonim et al. 1997; Stewart et al. 1997; Deloukas et al. 1998). Variations of these conversion factors are expected along the chromosomes as observed in the case of chromosome 16 for instance (Doggett et al. 1995), mostly in the centromeric and most telomeric regions. In addition, and because the GB4 panel was used in several independent reports involving different sets of markers, the cR3000-to-kb conversion factor varies depending the origin of the GB4-based map.
Here, application of these conversion factors (see references in Materials and Methods) would suggest that the interval of localization of the GENX-769 marker in Gene Map ’96 (4 cM) is ∼2.5-Mb long, whereas the same interval in Gene Map ’98 is 1.4-Mb (7.4 cR3000) long. The Whitehead data with a slightly larger interval on the genetic map (5 cM) correspond to a 7 cR3000, that is, 2.3-Mb interval. Finally, the most precise localization of the GENX-769 marker is provided by the Kazusa and Stanford data, which indicate the approximate position of the GENX-769 marker in the intervals (1.61 cR3000, i.e., ∼324 kb from the CHLC.GATA-86C08 marker, 10 cR10000, i.e., ∼340 kb from the AFMa191ya9 marker, respectively). Physical resources such as cosmid sequence-ready maps, available in the region (Doggett et al. 1995), and sequence data are awaited to provide an exact localization of this gene.
In addition to comparing the data pertaining from various maps, Figure 4 allows easy searching through available mapping resources to find all other ESTS markers corresponding to other potential genes assigned to the same interval as the GENX-769 markers. For instance, a query of Gene Map ’96 using the AFMa304wa9 (D16S3061)–AFM135xg5 (D16S520) interval yields a list of 38 ESTS markers localized to this interval in this map.
The integrated mapping figure (Fig. 4) also provides tools to gain access to physical genomic ressources, such as YAC and cosmid contigs positioned on the genome owing to somatic hybrid cell line DNA. Browsing through the chromosome 16 data (Table 4) is facilitated by knowing the name of the closest microsatellite markers in various maps. In the case of the GENX-769 gene, the D16S520 marker points to a list of YAC and cosmid contigs that could prove useful to study the structure of the GENX-769 gene.
Table 4.
Uniform URLs on the World Wide Web
| Database | URL |
|---|---|
| Sequence | |
| UniGene (NCBI) | http://www.ncbi.nlm.nih.gov/UniGene/ |
| GenBank | http://www.ncbi.nlm.nih.gov/Entrez/nucleotide.html |
| SWISS-PROT | http://www.expasy.ch/sprot/ |
| Washington University | http://genome.wustl.edu/est/esthmpg.html |
| Map | |
| RHdb (EBI) | http://www.ebi.ac.uk/RHdb/ |
| Gene Map ’96 | http://www.ncbi.nlm.nih.gov/SCIENCE96/ |
| Whitehead Institute/MIT | http://www.genome.wi.mit.edu/ |
| Whitehead Institute map | http://carbon.wi.mit.edu:8000/cgi-bin/contig/phys_map/ |
| TIGR | http://www-tigr.org/tdb/hgi/hgi.html |
| SANBI/STACK | http://ziggy.sanbi.ac.za/stack |
| Genome Data Base | http://www.gdb.org/ |
| Chromosome 16 data | http://www-ls.lanl.gov/data/map16.txt/ |
| GenAtlas | http://bisance.citi2.fr/GENATLAS/ |
| OMIM | http://www3.ncbi.nlm.nih.gov/Omim/ |
| Stanford | http://www-shgc.stanford.edu/ |
| General | |
| IMAGE | http://www-bio.llnl.gov/bbrp/image/image.html |
| LENS | http://agave.humgen.upenn.edu/lens/ |
| Infobiogen | http://www.infobiogen.fr/ |
From Expression to Sequence to Map: GENX-200991
Expression
GENX-200991 was found to be expressed at a relatively high level in adult and fetal brains and at a very low level in adult liver. Moreover, of the large number (153) of cDNA clones derived from the GENX-200991 mRNA and registered in GenBank, 150 or 98% are from brain, 2 are from pancreas, and 1 is from lung libraries, establishing the high degree of brain-specific expression of this gene.
The GENX-200991 gene is expressed at a higher level in temporal, frontal, and occipital lobes, cortex, hippocampus, and amygdala than in other parts of brain. Expression is very low in spinal cord as compared to other parts of the central nervous system.
In situ hybridization revealed a remarkable concentration of neurons expressing the GENX-200991 mRNA in the hippocampal formation. Labeled cells belonged to the pyramidal layer and particularly strongly labeled neurons were found in the hilar region of the dentate gyrus.
Labeled cells were abundant throughout the cerebral cortex: in the frontal, parietal, and perirhinal regions. Although a complete mapping of the entire central nervous system was not realized, serial sectioning revealed distribution of the GENX-200991 transcript in different other areas of the thalamus: habenula and paraventricular central medial, centrolateral, and thalamic nuclei. Hybridizing neurons were also found within the granular cell layer of the cerebellum. The expression of the GENX-200991 gene in brain structures that are sensory relay centers of the olfactory pathway (habenulae, hippocampal archeocortex, paraventricular, and central thalamic nuclei), in the limbic system (amygdala and hippocampus), which is thought to mediate the affective component of odors, and in the thalamus–neocortex projection involved in conscious perception of smell suggest a possible role of the encoded protein in the physiology of olfaction. In this context, expression in the cerebellum could be related to the integration of the inputs from the hippocampus through afferent projections, then relayed through the thalamus to the neocortex.
