SUMMARY
Huntington’s disease like-2 (HDL2) is a phenocopy of Huntington’s disease caused by CTG/CAG repeat expansion at the Junctophilin-3 (JPH3) locus. The mechanisms underlying HDL2 pathogenesis remain unclear. Here we developed a BAC transgenic mouse model of HDL2 (BAC-HDL2) that exhibits progressive motor deficits, selective neurodegenerative pathology and ubiquitin-positive nuclear inclusions (NIs). Molecular analyses reveal a novel promoter at the transgene locus driving the expression of a CAG repeat transcript (HDL2-CAG) from the strand antisense to JPH3, which encodes an expanded polyglutamine (polyQ) protein. Importantly, BAC-HDL2 but not control BAC mice accumulate polyQ-containing NIs in a pattern strikingly similar to those in the patients. Furthermore, BAC mice with genetic silencing of the expanded CUG transcript still express HDL2-CAG transcript and manifest polyQ pathogenesis. Finally, studies of HDL2 mice and patients revealed CBP sequestration into NIs and evidence for interference of CBP-mediated transcriptional activation. These results suggest overlapping polyQ-mediated pathogenic mechanisms in HD and HDL2
INTRODUCTION
Huntington’s disease like-2 (HDL2) is an autosomal dominant neurodegenerative disorder that has a broad phenotypic overlap with Huntington’s disease (HD) (Margolis et al., 2001). Similar to HD, HDL2 is characterized by adult-onset of symptoms including chorea, dystonia, rigidity, bradykinesia, psychiatric symptoms, and dementia, eventually leading to premature death about 10–15 years after disease onset (Margolis et al., 2005). HDL2 accounts for a small subset of patients with clinical manifestations of HD who do not have the HD mutation, an expanded CAG repeat encoding polyglutamine (polyQ) repeat in huntingtin (Margolis et al., 2004; Margolis et al., 2001; Schneider et al., 2007).
The neuropathology of postmorterm HDL2 brains is strikingly similar to that of HD (Greenstein et al., 2007; Rudnicki et al., 2008). Both exhibit robust and selective striatal and cortical atrophy, with neurodegeneration primarily targeting the striatal medium spiny neurons (MSNs) and a subset of cortical pyramidal neurons (ibid). Moreover, both HD and HDL2 brains contain intranuclear inclusions (NIs) that are ultrastructurally similar and are immunostained with ubiquitin and 1C2 (an antibody against the expanded polyQ epitope but can also recognize polyleucine; Trottier et al., 1995; Dorsman et al., 2002). The pattern of NI distribution in HD and HDL2 is similar but not identical. They both have a higher density in the cortex and amygdala than in the striatum, and NIs are rarely observed in the cerebellum or midbrain (Greenstein et al., 2007; Rudnicki et al., 2008). However, unlike those in HD, NIs in HDL2 are more frequent in the upper cortical layers II/III than deep cortical layers, and they are absent in pons and medulla (ibid). Another key pathological differences between the two disorders is that NIs in HD, but not those in HDL2, contain mutant huntingtin (mhtt) (Margolis et al., 2001). Therefore, the pathogenic origin of NIs in HDL2 remains to be uncovered.
HDL2 is caused by a CTG/CAG expansion on chromosome 16q24.3 (Holmes et al., 2001). The expanded CTG/CAG repeats in HDL2 patients range from 40–59, with normal individuals carrying 6–28 repeats. HD and HDL2 not only have comparable ranges of CTG/CAG repeat expansions, but also similar slopes in their inverse relationships between the repeat length and age of onset for movement disorders (Margolis et al., 2004). The CTG repeat expansion is located within the variably spliced exon 2A of JPH3, which is not part of the main transcript encoding JPH3 protein (Holmes et al., 2001). On the sense strand, three splice variants that include exon 2A have been described, placing the expanded CTG repeat in polyleucine or polyalanine open reading frames (ORFs), or in the 3′ untranslated region (UTR).
Currently, the molecular pathogenic mechanisms for HDL2 remain an enigma (Margolis et al., 2005; Orr and Zoghbi, 2007). Three possible mechanisms have been postulated. First, the expansion of the CUG repeat may reduce JPH3 mRNA expression leading to a partial loss-of-function for JPH3 protein, which normally tethers the plasma membrane to the endoplasmic reticulum to facilitate crosstalk between cell-surface and intracellular ion channels (Nishi et al., 2002; Takeshima, 2001). In support of this theory, JPH3 knockout mice exhibit motor impairment but such mice do not appear to accumulate NIs or exhibit neurodegeneration (Nishi et al., 2002). A second possible pathogenic mechanism, similar to that demonstrated for myotonic dystrophy type-1 and 2 (DM1 and DM2), is that the expanded CUG or CCUG repeat RNA form RNA foci which can sequester an RNA binding protein muscleblind-like 1 (MBNL1) and interfere with its function in regulating alternative splicing (Ranum and Cooper, 2006; Kanadia et al., 2003). Supporting this possibility, Rudnicki and colleagues (Rudnicki et al., 2007) showed CUG RNA foci in HDL2 brains, and the ability of mutant HDL2-CUG RNA transcripts to interfere with the splicing of MBNL1 targets in cultured cells (Rudnicki et al., 2007). However, the expanded CUG RNA in DM1 was not known to elicit NIs or apparent neurodegeneration. Moreover, CUG RNA foci in HDL2 patients do not frequently co-localize with NIs (ibid), suggesting distinct pathogenic origins for these entities.
To gain insights into the pathogenesis of an HD phenocopy, we developed a series of Bacterial Artificial Chromosome (BAC)-mediated transgenic mouse models of HDL2 (BAC-HDL2) that contain an expanded CTG/CAG repeat in the human JPH3 BAC, as well as control BAC mice with a non-expanded CTG/CAG repeat. BAC-HDL2 but not control BAC mice recapitulate motor, neuropathological and molecular phenotypes similar to those in the patients. Importantly, molecular analyses revealed a novel promoter driving the expression of an expanded CAG repeat containing transcript emanating from the strand antisense to JPH3. This mutant HDL2-CAG transcript can mediate polyQ protein toxicity (e.g. sequestration and interference of CBP-mediated transcription), hence providing a molecular pathogenic link between HD and HDL2.