Sequence
The GENX-200991 gene is the human gene encoding an olfactomedin-related protein localized in the endoplasmic reticulum. A full-length cDNA was isolated recently from rat (Nagano et al. 1998) but not yet from human cDNA. This protein plays a role in the maintenance, growth, or differentiation of chemosensory cilia on the apical dendrites of olfactory neurons, a major component of the extracellular matrix of the olfactory neuroepithelium. Such a similarity reinforces the functional hypothesis suggested by the expression pattern described above. Of the 263 sequences included in the GENX-200991 cluster, 258 were found in two UniGene clusters: Hs.25999 and Hs.74376.
Map
The GENX-200991 gene lies in a 19-cM interval at 9q34.1–q34.3 in the region associated with an orphan pathology of the nervous system, ALS4, a familial form of amyotrophic lateral sclerosis (ALS). The large 19-cM mapping interval is mainly attributable to the fact that the location for one marker varies between the different maps. Marker NIB29 is assigned to three different intervals in Gene Map ’96, Gene Map ’98, and the Whitehead ’97 map. These discrepancies may reflect some experimental difficulties inherent to the marker itself, especially in the case of Gene Map ’98 in which its location is reported with low confidence. Thus, it appears reasonable to only take into account those concordant data obtained with the other markers SHGC-20581, STSG-63472, and Cda0af08. The localization of the GENX-200991 gene could thus be reduced to a 12-cM (∼12 Mb) interval at 9q34.3, between the AFMb001ve9 and AFM073yb11 markers. The distance to the centromeric marker is estimated in the G3 Stanford map to 28 cR10000 (∼840 kb).
From Expression to Sequence to Map: GENX-465
Expression
Northern blots demonstrated a low level of expression of the GENX-465 mRNA, in both adult and fetal brain. Expression is very low and almost similar in all the regions of brain tested.
In situ hybridization revealed weak expression of the GENX-465 mRNA in different brain regions. A diffuse pattern of labeled cells was observed in the caudate putamen and cerebral cortex areas. In the hippocampus, moderately labeled neurons were distributed in the granular cell layer of the dentate gyrus and in the pyramidal cell layer of Ammon’s horn. In contrast with these diffuse patterns, a distinct hybridization signal was observed in the Purkinje cell layer in the cerebellar cortex. Scattered isolated cells were labeled in the molecular layer and a faint signal was observed in the granular layer. Based on these observations, it is worth exploring if the receptor encoded by the GENX-465 gene has something to do with the inhibitory control of motor behavior or emotional perception exerted by Purkinje cells.
Sequence
The GENX-465 gene is related to the rat calcium-independent α-latrotoxin receptor gene encoding latrophilin. α-Latrotoxin is a neurotoxin that stimulates neurotransmitter release from all synapses. Latrophilin represents the calcium-independent receptor and/or molecular target for α-latrotoxin (Davletov et al. 1996). It is a novel member of the secretin family of G protein-coupled receptors that are involved in secretion (Kraspoperov et al. 1997; Lelianova et al. 1997). Of 30 (72%) cDNA clones registered in GenBank, 24 are from brain libraries, and the remaining are from retina (4) and placenta (2) libraries. Among 54 cDNA sequences included in the GENX-465 cluster, 14 were not present in UniGene cluster Hs.21917.
Map
Mapping data for the GENX-465 gene involve three ESTS markers and spread at 4q12 along a 7-cM (∼5.8 Mb) interval between the AFM356tc5 and AFM036yb2 markers. No orphan pathology of the nervous system was found to be associated with that genomic region in the GENATLAS data base.
The integration of the data concerning the mapping of the GENX-465 gene reveals that Gene Map ’98 assigns one GENX-465-specific ESTS to a more centromeric interval when compared to Gene Map ’96. The assignment of another GENX-465 marker reported previously in Gene Map ’96, is conserved in Gene Map ’98 but with low confidence. Interestingly however the very precise localization of the GENX-465-specific marker in the G3 Stanford map (8 cR10000, i.e., ∼190 kb from the AFM297za5 microsatellite marker) is in good concordance with the assignment reported in Gene Map ’96 but not with the most confident Gene Map ’98 assignment. This illustrates one of the numerous problems encountered when trying to integrate mapping data. Physical mapping and sequence data should help in resolving such ambiguities ultimately.
From Expression to Sequence to Map: GENX-4858
Expression
Expression of the GENX-4858 mRNA measured by Northern blot analysis is very high in amygdala, high in occipital pole, cerebellum, hippocampus, and caudate nucleus and low in subthalamic nucleus and corpus callosum, whereas it is very low in spinal cord.
In situ hybridization of coronal sections of adult rat brain revealed a wide distribution in various brain structures. Strong hybridization signals were detected in the cerebral cortex: neocortex, entorhinal, and perirhinal cortex. In the hippocampal formation, dense accumulation of silver grains was also detected in Ammon’s horn and dentate gyrus. Positive hybridization was observed in the amygdaloid complex and in the caudate putamen. Most of the thalamic nuclei and the hypothalamus were labeled diffusely. The granular layer in the cerebellum was labeled strongly. A moderate labeling was observed in deep cerebellar nuclei. Within the pons, mRNAs were detected in the vestibular nuclei, spinal trigeminal nucleus, cochlear and facial nuclei, and reticular formation. The wide pattern of expression of this gene in numerous structures suggest a general rather than specific role in the maintenance of brain functions.
Sequence
The GENX-4858 sequence matches exactly two registered human genomic sequences of chromosome 22, indicating that it contains a newly identified gene, preferentially transcribed in brain as shown above and confirmed by the observation that 28 out of 30 (93%) cDNA clones are derived from brain libraries and 2 from a placenta library. Thirty-six out of 39 sequences of the GENX-4858 cluster are found in UniGene cluster Hs.8073.
Map
A single marker (WI-6209) for the GENX-4858 gene can be found in four distinct reports in the mapping data bases, locating the gene at 22q13. The integration of these data proved very tedious as none of the intervals described on the various maps had common flanking markers. Ordering the intervals with respect to each other was performed thanks to the genetic markers contained in that region and to the localization of some of the framework markers of a given map inside specific intervals of the other maps. It is worth noting that the interval indicated by Gene Map ’98 (17 cR3000 or ∼3.9 Mb) is the largest among all data and corresponds to 7 cM (∼3.7 Mb) on the genetic map. A more precise localization is provided by Gene Map ’96 (2 cM, ∼1 Mb), between the AFM261xd9 and AFMb357yc9 markers. In addition, the GENX-4858 gene is present on a 250-kb YAC from the Whitehead contig WC-983, together with a genetic marker WI-581. Unfortunately this marker has not been used in any of the RH maps. Integration with the other mapping data therefore involves an overlapping larger (1720 kb) YAC of the contig, which contains both the GENX-4858 marker and the α-N-acetyl-galactosaminidase precursor gene (NAGA) which has been assigned to the AFM261xd9–AFM164th8 interval in Gene Map ’96.
From Sequence to Map to Expression: GENX-4044
Sequence
The GENX-4044 gene appears to be related to the mouse and the cattle cyclin-dependant kinase regulatory subunit p35 gene that was described to be expressed in neurons in which it phosphorylates both high-molecular-weight neurofilaments and microtubule-associated protein Tau (Ohshima et al. 1995, 1996). From the 44 cDNA sequences included in the GENX-4044 cluster, only 20 are found and split into the UniGene Hs.90202 and Hs.93597 clusters.
Map
Various results obtained with four independent ESTS markers lead to the localization of the GENX-4044 gene in an interval of 19 cM around the centromere of chromosome 17.
The interval is smaller if the distance is estimated marker by marker. For example the localization is at 3 cR10000 (∼90 kb) from a framework marker with the G3 panel, and in an interval of 3.9 cR3000 (∼660 kb) using the GB4 panel in the Gene Map ’98. The improvement provided by this latter map through the merging of the GB4 mapping data from different centers (Deloukas et al. 1998) is obvious here as two markers that were assigned to two different intervals (5 and 9 cM) separated by a 5-cM interval in Gene Map ’96 are now found in a single 3.9 cR3000 interval in Gene Map ’98, corresponding to a 2 cM (∼880 kb) interval between the AFMb322wg5 and AFM179xg11 microsatellite markers. Unfortunately, the precise localization proposed by the G3 Stanford map is divergent, i.e., outside of this interval.
Finally, the GENX-4044 gene was located on a YAC contig that also contains the microsatellite markers delineating Gene Map ’98 interval. The approximate length of the interval can thus be estimated since at least two overlapping YACs of 1240 and 1300 kb are involved, suggesting a distance smaller than 2.5 Mb (sum of the two YACs).
The 17p11.2–q21.1 region in which the GENX-4044 gene maps, is associated with two brain orphan pathologies: the neurosensory recessive deafness 3 (DFNB3, a putative homolog of mouse shaker 2) and a frontotemporal dementia with parkinsonism with variable phenotypes (DDPAC).
Expression
The GENX-4044 gene is expressed highly in adult and fetal brains. Its expression is almost similar in all the regions of brain tested. Thirty-one out of 34 (91%) of the cDNA clones contained in the GENX-4044 cluster are from brain libraries.
Concluding Remarks
To take advantage of the vast and increasing amount of diverse types of sequence, map, and expression data collected using the IMAGE Consortium resources, we have developed the Genexpress IMAGE Knowledge Base as a prototype model of a resource for functional and computational genomics.
It is based in part on cumulative annotations and clustering of IMAGE cDNA clones, the sequences and the genic ESTS markers derived from them, registered in the Genexpress Index2 (Mariage-Samson et al., in prep.). Comparison with other clustering efforts conducted at NCBI, TIGR, and SANBI data libraries, based on the limited number of examples discussed in the present paper, underlines their complementarity. However these other approaches attempt to solve the clustering problems by purely algorithmic means, enabling complete recalculation each time, a task that requires large computational power. The rapidly increasing number of sequence databases that have to be cross-checked (there are currently almost 200 of them listed on the Infobiogen server) makes it a formidable challenge, even with the projected increases in computing power. In contrast, we have relied on a multistep approach, with intermediate validation steps requiring human intervention and therefore enabling input of biological expertise. This follows the model that was developed by Amos Bairoch and his team to establish SWISS-PROT (Bairoch and Apweiler 1997, 1998), recognized as the best annotated and curated database of proteins, although it lags behind the vast reservoir of putative proteins registered in TREMBL (translations of EMBL/GenBank./DDBJ nucleic acid sequences). As a consequence, the development of the Genexpress Index has the same potential advantages, and faces the same limitations of keeping in pace with the increasing number of cDNA sequences of GenBank.