RESULTS
Generation and Characterization of a BAC Transgenic Mouse Model of HDL2
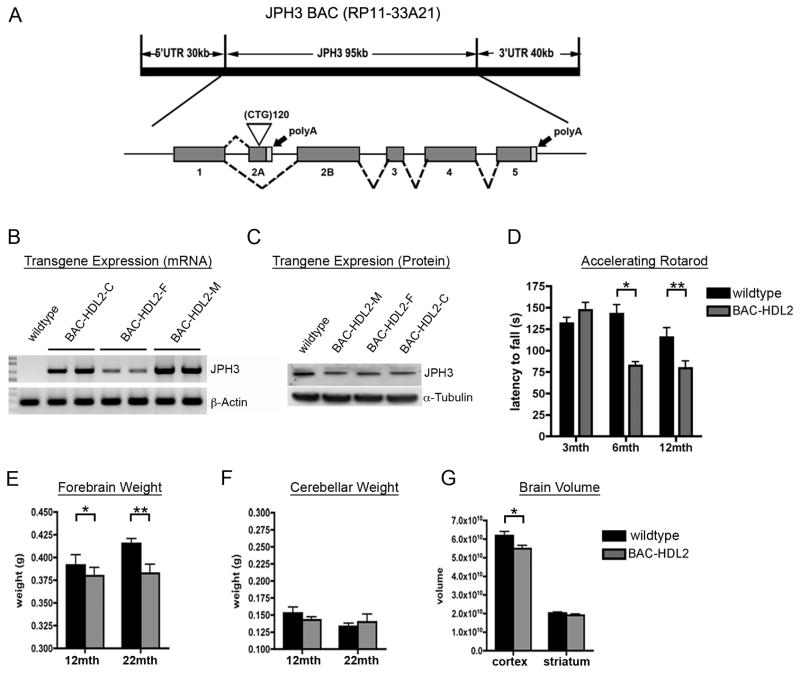
Since BACs preserve the intact human genomic context and have been successfully used to develop transgenic mouse models for other neurodegenerative disorders including HD (Gong et al., 2002; Yang et al., 1997; Gray et al., 2008; Gu et al., 2009), we undertook a BAC transgenic approach to develop a mouse model for HDL2. We selected a human BAC (RP11-33A21) that contains the intact 95 kb JPH3 genomic locus in addition to approximately 30 kb 5′- and 40 kb 3′-genomic flanking sequences. The BAC was engineered to contain an expanded CTG/CAG track of 120 repeats in the exon 2A of JPH3, preserving the repeat ORFs in both the sense and antisense strands compared to those in the patients (Figure 1A). In designing the BAC-HDL2 construct, we purposely chose a longer stretch of CTG/CAG repeats (~120 repeats) than what is found in patients (i.e. 40–59) because prior experience in modeling other trinucleotide repeat disorders such as SCA1 and HD suggests that longer repeat lengths are needed to accelerate the disease process such that disease manifestation occurs within the short lifespan of a mouse (Zoghbi and Botas, 2002).
Figure 1.
Generation and Characterization of BAC-HDL2 animals. (A) A schematic representation of the human JPH3 locus. A BAC containing the intact JPH locus (RP11-33A21) was modified in order to insert ~120 CTG repeats into the alternatively spliced exon 2A (white triangle). (B) RT-PCR performed using primers specific to human JPH3 exons 2B and 4. (C) JPH3 Western blot performed on brain lysates isolated from BAC-HDL2C/M/F lines and wildtype control. (D) Accelerating rotarod testing performed at 3, 6 and 12 months of age (3 month: wild-type N=22, BAC-HDL2-C N=16; 6 month: wild-type N=14, BAC-HDL2-C N=11; 12 month: wild-type N=10, BAC-HDL2-C N=11). (E) BAC-HDL2-C line forebrains weigh significantly less than their wild-type littermates at both 12 and 22 months of age. (F) No significant differences detected in cerebellum weights between HDL2-C and its wild type littermates at 12 and 22 months. (G) Stereological brain volume measurements of BAC-HDL2-C line cortex and striatum revealed a significant decrease in the cortical volume at 18–22 months of age. (BAC-HDL2-C: striatum, N=6, cortex, N=6; wildtype, striatum, N=6, cortex, N=6). Values are mean ± SEM (*p< 0.05, **p< 0.01, Student’s t test).
The engineered mutant BAC was microinjected into inbred FVB/N mouse embryos to generate transgenic founders. A total of ten BAC-HDL2 founders were obtained and five were bred for germline transmission. Three of the BAC-HDL2 lines (C, F and M) integrated 1–4 copies of the BAC transgene (data not shown). Direct sequencing was used to determine the precise repeat length: C line has 116 CTG/CAG repeats, F line has 122 repeats, and M line has integration at a single locus of BAC with 119 and 13 repeats. Since both C and F lines have only the expanded repeats, we focused our phenotypic studies on these two independent lines. We next assessed whether JPH3 mRNA and JPH3 protein are overexpressed in these models. As demonstrated in Figure 1B, reverse transcriptase PCR (RT-PCR) analysis that specifically amplified exon 2B to exon 4 of JPH3 readily detects transgene expression in the brain of BAC-HDL2 lines. Semi-quantitative RT-PCR (sqRT-PCR) analyses revealed that the level of overexpression was about 100% of endogenous murine Jph3 in the C line and about 20% in the F line (Supplemental Figure S1). Intriguingly, when we analyzed BAC-HDL2 lines for JPH3 protein levels, we did not detect any significant overexpression (Figure. 1C and Supplemental Figure S1). Nonetheless, the latter observation is consistent with the finding in DM1 that the expanded CUG repeat may impair DMPK protein expression via a cis-mechanism of reduced RNA nuclear export (Ranum and Cooper, 2006). Because of the higher level of transgene expression in BAC-HDL2-C line, the majority of our phenotypic analyses are focused on this line (hereafter referred to as BAC-HDL2). However, several pathogenic phenotypes were also independently confirmed using the F line (BAC-HDL2-F).
BAC-HDL2 mice exhibit age-dependent motor deficits and neurodegenerative pathology
HDL2 patients are characterized clinically by a middle-age onset of movement disorders including motor incoordination (Margolis et al., 2005), and neuropathologically by the selective atrophy of the striatum and cortex (Greenstein et al., 2007; Rudnicki et al., 2008). To evaluate whether our model recapitulates aspects of these disease features, we studied a cohort of BAC-HDL2 mice and wildtype littermates using the behavioral and neuropathological assays that have been established for HD mice (Gray et al., 2008). To assess for evidence of age-dependent motor deficits, we used accelerating rotarod assay and observed significant impairment in BAC-HDL2 mice compared to wildtype controls at both 6 and 12 but not at 3 months of age (Figure 1D). Statistical analyses using a general linear model with repeated measure two way ANOVA revealed an effect of time (F2, 30=8.728, p=0.001) and genotype (F1,15=4.651, p=.048) on rotarod performance, as well as a significant time/genotype interaction (Figure 1E) (F2,30=14.822, p<0.001). One way ANOVA analysis revealed that latency to fall significantly decreased in BAC-HDL2 mice over time (F2,37=19.047, p<0.001), while no such change was observed in wildtype littermates. These results reveal that BAC-HDL2 mice exhibit progressive motor deficits when compared to their wildtype littermate controls.
To assess whether BAC-HDL2 mice also exhibited evidence of neurodegenerative pathology, we weighed the forebrain and cerebellum at 12 and 22 months of age, an assay that is sensitive to detect selective forebrain atrophy in BACHD mice (Gray et al., 2008). As shown in Figure 1E and 1F, we observed a significant decrease in forebrain weight but not cerebellar weight in BAC-HDL2 mice as compared to wildtype controls. No difference in forebrain weight was detected at 3 months of age (data not shown). To more precisely quantify forebrain atrophy, we performed unbiased stereology using 18–22 month old BAC-HDL2 and control brains, and found a significant reduction of cortical but not striatal volumes in BAC-HDL2 mice compared to the controls (Figure 1G). The latter finding may reflect a more slowly progressive neurodegenerative process in BAC-HDL2 striata. In summary, our behavioral and neuropathological studies reveal that BAC-HDL2 mice exhibit age-dependent motor deficits and neurodegenerative pathology consistent with those in HDL2.
BAC-HDL2 Mice Recapitulate Key Molecular Pathological Features of HDL2
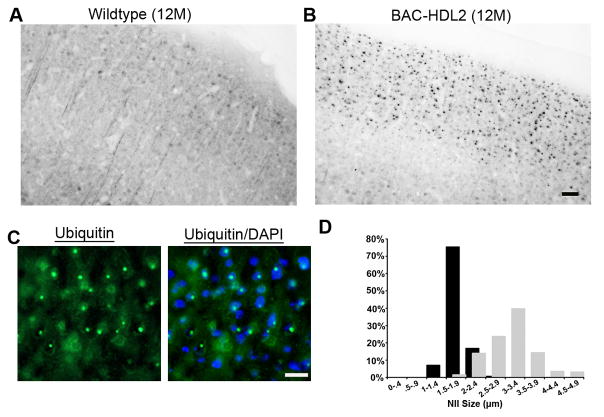
We next addressed whether BAC-HDL2 mice also recapitulate the two molecular pathological hallmarks of HDL2, ubiquitin-positive NIs and CUG RNA foci that co-localize with MBNL1 (Greenstein et al., 2007; Rudnicki et al., 2007; Rudnicki et al., 2008). As shown in Figure 2, we could readily detect prominent ubiquitin immunoreactive inclusion bodies in BAC-HDL2 mice but not wildtype controls at 12 month of age. Double fluorescent staining with an anti-ubiquitin antibody and DAPI demonstrated that the inclusion bodies were exclusively localized in the nucleus hence were NIs (Figure 2C). Moreover, double immunostaining for ubiquitin and NeuN revealed that NIs were exclusively localized within the neurons in BAC-HDL2 brains (data not shown). The brain regional distribution of the NIs in BAC-HDL2 is remarkably similar to that in the patients (Supplemental Fig. S2, Panel A; Greenstein et al., 2007; Rudnicki et al., 2008). Ubiquitin-positive NIs were most abundant in the upper cortical layers, hippocampus (data not shown) and amygdala, with relatively lower levels detected in the deep cortical layers and striatum. NIs were not detected in the cerebellum (Supplemental Figure S2, Panel A), substantia nigra, thalamus, or brain stem (data not shown).
Figure 2.
Ubiquitin immunoreactive nuclear inclusions in BAC-HDL2 brains. (A,B) Antigen retrieval followed by immunohistochemical staining with 1C2 antibody. 12 month old BAC-HDL2-C brain sections revealed numerous large inclusion bodies in the cortical layers II/III (B). No such staining was observed in the respective brain regions in the wildtype animals (A); (C) Ubiquitin immunoreactive inclusion bodies in BAC-HDL2-C brain are co-localized with the nuclear marker DAPI, hence they are nuclear inclusions (NIs). (D) Quantification of NI size at 3 and 12 months of age with summary histograms shown [mean=1.8 μm vs. 3.13 μm, for 3 and 12 months respectively; *p<.001, Student’s t test; BAC-HDL2-C: motor sensory (MS) cortex layers I/II/III, 3 month, (155 NIs measured in two mice), 12 month (194 NIs measured in two mice)]. Scale bar, 50 μm.
We next investigated whether the formation of NIs is progressive in BAC-HDL2 brains. NIs were absent in BAC-HDL2 brains at 1 month of age, but could be readily detected in the cortex and hippocampus starting at 3 months of age (data not shown). The size of NIs in BAC-HDL2 cortical neurons increases from an average diameter of 1.8 μm at 3 months of age to 3.13 μm by 12 months of age (Figure 2D). In summary, neuropathological analyses revealed that BAC-HDL2 mice recapitulate the progressive and brain-regional specific formation of ubiquitin positive NIs, a key pathological hallmark of HDL2.
The second pathological hallmark for HDL2 is the formation of CUG repeat containing RNA foci that are independent of the NIs (Rudnicki et al., 2007). To assess whether six month old BAC-HDL2 mice might also recapitulate such phenotype, we performed Fluorescent In Situ Hybridization (FISH) using an established protocol (Rudnicki et al., 2007). We could readily detect CUG RNA foci in BAC-HDL2 but not wildtype cortical sections (Supplemental Figure S2, Panel B). Similar to those in HDL2 patients, the RNA foci were rarely co-localized with NIs and were co-localized with Mbnl1 (Supplemental Figure S2, Panel B). Thus, we concluded that BAC-HDL2 mice also recapitulate the phenotype of CUG RNA foci, another molecular pathological marker for HDL2.
Evidence for Polyglutamine Protein Pathology in BAC-HDL2 Brains
One intriguing finding in HDL2 neuropathology is the immunoreactivity of NIs with 1C2, a monoclonal antibody that has relatively high specificity to all expanded neuropathogenic polyQ proteins (Trottier et al., 1995), but it can also recognize some normal long polyQ proteins such as TBP as well as some other amino acid stretches such as polyleucine (Dorsman et al., 2002). Because of this latter possibility, the precise molecular nature of the 1C2 immunoreactivity within NIs in HDL2 remains to be clarified.
We next asked whether the NIs in BAC-HDL2 mice, like those in HDL2 patients, could be immunostained with 1C2. Using a sensitive antigen-retrieval technique (Osmand et al., 2006), we were able to detect 1C2 immunoreactive NIs in 12 month old BAC-HLD2 brains that are unlike the faint diffuse nuclear staining found in the wildtype controls (Supplemental Figure S3, Panel A). Such 1C2 (+) NIs were not detected at 1 month of age, but could be detected at 3 months of age and became progressively enlarged at 6 and 12 months of age (Supplemental Figure S3, Panels A and C). Finally, double immunofluorescent staining revealed that 1C2 immunoreactive NIs co-localized with ubiquitin-positive NIs (Supplemental Figure S3, Panel B), suggesting the composition of NIs in BAC-HDL2 mice is quite similar to those described in HDL2 patients.