The collection and integration of available gene mapping data is another demanding task, requiring browsing through multiple heterogeneous public electronic databases and genome center Web sites. We did not attempt to propose a unique precise assignment for each gene, but rather provided a visual representation of the current state of knowledge enabling to keep track of the origin of the data used and review it again if needed. The tools are thus provided to further investigate additional chromosome-specific resources, and to identify additional genes located in the same genomic region as putative candidates to study in relation with physiology and disease.
With regard to expression profiles, the situation is very different. There is no established model for an international public repository of expression profiles. However, there are several efforts to develop such expression databases in relation with model organisms such as the mouse (Ringwald et al. 1997) or Drosophila (Bellen and Smith 1995) and one was proposed very recently (Emolaeva et al. 1998). Therefore it can be anticipated that in the future, Northern blots, RT–PCR and in situ hybridization will be reported in a standard format enabling registration in public electronic databases and will have an accession number attached to them in much the same manner as sequence and mapping data. This will make it possible for large number of individual scientists and laboratories to contribute additional expertise and knowledge on a scale unattainable by even the largest dedicated genome centers.
In this context, the collection of quantitative hybridization data with cDNA arrays provides a unique opportunity to bridge the gap between the genome and proteomes by a comprehensive analysis of transcriptomes. Hybridization signals can be measured over the three to four orders of magnitude that correspond to the usual distribution of mRNA abundances. Therefore, if one considers both coverage, as represented by the number of targets in the cDNA array, and deepness of analysis, as measured by the hybridization signal intensity reflecting mRNA abundance, it is possible to collect millions of biologically meaningful bits of information in single experiments that can be performed in a matter of hours or days. A similar throughput can be matched only by massive parallelization of cDNA sequencing that is only partially achieved in the SAGE technique (Velculescu et al. 1995). Exploitation and understanding of the results is another matter. It remains a major challenge, as shown by studies performed in yeast, the only eukaryotic organism in which expression profiles have been collected on a genome scale so far (DeRisi et al. 1997; Velculescu et al. 1997; Wodicka et al. 1997; Cho et al. 1998; Gray et al. 1998).
Through the different examples discussed in this paper, we have illustrated the potential represented by the integration of the different types of data collected. We believe that advanced programming for each of the sequence, map, and expression modules, combined with simple and powerful graphical interfaces and communication systems using alert agents will provide the means and the necessary compromises that are needed for the further development of our IMAGE Knowledge Base to provide an up-to-date, integrated vision of current biological knowledge on the scale of entire transcriptomes with the participation of the community of biologists.
METHODS
Expression Profiling by Semiquantitative cDNA Array Hybridization
The detailed procedures are those described in Piétu et al. (1996).
Human Brain cDNA Array and High-Density Filters
The cDNA clones are from the normalized infant brain cDNA library cloned in the Lafmid vector (Soares et al. 1994). The array used in this study consisted of 7451 from the first 9216 cDNA clones sequenced by the Genexpress team (Auffray et al. 1995) and subjected to clone and sequence-clustering analyses (Houlgatte et al. 1995). A total of 1765 clones were excluded because they were shown to be chimeric, derived from mitochondrial transcripts, or associated with bad-quality sequences.
The cDNA clone inserts were amplified by PCR from their original 96-well plates using primers flanking the cloning site of the Lafmid vector. One to two nanograms of the PCR products were spotted onto 8- × 12-cm Nylon filters (Hybond N+, Amersham, UK) using a Flexys robot from Hybaid (Cambridge, UK) at a density of 25 microtiter plates per filter (2400 clones) in a 5 × 5 format.
Preparation and Labeling of Complex cDNA Probes
Single-strand cDNA probes were derived from various poly(A)+ mRNA preparations from human brain, skeletal muscle, heart, placenta, testis, and thymus (Clontech, Palo Alto, CA). Poly(A)+ mRNA (250 ng) was reverse transcribed using Superscript II reverse transcriptase (RNase H−) (GIBCO-BRL, Gaithersburg, MD), as described in the manufacturer’s protocol, using random hexamers for priming. Labeling was performed simultaneously by incorporation of [33P]dATP (3000 Ci/mmole, Amersham, UK).
Hybridization and Signal Quantitation
Duplicate filters were hybridized with a first preparation of each probe as well as with Alu and LINE repetitive probes labeled by random priming (see below); then the entire set of hybridizations was repeated with novel probe preparations, (except for the heart probe) so that for each clone/probe combination, four hybridization data points were collected.
Hybridization signals were captured by exposure to Phosphor screens followed by scanning with the PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Identification and quantitation of the spots were performed using the Xdots Reader sofware (Cose, Le Bourget, France). The intensity values of each individual hybridization signal were normalized using the four sets of 2400 values collected for each filter. The resulting normalized intensities (Ri) are represented by the number of standard deviations separating the signal from the population of weak signals following a normal distribution, allowing assessment of their significance in differential hybridization comparisons (see Piétu et al. 1996 for details).