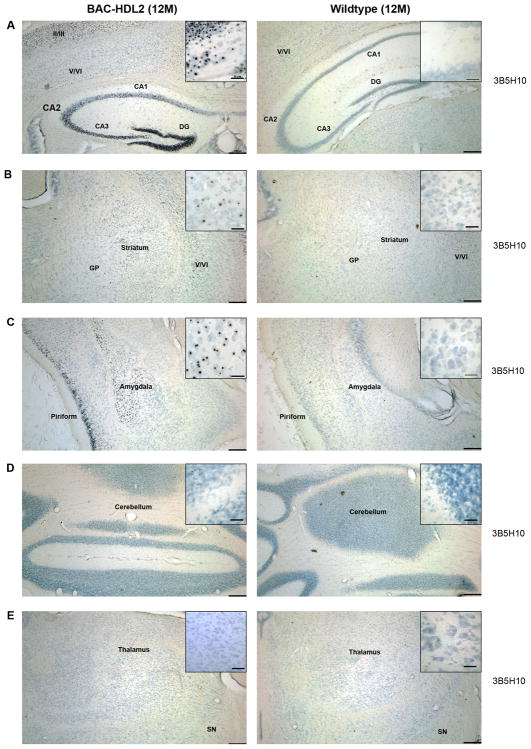
To provide further evidence that BAC-HDL2 NIs contain an expanded polyQ protein, we used another monoclonal antibody, 3B5H10, which has been shown to be specific to the expanded polyQ epitope in all known polyQ disorders (Brooks et al., 2004). Immunostaining with 3B5H10 after antigen retrieval revealed that NIs in 12 month old BAC-HDL2 cortices and striatum were prominently stained with this expanded polyQ specific antibody (Figure 3). No such 3B5H10 (+) NIs were detected in the brains of wildtype control littermates at 12 months of age (Figure 3). Importantly, the distribution of 3B5H10 immunoreactive NIs in BAC-HDL2 brains is strikingly similar to that of patients, with prominent levels of NIs in the cortex (the upper cortical layers more than the deep cortical layers), hippocampus and amygdala, decreased abundance in the striatum and very few if any NIs detected in the cerebellum, thalamus and brain stem (Figure 4 and data not shown).
Figure 3.
The distribution of 3B5H10 immunoreactive NIs in BAC-HDL2 brains recapitulates that seen in HDL2 patients. Immunostaining with a monoclonal antibody against expanded polyQ (3B5H10) in 12 month old BAC-HDL2 and wildtype brain sections. Prominent 3B5H10 stained aggregates are detected in the mutant but not wildtype mouse brains in the upper cortical layers and hippocampus (A), striatum (B), amygdala (C). Very few if any 3B5H10 immunoreactive aggregates were detected in the cerebellum (D) or thalamus (E). Scale bar, 200 μm; Inset scale bar, 25 μm.
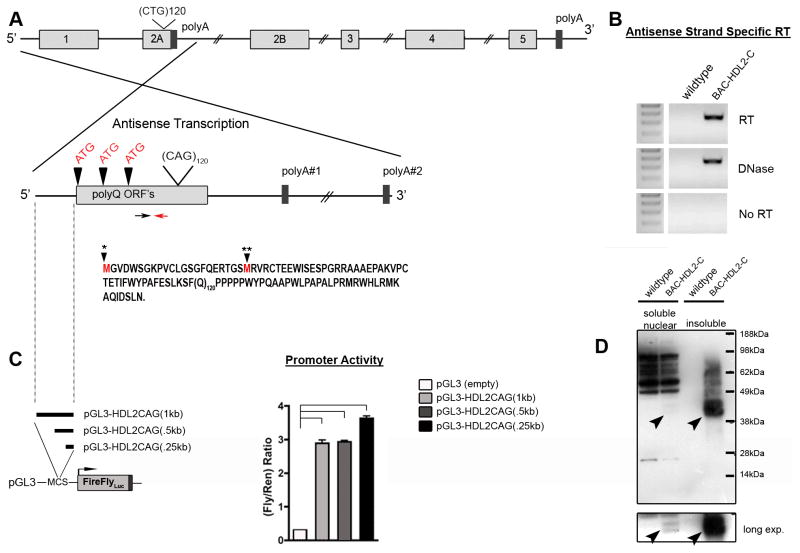
Figure 4.
Expression of mutant HDL2-CAG RNA and proteins in BAC-HDL2 brains. (A) Graphic representation of potential HDL2-CAG transcript(s) containing the expanded CAG repeat with polyQ ORF’s (vertical arrowheads). The two closest polyA signals are shown. A red arrow marks the location of the primer used to drive reverse-strand-specific cDNA synthesis. The black arrow marks the location of the forward primer used for PCR after cDNA synthesis. Defined HDL2-CAG mutant transcripts include two potential ORF’s through the CAG repeat (black arrowhead with star marks) based on the differential usage of the two ATG translational initiation codons that can be amplified (the first ATG codon is not in any 5′ RACE product); (B) RT-PCR analyses provide evidence for the expression of HDL2-CAG transcripts emanating from the JPH3 antisense strand in BAC-HDL2-C animals. (C) DNA upstream of HDL2-CAG (position defined by dotted line) was cloned in front of luciferase in order to detect promoter activity in primary cortical neurons (*p<0.01 for pGL3-HDL2CAG1kb, -HDL2CAG.5kb and -HDL2CAG.25kb compared to empty vector (pGL3-empty); values are mean ± SEM). (D) Western blot analysis using 3B5H10 antibody was performed on soluble cytoplasmic and nuclear soluble fractions (i.e. cyto sol or nuc sol) as well as insoluble cellular fractions (i.e. insol; crude inclusion fraction) extracted from the forebrain of BAC-HDL2-C and wildtype mice at 18 months of age. Black arrowheads point to novel disease associated polyQ protein bands found specifically in BAC-HDL2-C line brains but not in wildtype littermate controls. This novel polyQ protein (termed HDL2-CAG) markedly accumulates in the insoluble fraction in the aged mutant brains.
Taken together, our neuropathological studies with both 1C2 and 3B5H10 antibodies demonstrated that the NIs found in BAC-HDL2 brains recapitulate the patterns seen in HDL2 patients. Furthermore, an expanded polyQ protein is likely to be a component of such NIs.
JPH3 BAC Transgenic Mice With Non-expanded CTG/CAG Repeats Do Not Exhibit Evidence of Disease Pathogenesis
Since pathogenesis of HDL2 has been linked to the expansion of CTG/CAG repeats at the human JPH3 locus (Holmes et al., 2001), we next addressed whether disease pathogenesis in the BAC-HDL2 model, particularly the formation of NIs, could also be dependent on the repeat expansion in the BAC transgene. To test this hypothesis, we used two different wildtype JPH3 BAC transgenic control mice. The first control model was generated in the FvB/N background using the same wildtype BAC (RP11-33A21) as the one used to generate BAC-HDL2, except the CTG/CAG repeat length was 14. This control wildtype BAC construct was engineered to insert an enhanced green fluorescent protein (EGFP) within exon 1 of JPH3 (Supplemental Figure S4, Panel A). The transgenic mouse line (termed BAC-JPH3) expressed EGFP as well as the non-expanded CUG- and CAG transcripts but not JPH3 protein (Supplemental Figure S1). This mouse line is an ideal control to address whether overexpression of non-expanded CUG- or CAG-containing mRNA transcripts from JPH3 transgene locus could induce toxicity in vivo. To assess whether overexpression of any wildtype proteins encoded by the JPH3 sense strand transcripts could induce disease pathogenesis, we employed a second control mouse model. This control, utilized a different BAC (CTD-2195P9) encompassing the intact human JPH3 genomic locus with 14 CTG/CAG repeats, and was created and maintained in the C57/BL6 background (BAC-JPH3b6; Supplemental Figure S4, Panel B). Expression analyses revealed BAC-JPH3 and BAC-JPH3b6 mice express JPH3 mRNA at levels comparable to that found in BAC-HDL2-F line mice (Supplemental Figure S1; data not shown).
Phenotypic studies of both BAC-JPH3 and BAC-JPH3b6 control mice did not reveal any evidence of disease pathogenesis. First, BAC-JPH3 mice did not exhibit any rotarod deficits at 3 or 6 months of age, and their brains did not show 3B5H10 immunoreactive polyQ NIs at 14–18 months of age (Supplemental Figure S4, Panel B). Second, brain sections from 18–22 month old BAC-JPH3b6 mice were not immunostained for ubiquitin or polyQ NIs (Supplemental Figure S4, Panel B). To ascertain that the lack of NI phenotype in BAC-JPH3 control mice was not due to the relatively low level of transgene expression compared to the BAC-HDL2 line (~20%), we also assessed NI formation in BAC-HDL2-F line, which has comparable levels of transgene expression as the BAC-JPH3 control mouse lines. As shown in Supplemental Figure S4 (Panel C), 3B5H10 immunoreactive NIs were readily detected in the cortex and hippocampus of 22 month old BAC-HDL2-F mice. Together, our analyses demonstrate that disease pathogenesis in HDL2 mice dependent on CTG/CAG repeat expansion in the BAC transgene.
A Novel Promoter Drives the Expression of an Antisense CAG Transcript Encoding an Expanded PolyQ Protein in BAC-HDL2 Mice
We next sought to address the molecular origin of the polyQ immunoreactivity within the NIs in BAC-HDL2 brains. Previously postulated sources of 1C2 immunoreactive NIs in HDL2 patients includes: expression of a novel polyQ protein emanating from the strand antisense to the JPH3 locus, expression of an expanded polyleucine protein encoded by a CUG transcript, or sequestration of proteins with a long but nonpathogenic polyQ stretches such as TBP (Holmes et al., 2001; Margolis et al., 2005; Rudnicki et al., 2007). Studies conducted with limited patient brain samples could not definitively identify the source for the 1C2 immunoreactive NIs in HDL2 brains. In this study, we tested the hypothesis that NIs in HDL2 are due to the expression of a novel polyQ protein encoded by a previously unrecognized JPH3-antisense transcript containing an expanded CAG repeat.
Bioinformatic analyses performed on the antisense strand of the human JPH3 locus revealed three ORFs that included the CAG-encoded polyQ stretch as well as several predicted downstream polyA signals (Figure 4A). To find evidence for the expression of such CAG transcripts, we used antisense-strand-specific and human-transcript-specific RT-PCR analyses (see Supplemental Experimental Procedures). These analyses readily detected the expression of antisense transcripts in BAC-HDL2 mouse brains but not in wildtype controls (Figure 4B). In order to define the 5′ and 3′ regions of the transcript, we performed Rapid Amplification of cDNA Ends (RACE). We were able to identify 5′ RACE products encompassing the proximal two ATG codons in the polyQ ORF and 3′ RACE revealed a polyA signal ~4kb from the repeat (data not shown). Similar antisense CAG transcripts were also detected in BAC-JPH3 control mice (see Figure 5D and 5E). This novel transcript, which we named HDL2-CAG, contains two translation-initiation codons (ATG) in-frame with the polyQ encoding CAG repeat. This protein contains a predicted ORF with 54 amino acids prior to the polyQ repeat and 27 amino acids following the repeat (Figure 4A).
Figure 5.
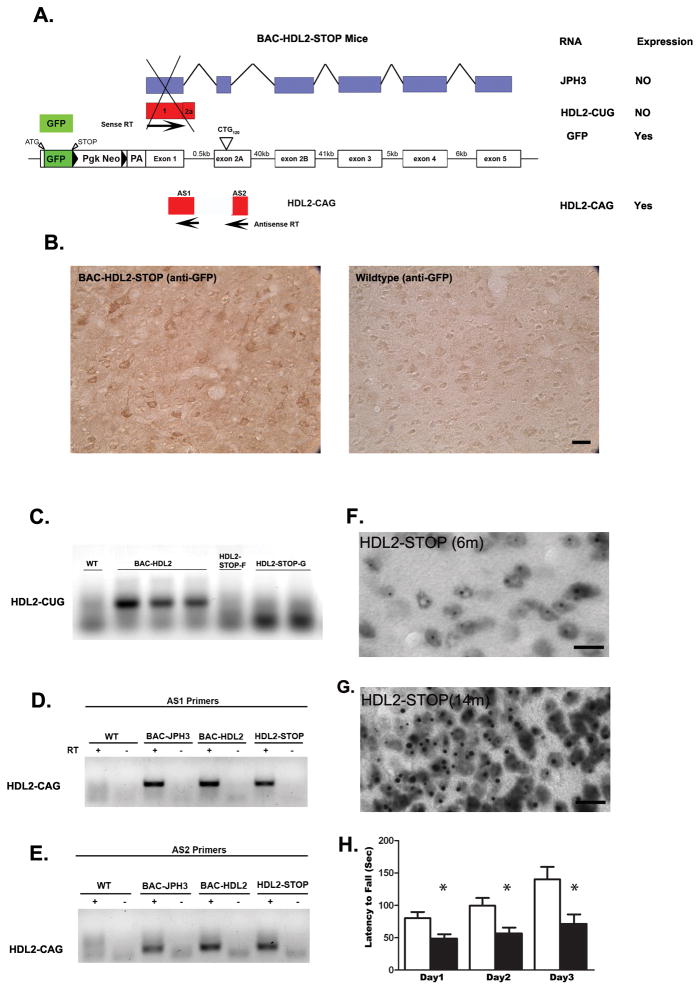
Antisense HDL2-CAG transcript and accumulation of polyQ are independent of the expression of sense strand of JPH3 gene. (A) A graphic representation of the construct of BAC-HDL2-STOP mice. A transcriptional STOP sequence consisted of GFP followed by triple polyadenylation signal was placed in front of the translation initiation codon of JPH3 in the exon1 of the JPH3 on the BAC transgene. This mouse line was designed to express GFP from JPH3 promoter, but there is no transcription and translation of any other mRNA in the JPH3 sense strand, including any of the transcripts that include the expanded CUG repeat. (B) Anti-GFP antibody readily detected the expression of GFP protein in the cortical neurons in BAC-HDL2-STOP mice but not WT controls. (C). JPH3-sense-strand specific RT-PCR to amplify HDL2-CUG transcript revealed the expression of such transcript in BAC-HDL2-C mice, but not in two lines of BAC-HDL2-STOP mice (lines F and G). (D) & (E). Two independent strand-specific RT-PCR primer sets (AS1 and AS2 primers) specific to the HDL2-CAG transcripts readily detected the antisense CAG transcript in JPH3-FvB control mice, BAC-HDL2 mice, and BAC-HDL2-STOP mice. (F and G). 3B5H10 immunostaining reveals progressive accumulation of polyQ NIs in cortical neurons of BAC-HDL2-STROP mice at 6 and 12 months of age, but such NIs were not detected in the same brain region in the wildtype littermates (data not shown). (H). Rotarod deficits in 12 month old BAC-HDL2-STOP mice compared to wildtype controls. Significant deficits were detected at all three testing days using a Students test (with *, P< 0.05). Repeat-measure ANOVA analysis revealed a significant effect of time (F(2,8) = 9.250; p < 0.0001), genotype (F(2,8) = 9.331; p = 0.009), and interaction of time and genotype (F(2,8) = 3.026; p < 0.0001). Scale bar, 50 μm.