Northern Blot Analyses
Northern blots containing 2 μg of poly(A)+ mRNA from 8 adult tissues, 4 fetal tissues, or 15 distinct regions from brain were purchased from Clontech (MTN blots). For each cDNA clone, plasmid DNA was purified by the Wizard protocol (Promega). After digestion of the plasmid by the restriction enzymes HindIII and NotI, the inserts were purified from agarose gel by centrifugation on Amicon Microcon-50 Micropure Nebulizer separator system (Grace). Twenty-five nanograms of purified insert was labeled with [33P]dATP (Amersham, 3000 Ci/mmole) by the random priming method according to the manufacturer’s protocol (GIBCO-BRL). Hybridization was performed as described by the manufacturer using ExpressHyb hybridization solution (Clontech) and 20 × 106 cpm of radiolabeled probe. Northern blots were analyzed after overnight exposure using the PhosphorImager. Human actin and ubiquitin probes were used to control the amount of RNA in each lane of the membranes.
In Situ Hybridization
Preparation of Tissue Sections
Wistar Rats (3-week-old males) were obtained from Iffa Credo (Domaine des Oncins, France). Coronal 10-μm brain section were cut in a cryostat (Frigocut 2800-N Leica, Rueil-Malmaison, France) and fixed in 4% paraformaldehyde for 20 min. Some sections were counterstained with toluidine blue for histological observations.
DNA Probes for ISH Analysis
Antisense and sense (control) single-stranded (ss) DNA probes were generated by two-step linear PCR amplification from a specific cDNA clone for each gene transcript (Table 3). The first step of the PCR procedure involves a conventional exponential amplification of the cDNA inserts from the plasmid templates. The double-stranded (ds) DNA was purified from agarose gels as described above. In the second step, the 33P-labeled ssDNA probes were prepared by linear PCR performed in presence of the sense or antisense oligonucleotides (Table 3). Thirty cycles of linear PCR were carried out with 20 pmoles of a single primer, 100 pmoles of dCTP, dGTP, and dTTP, 10 ng of dsDNA template in 50 μl of 1× Taq buffer, 5 μl (10 pmoles) [33P]dATP (sp. act. >3000 Ci/mmole, Amersham) and 1 unit of Taq DNA polymerase (Amersham) using the Perkin Elmer 9600 DNA thermal cycler. Each cycle was made of three consecutive periods of 30 sec for denaturation at 94°C, annealing at 55°C, and extension at 72°C. Unincorporated nucleotides were separated from the labeled probes by gel filtration using a Sephadex G-50 column (Boehringer, Mannheim, Germany). The specific activity of the probes was 5 × 109 cpm/μg.
Table 3.
Probes for in Situ Hybridization
| GENX | Genexpress clone (1NIB) name | Size (hp) | Probe | |
|---|---|---|---|---|
| 465 | c-0ob03 | 235 | sense | 5′-TACTCCACAGGCTCACAGAG-3′ |
| antisense | 5′-AGCCACGGTCTATGATTTGC-3′ | |||
| 769 | c-08e08 | 169 | sense | 5′-TCTCTTTCTCAAGCTCCACAC-3′ |
| antisense | 5′-ACAGGCGTGAGGCATCACAC-3′ | |||
| 4858 | c-03a10 | 200 | sense | 5′-CCTGGCACCCCTAAACCAT-3′ |
| antisense | 5′-GAGTGAGAAGCAGCGCTTGA-3′ | |||
| 200991 | c-0oa03 | 231 | sense | 5′-ACACGGACAATTTCACCTCC-3′ |
| antisense | 5′-TCTCGTCCACCATGAGGTCG-3′ |
Hybridization
The probes were diluted in the hybridization buffer [50% vol/vol deionized formamide, 0.6 m NaCl, 10 mm Tris-HCl at pH 7.4, 1 mm EDTA, 1× Denhardt’s solution, 100 mm DTT, 250 μg/ml hydrolyzed salmon sperm DNA, 250 μg/ml yeast tRNA and 5% (wt/vol) dextran sulfate]. The hybridization cocktail was denatured at 85°C, prior to its application to sections that had been pretreated with 0.02 m HCl for 10 min and 2 mg/ml proteinase K for 8 min at room temperature to increase probe access. After incubation overnight at 52°C, the slides were first washed at 60°C in 4× SSC for 20 min, followed by a final wash at 0.5× SSC at room temperature. Autoradiography was performed for 10 days using the Ilford K5 emulsion. The slides were visualized and photographed with Leica Wild M 3B or Leitz Laborlux S microscopes.
Clustering of cDNA Clones, Sequences, and Genic Markers
The 7451 clones of the human brain cDNA array correspond to 5058 clusters of clones, sequences, and genic ESTS markers assembled in the Genexpress Index, or Index1, as described (Houlgatte et al. 1995). To extend the annotation of these 5058 clusters, referred to as GENX clusters, we relied on a second generation, updated and upgraded version of Index1, Index2, which contains 61,000 GENX clusters (Mariage-Samson et al., in prep.). Index2 features information on cDNA libraries, authors, localization at the 5′ or 3′ end of the transcript and clone name, extracted from GenBank release 105.0; name, GDB entry, clone source, clone name, insert size, and localization at the 5′ or 3′ end of transcript extracted from dbEST release 03-10-98; definition and comments on RNAs and encoded proteins extracted from GenBank release 105.0 and SWISS-PROT release 35.0, respectively.
Because some GENX clusters from Index1 were merged based on newly identified links or split because of subsequent identification of erroneous links, some GENX numbers of Index2 differ from those of Index1. As a result, the 5058 GENX of Index1 became 4469 in Index2 (see Table in the web site).
All validated information is included in a GENX cluster that can be accessed by clicking on the GENX number in the table. For each GENX cluster, the Fragment Assembly Project module of the GCG package is used to create contigs and then a graphical display is generated. It is followed by text sections featuring clone, sequence, and map information that contains pointers to the appropriate entries in the public electronic databases and genome center Web sites, and ends with full contig sequence alignments.
Integrated Gene Mapping
RH data have been produced by several genome centers using the Genebridge4 (GB4) or the G3 RH panel. The GB4 panel consists of 93 hamster cell lines retaining random fragments of the human genome of ∼10 Mb after exposure to 3000 rads of X-rays (Walter et al. 1994; Gyapay et al. 1996). This panel provides an average resolution of 270 kb/cR based on data available on the Whitehead Institute server. The G3 panel is made of 83 hamster cells lines retaining fragments of ∼4 Mb after exposure to 10,000 rads, providing an average resolution of 30 kb/cR (Stewart et al. 1997).
Available mapping data concerning a given GENX cluster were retrieved from the appropriate Web sites according to the following procedure. First, the sequence accession numbers associated with each GENX cluster were extracted from Index2 and the corresponding UniGene entries (Build#33), then cross-checked against the dbSTS database (release 02-19-98) to identify aliases. Second, the completed list of sequence accession numbers was cross-checked against the Radiation Hybrid Data Base (RHdb release 10.0) to retrieve all RH markers associated with a given GENX cluster. The RH markers and their aliases, as well as the type of RH panel and map used were extracted from the RHdb pages and from the corresponding Amplimer pages of the Genome Data Base (GDB release 6.4). Mapping data obtained with the G3 RH panel were collected from the Stanford Human Genome Center server or generated de novo using the same procedures (Stewart et al. 1997). Mapping data obtained with the GB4 RH panel were collected from Gene Map ’96 (Schuler et al. 1996), Human Gene Map ’98 (Deloukas et al. 1998), the Kazusa DNA Research Institute (Nagase et al. 1998), and together with physical mapping data when available from the Whitehead Institute (Hudson et al. 1995).
Integration of the gene mapping data with genetic linkage data was performed by reference to the Généthon (Dib et al. 1996) and CHLC (Sheffield et al. 1995) maps, using the microsatellite marker delineating the intervals. Cytogenetic localizations of the intervals and the associated human pathologies were extracted from GENATLAS.
Conversion of the various scales to a kilobase scale is indicated in the text only and was performed using the mean factors indicated by the various authors or deduced form their data (Gyapay et al. 1996, for the cM-to-Mb conversion; Stewart et al. 1997, for the cR10000-to-kb conversion; Slonim et al. 1997, for the cR3000-to-kb conversion in the Whitehead map; Deloukas et al. 1998, for the cR3000-to-kb conversion in Gene Map ’98).
The URLs for the public electronic databases and genome center Web sites are listed in Table 4. A dedicated web site was implemented at CNRS (http://idefix.upr420.vjf.cnrs.fr/EXPR/welcome.html) by Brainstorm, Paris for the presentation of the data.
Acknowledgments
This work was supported by the CNRS and grants from the European Union to C.A. (GENE-CT-93-0089) and BIOMED2 programs (EURO-IMAGE Consortium, BMH4-CT-97-2284) to C.A. and W.A. C.D. was supported by a fellowship from Groupements Parkinsoniens, E.E. and C.M. by Rhône-Poulenc Rorer, and Y.V. by Genome Express.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pietu@infobiogen.fr; FAX (33-1) 49583509.
REFERENCES
- Adams MD, Kervlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995;377:3–174. [PubMed] [Google Scholar]
- Anderson NL, Anderson NG. Proteome and proteomics: New technologies, new concepts and new words. Electrophoresis. 1998;11:1853–1861. doi: 10.1002/elps.1150191103. [DOI] [PubMed] [Google Scholar]
- Antequera F, Bird A. Predicting the total number of human genes. Nat Genet. 1994;8:114. doi: 10.1038/ng1094-114a. [DOI] [PubMed] [Google Scholar]
- Auffray C, Béhar G, Bois F, Bouchier C, Da Silva C, Devignes MD, Duprat S, Houlgatte R, Jumeau MN, Lamy B, et al. IMAGE: Integrated molecular analysis of the human genome and its expression. C R Acad Sci. 1995;318:263–272. [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database: Its relevance to human molecular medical research. J Mol Med. 1997;75:312–316. [PubMed] [Google Scholar]
- ————— The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1998. Nucleic Acids Res. 1998;26:38–42. doi: 10.1093/nar/26.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Smith RF. FlyBase: A virtual Drosophilia cornucopia. Trends Genet. 1995;11:456–457. doi: 10.1016/s0168-9525(00)89145-3. [DOI] [PubMed] [Google Scholar]
- Burke J, Wang H, Hide W, Davison DB. Alternative gene form discovery and candidate gene selection from gene indexing projects. Genome Res. 1998;8:276–290. doi: 10.1101/gr.8.3.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TC, Gabrielian AE, Landsman D, Lockhart DJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;1:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Davletov BA, Shamotienko OG, Lelianova VG, Grishin EV, Ushkaryov YA. Isolation and biochemical characterization of a Ca2+-independent alpha-latrotoxin-binding protein. J Biol Chem. 1996;271:23293–23245. doi: 10.1074/jbc.271.38.23239. [DOI] [PubMed] [Google Scholar]