Since BAC-HDL2 mice express the novel HDL2-CAG transcript, we asked whether the genomic sequence preceding the polyQ ORF could possess promoter activity in primary neurons. To test this possibility, we subcloned three genomic DNA fragments consisting of 0.25 kb, 0.5 kb, and 1 kb of genomic DNA sequence preceding the HDL2-CAG ORF into a luciferase reporter construct (Figure 4C). The resulting constructs were transfected into primary cortical neurons to test their ability to drive luciferase transcription. Surprisingly, all three genomic fragments exhibited robust promoter activity in this assay (Figure 4C), suggesting that the promoter driving HDL2-CAG expression is located immediately preceding the polyQ ORF.
We next sought to provide direct evidence for the expression of a novel expanded polyQ protein consistent with the size of HDL2-CAG protein in BAC-HDL2 brains. We first experimentally determined the size of both mutant and wildtype HDL2-CAG protein by performing in vitro experiments where we expressed Flag-tagged HDL2-CAG protein with 120-(HDL2-CAG120) or 14-CAG (HDL2-CAG14) repeats in HEK293 cells (Supplemental Figure S5). Western blot analyses using 1C2 and 3B5H10 antibodies revealed that HLD2-CAG120 protein in such transfected cells migrates as a doublet between 40–45kDa (Supplemental Figure S5). HDL2-CAG14, which migrates at ~16kDa, is detected with the anti-Flag antibody and only marginally by the 1C2 antibody. Since 3B5H10 can selectively detect HDL2-CAG120 protein in vitro, we performed Western blot analyses using fractionated brain extracts from 22 month old BAC-HDL2 mice and wildtype controls. Immunoblotting using 3B5H10 antibody detected a doublet that migrated between 40–45kDa in BAC-HDL2 but not wildtype control brains, consistent with the size of HDL2-CAG120 protein in in vitro experiments (Figure 4D). Interestingly, mutant HDL2-CAG protein can be robustly detected in insoluble nuclear fractions once the preparation has been solubilized by boiling in 2% SDS, while only a small but still detectable amount of mutant HDL2-CAG can be found in the soluble nuclear fraction in the mutant but not wildtype mouse brains (Figure 4D, long exposure).
In summary, we have demonstrated evidence for the expression of a novel CAG repeat containing transcript in BAC-HDL2 mice, emanating from the strand antisense to the JPH3 genomic locus. This expanded CAG transcript is driven by a promoter located immediately upstream of the polyQ ORF and is translated into an expanded polyQ protein in vivo.
Genetic Silencing of HDL2-CUG Transcripts Does Not Prevent the Expression of HDL2-CAG Transcripts and Polyglutamine Pathogenesis
Since genomic DNA immediately 5′ to the HDL2-CAG ORF exhibits robust promoter activity, it raises the possibility that expression of the HDL2-CAG transcript and the resulting polyQ pathogenesis may be independent of the expression of JPH3 sense strand transcripts and their protein products. To test this idea, we created a new transgenic mouse model with a BAC construct replacing the JPH3 exon1 with GFP sequence followed by a transcriptional STOP sequence (Soriano, 1999), but still containing the expanded CTG/CAG repeats (~120 repeats) on the BAC (Figure 5A). The STOP sequence, consisting of a floxed neo cassette followed by triple polyA signals, is a classic DNA sequence used to terminate transcription (Soriano, 1999; Srinivas et al., 2001). The resulting two mouse lines (F and G of BAC-HDL2-STOP mice) should express only GFP driven by the JPH3 promoter but no other sense strand CUG repeat or JPH3 transcripts should be expressed (Figure 5A). On the other hand, the STOP sequence should not interfere with the transcription of the antisense HDL2-CAG transcripts; hence the model is still predicted to manifest polyQ pathogenesis.
To confirm the silencing of the sense strand transcripts, we first showed the expression of GFP protein in the BAC-HDL2-STOP, but not wildtype brains, by immunohistochemistry (Figure 5B). Using sense strand-specific RT-PCR, we were able to confirm that HDL2-CUG transcripts are indeed silenced in the BAC-HDL2-STOP mice (both F and G lines) as compared to the BAC-HDL2 mice (Figure 5C). Conversely, RT-PCR performed using two separate antisense strand-specific primers readily detected HDL2-CAG transcripts in the brains of BAC-HDL2-STOP mice as well as BAC-HDL2 and BAC-JPH3 mice (Figure 5D and 5E). These analyses confirmed that the STOP sequence successfully silenced the expression of JPH3 and HDL2-CUG transcripts while leaving HDL2-CAG expression unperturbed in BAC-HDL2-STOP mice.
We next asked whether BAC-HDL2-STOP mice would develop NIs in vivo. Immunohistochemistry using 3B5H10 readily detected the progressive formation of polyQ NIs between 6 and 14 months of age in BAC-HDL2-STOP mice (G line) but not wildtype controls (Figure 5F–G; data not shown). Similar to those in BAC-HDL2, the NIs in BAC-HDL2-STOP mice were particularly abundant in the cortex and hippocampus, and diffuse nuclear accumulation could also be detected in the striatum (Supplemental Figure S6, Panel A).