- de Saizeu A, Certa U, Warrington J, Gray C, Keck W, Mous J. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat Biotechnol. 1998;16:45–48. doi: 10.1038/nbt0198-45. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tome P, Hui L, Matise TC, McKusick KB, Beckman JS, et al. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JF. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Vishwanath RI, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Dib C, Fauré S, Fizames C, Samson D, Drouot N, Vignal A, Millaseau P, Marc S, Hazan J, Seboun E, et al. A comprenhensive genetic map of the human genome based on 5264 microsatellites. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- Doggett, N.A., L.A. Goodwin, J.G. Tesmer, L.J. Meinck, D.C. Bruce, M.R. Clark, M.R. Altherr, A.A. Ford, H.C. Chi, B.L. Marrone et al. 1995. An integrated physical map of human chromosome 16. Nature 377 (suppl.): 335–365. [DOI] [PubMed]
- Dujon B. European functional analysis network (EUROFAN) and the functional analysis of the Saccharomyces cerevisia genome. Electrophoresis. 1998;19:617–624. doi: 10.1002/elps.1150190427. [DOI] [PubMed] [Google Scholar]
- Ermolaeva O, Rastogi M, Pruitt KD, Schuler GD, Bittner ML, Chen Y, Simon R, Meltzer P, Trent JM, Boguski M. Data management and analysis for gene expression arrays. Nat Genet. 1998;20:19–23. doi: 10.1038/1670. [DOI] [PubMed] [Google Scholar]
- Fields C, Adams MD, White O, Venter JC. How many genes in the human genome? Nat Genet. 1994;7:345–346. doi: 10.1038/ng0794-345. [DOI] [PubMed] [Google Scholar]
- Gray NS, Wodicka L, Thunnissen AM, Norman TC, Kwon S, Espinoza FH, Morgan DO, Barnes G, LeClerc S, Meijer L, et al. Exploiting chemical libraries, structure and, genomics in the search for kinase inhibitors. Science. 1998;281:533–538. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- Gyapay G, Schmitt K, Fizames C, Jones H, Vega-Czarny N, Spillett D, Muselet D, Prud’Homme JF, Dib C, Auffray C, et al. A radiation hybrid map of the human genome. Hum Mol Genet. 1996;5:339–346. doi: 10.1093/hmg/5.3.339. [DOI] [PubMed] [Google Scholar]
- Hatzfeld M, Nachtsheim C. Cloning and characterization of a new armadillo family member, p0071, associated with the junctional plaque: Evidence for a subfamilly of a closely related proteins. J Cell Sci. 1996;109:2767–2778. doi: 10.1242/jcs.109.11.2767. [DOI] [PubMed] [Google Scholar]
- Hillier L, Lennon G, Becker M, Bonaldo MF, Chiapelli B, Chissoe S, Dietrich N, DuBusque T, Favello A, Gish W. Generation and analysis of 280,000 human expressed tags. Gen Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- Houlgatte R, Mariage-Samson R, Duprat S, Tessier A, Bentolila S, Lamy B, Auffray C. The Genexpress Index: A resource for gene discovery and genic map of the human genome. Gen Res. 1995;5:272–304. doi: 10.1101/gr.5.3.272. [DOI] [PubMed] [Google Scholar]
- Hudson TJ, Stein LD, Gerety SS, Ma J, Castle AB, Silva J, Slonim DK, Baptista R, Kruglyak L, Xu S, et al. An STS-based map of the human genome. Science. 1995;270:1945–1954. doi: 10.1126/science.270.5244.1945. [DOI] [PubMed] [Google Scholar]
- Humphery-Smith I, Cordwell SJ, Blackstock WP. Proteome research: Complementary and limitations with respect to the RNA and DNA worlds. Electrophoresis. 1997;8:1217–1242. doi: 10.1002/elps.1150180804. [DOI] [PubMed] [Google Scholar]
- Krasnoperov VG, Bittner MA, Beavis R, Kuang Y, Salnikow KV, Chepurny OG, Little AR, Plotnikov AN, Wu D, Holz RW, et al. Alpha-latrotoxin stimulates exocytosis by the interaction with a neuronal G-protein-coupled receptor. Neuron. 1997;18:925–937. doi: 10.1016/s0896-6273(00)80332-3. [DOI] [PubMed] [Google Scholar]
- Lashkari DV, DeRisi JL, McCusker JH, Namath AF, Gentile C, Hwang SY, Brown PO, Davis RW. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc Natl Acad Sci USA. 1997;94:13057–13062. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelianova VG, Davletov BA, Sterling A, Rahman MA, Grishin EV, Totty NF, Ushkaryov YA. Alpha-latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. J Biol Chem. 1997;272:21504–21508. doi: 10.1074/jbc.272.34.21504. [DOI] [PubMed] [Google Scholar]
- Lennon G, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E consortium: An integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Folletie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Nagano T, Nakamura A, Mori Y, Maeda M, Takami T, Shisaka S, Takagi H, Sato M. Differentially expressed olfactomedin-related glycoproteins (pancortins) in the brain. Mol Brain Res. 1998;53:13–23. doi: 10.1016/s0169-328x(97)00271-4. [DOI] [PubMed] [Google Scholar]
- Nagase T, Ishikawa K, Miyajima N, Tanaka A, Kotitani H, Nomura N, Ohara O. Prediction of the coding sequence of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5:31–59. doi: 10.1093/dnares/5.1.31. [DOI] [PubMed] [Google Scholar]