We next addressed whether the selective expression of the mutant HDL2-CAG transcripts, but not HDL2-CUG or JPH3 transcripts, is sufficient to elicit motor deficits and/or neurodegenerative pathology. As shown in Figure 5H, BAC-HDL2-STOP mice exhibit a significant accelerating rotarod deficit at 12 months of age (N=8 per genotype; p < 0.05 for each of the three testing days using Student’s t test). Repeated measures ANOVA analysis reveals a significant effect of time (F(2,8) = 9.250; p < 0.0001), genotype (F(2,8) = 9.331; p = 0.009), and interaction of time and genotype (F(2,8) = 3.026; p < 0.0001), suggesting that mutant mice exhibit both motor performance and motor learning deficits in the rotarod test. To assess whether BAC-HDL2-STOP transgenic mice also show evidence of neurodegenerative pathology similar to that in BAC-HDL2 mice, we weighed the forebrains and cerebella of the mutant and WT mice at 12 months of age (N=8 per genotype). We did not detect any significant reduction of forebrain or cerebellar weights in the mutant mice at this age (Supplemental Figure S6, Panel B). These results show that the selective expression of mutant HDL2-CAG transcripts but not HDL2-CUG transcripts is sufficient to elicit neuronal dysfunction (e.g. rotarod deficits) but not yet sufficient to induce neurodegeneration at 12 months of age.
In conclusion, the BAC-HDL2-STOP model provides definitive mouse genetic evidence that selective expression of HDL2-CAG transcript without co-expression of JPH3 or HDL2-CUG transcript is sufficient to elicit polyQ pathogenesis and neuronal dysfunction in vivo.
Sequestration of CREB Binding Protein (CBP) and Functional Interference of CBP-mediated transcription in HDL2 Mice and Patients
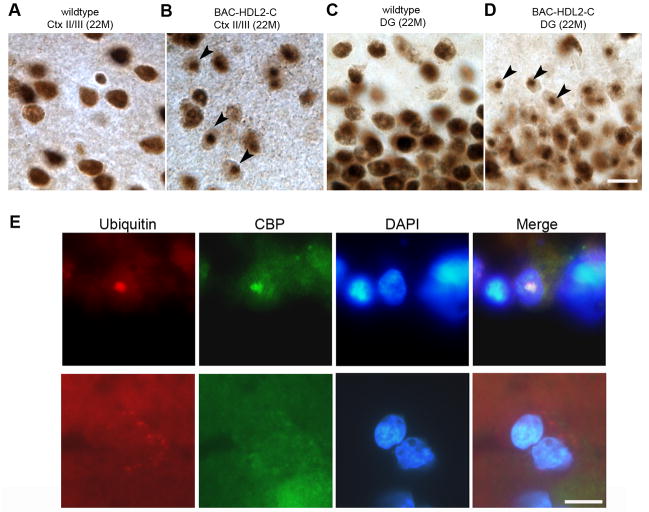
We next explored whether NIs in BAC-HDL2 could exhibit other molecular features similar to polyQ disorders including HD (Orr and Zoghbi, 2007). One such molecular marker that has been observed in several polyQ disorders (e.g. HD, SBMA and SCA3) is the sequestration of polyQ-domain containing nuclear transcription factors in NIs, such as the potent transcription co-activator CBP (Kazantsev et al., 1999; Nucifora et al., 2001; McCampbell et al., 2000). We tested this possibility by immunohistochemical staining for CBP using 22 month old BAC-HDL2 and control brain sections. In wildtype brains, we detected the characteristic diffuse CBP staining in nuclei throughout the brain (Figure 6A and 6C). However, in BAC-HDL2 brains, we detected the presence of CBP-immunoreactive NIs and a corresponding reduction of diffuse nuclear CBP staining in the cortical and hippocampal neurons (Figure 6B and 6D). Occasionally, CBP-immunoreactive NIs could also be detected in the striatum (data not shown). The formation of CBP NIs is time-dependent, as they were initially detected at 12 months of age but not at 3 months of age (data not shown). Thus, the sequestration of CBP in NIs occurred after the formation of polyQ NIs (first detected at 3 months). Consistent with the hypothesis that CBP pathology is mediated by mutant HDL2-CAG protein, we did not detect any CBP inclusions in 18–22 month old BAC-JPH3 (Supplemental Figure S4, Panel A) or BAC-JPH3b6 controls (Supplemental Figure S4, Panel B). As expected, silencing sense strand transcript in BAC-HDL2-STOP mice did not prevent CBP inclusion formation (Supplemental Figure S6, Panel A).
Figure 6.
CBP sequestration in NIs in BAC-HDL2 Mice and HDL2 Patient Brains. (A–D) Two anti-CBP antibodies (A-22 and sc-583) were used to confirm the presence of CBP immunoreactive NIs in BAC-HDL2 mice (sc-583 is shown). Representative images of CBP staining of the superficial cortical layers (Cortical layers II/III; 6A and 6B) and hippocampal dentate gyrus (DG; 6C and 6D) for wildtype (6A and 6C) and BAC-HDL2 (6B and 6D) sections were shown. Selected NIs are highlighted with black arrowheads. (E) Double immunofluorescence staining using anti-CBP and anti-ubiquitin antibodies revealed the presence of CBP immunoreactive NIs that co-localize with ubiquitin staining in the nucleus (DAPI staining) in the cortical cells in HDL2 postmortem brain. Cortical neurons in control brains do not have CBP and ubiquitin-immunoreactive NIs, and CBP is diffusely distributed. Scale bar, 50 μm
To provide further evidence that the CBP pathology identified in BAC-HDL2 mice is relevant to HDL2 patients, we performed double immunofluorescent staining of HDL2 patient cortical tissue using anti-CBP and anti-ubiquitin antibodies. As shown in Figure 6E, we were able to detect CBP immunoreactive NIs that co-localized with ubiquitin immunoreactive NIs in the superior frontal gyri of two postmortem HDL2 brains but not in the brains of unaffected controls.
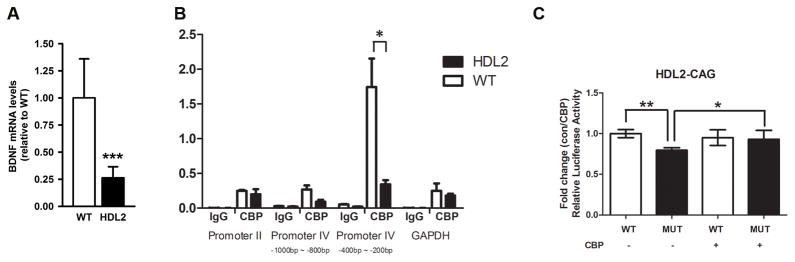
CBP is a histone acetyltransferase and a critical transcriptional co-activator (Chan and La Thangue, 2001), and CBP sequestration and transcriptional interference has been implicated in HD pathogenesis (Nucifora et al., 2001; Steffan et al., 2001). To assess for evidence of CBP functional impairment in HDL2, we decided to use CBP-mediated BDNF gene transcription as a model. BDNF is a critical trophic factor for striatal neurons, and its transcriptional down-regulation has been implicated in HD pathogenesis (reviewed by Zuccato et al., 2010). Moreover, our analyses of BDNF transcription using quantitative RT-PCR analyses confirmed a significant reduction of transcripts containing the entire BNDF coding region in 15-month-old BAC-HDL2 cortices compared to those in the wildtype controls (Figure 7A).