- Nguyen C, Rocha D, Granjean S, Baldit M, Bernard K, Naquet P, Jordan B. Differential gene expression in the murine thymus assayed by quantitative hybridization of arrayed cDNA clones. Genomics. 1995;29:207–216. doi: 10.1006/geno.1995.1233. [DOI] [PubMed] [Google Scholar]
- Ohshima T, Nagle JW, Pant HC, Joshi JB, Kozak CA, Brady RO, Kulkarni AB. Molecular cloning and chromosomal mapping of the mouse cyclin-dependent kinase 5 gene. Genomics. 1995;28:585–588. doi: 10.1006/geno.1995.1194. [DOI] [PubMed] [Google Scholar]
- Ohshima T, Kozak CA, Nagle JW, Pant HC, Brady RO, Kulkarni AB. Molecular cloning and chromosomal mapping of the mouse gene encoding cyclin-dependent kinase 5 regulatory subunit p35. Genomics. 1996;35:372–375. doi: 10.1006/geno.1996.0370. [DOI] [PubMed] [Google Scholar]
- Piétu G, Alibert O, Guichard V, Lamy B, Bois F, Leroy E, Mariage-Samson R, Houlgatte R, Soularue P, Auffray C. Novel gene transcripts preferentially expressed in human muscles revealed by quantitative hybridization of a high density cDNA array. Genome Res. 1996;6:492–503. doi: 10.1101/gr.6.6.492. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Ide S, Soares MB, Lennon GG. Sequence characterization and genetic mapping of the human VSNL1 gene, a homologue of the rat visinin-like peptide RNVP1. Genomics. 1995;29:273–275. doi: 10.1006/geno.1995.1244. [DOI] [PubMed] [Google Scholar]
- Ringwald M, Davis GL, Smith AG, Trepanier LE, Begley DA, Richardson JE, Eppig JT. The mouse gene expression database GXD. Cell Dev Biol. 1997;8:489–497. doi: 10.1006/scdb.1997.0177. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Heller R, Chai A, Brown P, Davis RD. Parrallel human genome analysis: Microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler GD. Pieces of the puzzle: Expressed sequence tags and the catalog of human genes. J Mol Med. 1997;75:694–698. doi: 10.1007/s001090050155. [DOI] [PubMed] [Google Scholar]
- Schuler GD, Boguski MS, Stewwart EA, Stein LD, Gyapay G, Rice K, White RE, Rodriguez-Tomé P, Aggarwal A. A gene map of the human genome. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- Sheffield VC, Weber JL, Buetow KH, Murray JC, Even DA, Wiles K, Gastier JM, Pulido JC, Yandava C, Sunden SL, et al. A collection of tri-and tetranucleotide repeat markers used to generate high quality, high resolution human genome-wide linkage maps. Hum Mol Genet. 1995;4:1837–1844. doi: 10.1093/hmg/4.10.1837. [DOI] [PubMed] [Google Scholar]
- Slonim D, Kruglyak L, Stein L, Lander E. Building human genome maps with radiation hybrids. J Comput Biol. 1997;4:487–504. doi: 10.1089/cmb.1997.4.487. [DOI] [PubMed] [Google Scholar]
- Soares MB, Bonaldo F, Jelenc P, Su L, Lawton L, Efstratiadis A. Construction and characterisation of normalized cDNA library. Proc Natl Acad Sci. 1994;91:9228–9232. doi: 10.1073/pnas.91.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EA, McKusick KB, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S, et al. An STS-Based radiation hybrid map of the human genome. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Hashida H, Zhao N, Misumi Y, Sakaki Y. High-density cDNA filter analysis of the expression profiles of the genes preferentially expressed in human brain. Gene. 1995;164:219–227. doi: 10.1016/0378-1119(95)00396-n. [DOI] [PubMed] [Google Scholar]
- VanBogelen RA, Abshire KZ, Moldover B, Olson ER, Neidhart FC. Escherichia coli proteome analysis using the gene-protein database. Electrophoresis. 1997;8:1243–1251. doi: 10.1002/elps.1150180805. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Volgestein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Zhou W, Volgeltein J, Basrai MA, Basset DE, Hieter P, Volgeltein B, Kinzler KW. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- Walter MA, Spillett DJ, Thomas P, Weissenbach J, Goodfellow PN. A method for constructing hybrid maps of whole genomes. Nat Genet. 1994;7:22–28. doi: 10.1038/ng0594-22. [DOI] [PubMed] [Google Scholar]
- Wilkins MR, Gasteiger E, Tonella L, Ou K, Tyler M, Sanchez JC, Gooley AA, Walsh BJ, Bairoch A, Appel RD, et al. Protein identification with N and C-terminal sequence tags in proteome projects. J Mol Biol. 1998;278:599–608. doi: 10.1006/jmbi.1998.1726. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Richards DR, Conway A, Goldstein AL, Kalman S, McCullough MJ, McCusker JH, Stevens DA, Wodicka L, Lockhart DJ, et al. Direct allelic variation scanning of the yeast genome. Science. 1998;281:1194–1197. doi: 10.1126/science.281.5380.1194. [DOI] [PubMed] [Google Scholar]
- Wodicka L, Dong H, Mittmann M, Ho M, Lockhart DJ. Genome-wide expression monitoring in Saccharomyces cerevisia. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- Zhao N, Hashida N, Takaashi N, Misumi Y, Sakaki Y. High density cDNA filter analysis: A novel approach for large scale quantitative analysis of gene expression. Gene. 1995;156:207–213. doi: 10.1016/0378-1119(95)00023-y. [DOI] [PubMed] [Google Scholar]