Figure 7.
CBP mediated transcriptional dysregulation in mouse and primary neuronal models of HDL2. (A) Quantitative RT-PCR analyses of the intact BDNF coding region reveal a significant reduction of BDNF transcripts in 15-month-old BAC-HDL2 mice relative to wildtype control mice (N=3 per genotype; ***p < 0.01, Student’s t-test). Values are mean ± SEM. (B). CBP occupancy of mouse BDNF promoter II, promoter IV (proximal region: 200–400bp from the transcription start site; distal region: 800–1000bp from the start site), BDNF coding region and GAPHD promoter were determined by ChIP-PCR in cortical samples derived from wildtype and BAC-HDL2 mice at 15 months of age (N=3 per genotype). ChIP-qPCR (IP/WCE,%) signals were normalized to whole cell extract (WCE). Immunoprecipitation with IgG was used for controls. Error bar represent S.E.M. determined from three independent experiments. The results suggest a significant and selective reduction of CBP binding to the proximal BDNF promoter IV (200–400bp) in cortical samples from mutant BAC-HDL2 mice compared to WT controls. Values are mean ± SEM (*p<0.01, Student’s t test). (C). Luciferase reporter assays were performed in rat cortical primary neurons with co-transfection of BDNF promoter IV-driven firefly luciferase reporter construct and vectors expressing either HDL2-CAG14 (wildtype HDL2-CAG protein with 14 glutamine repeat; labeled WT) or HDL2-CAG120 (mutant protein with 120 glutamine repeats; labeled HDL2). A separate set of transfection assays was performed with the same plasmids as above but the addition of plasmids overexpressing CBP (labeled CBP +). A renilla luciferase expression plasmid was used to normalize transfection efficiency in all the assays. Values are mean ± SEM (**p<0.01, *p<0.05, Student’s t test).
We next addressed whether such BNDF transcriptional deficit in BAC-HDL2 could in part be due to functional interference of CBP. Transcription of BDNF is initiated at multiple promoters (Hong et al., 2008). In HD, there is evidence that transcription is reduced at both BDNF promoter II (Zuccato et al., 2001) and promoter IV (Gambazzi et al., 2010; Gray et al., 2008). Relevant to the current study, the BDNF promoter IV is regulated by neuronal activity and targeted by CREB and CBP (Hong et al., 2008). We hypothesized that mutant HDL2-CAG may interfere with CBP function therefore could alter the transcription from BDNF promoter IV. To test this hypothesis, we used 15 month old BAC-HDL2 and wildtype cortical extracts to perform chromatin immunoprecipitation (ChIP) experiments to quantify the amount of CBP bound to proximal vs. distal regions of BDNF promoter IV, using BNDF promoter II as well as GAPDH promoter regions as controls (Martinowich et al., 2003). As shown in Figure 7B, we found a significant and selective reduction in the amount of CBP bound to the proximal region of BDNF promoter IV in BAC-HDL2 cortical samples compared to those from wildtype controls. Such difference was not observed in the distal region of BNDF promoter IV nor in the control BNDF promoter II or GAPDH promoter. Consistent with this finding, we also detected a decrease in histone H4 acetylation (H4Ac), an epigenetic marker of transcriptionally-active chromatin, in the BDNF promoter IV in the BAC-HDL2 cortical samples compared to the wildtype controls (Supplemental Figure S7). However, we did not observe any significant changes of other histone markers such as H3K4me3 or H3K27me3 (ibid).
Finally, to further explore in a primary neuron model whether CBP could functionally modify HDL2-CAG mediated transcriptional dysregulation of BDNF promoter IV, we co-transfected primary cortical neurons with a reporter plasmid containing BDNF promoter IV-driven firefly luciferase and plasmids expressing either mutant (120 CAG repeats) or wildtype (14 CAG repeats) FLAG-tagged HDL2-CAG proteins (Figure 7C; Supplemental Figure S5). At 24 hours post transfection, when we did not detect any significant cell death (data not shown), we found that mutant but not wildtype HDL2-CAG can induce significant reduction of firefly luciferase activity relative to the control renilla luciferase activity, suggesting expression of mutant but not wildtype HDL2-CAG can interfere with BDNF promoter IV transcriptional activities (Figure 7C). Importantly, co-transfection of CBP can rescue such polyQ-length dependent interference of transcription in this primary neuron model (Figure 7C).
In summary, our analyses of HDL2 models in primary neurons, mice and patients provide converging evidence to support polyQ-length dependent CBP sequestration and functional interference of CBP-mediated transcription in HDL2.
DISCUSSION
We have generated and characterized the first BAC transgenic mouse models of an HD-like disorder, HDL2. BAC-HDL2 mice recapitulate several key phenotypes found in HDL2 patients, including age-dependent motor deficits and selective forebrain atrophy. They also capture two molecular pathogenic hallmarks of HDL2: the progressive accumulation of ubiquitin-positive NIs, and the presence of CUG containing RNA foci that are not co-localized with NIs. Importantly, this model reproduces the brain region specific distribution of NIs seen in the patients, suggesting that the mechanism underlying the pathogenesis of NIs is likely reproduced in this mouse model. Furthermore, the disease phenotypes are not present in control BAC mice without the CTG/CAG repeat expansion.
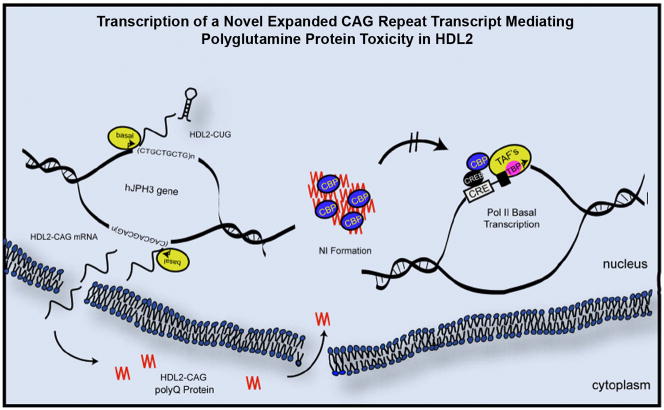
A significant finding of our study is the discovery of a novel molecular mechanism leading to polyQ pathogenesis in BAC-HDL2 mice (see schematics in Figure 8). Using our series of novel mouse models, we first demonstrated the expression of an expanded CAG-containing transcript from the strand antisense to JPH3, with its expression driven by a novel promoter. Second, immunohistochemistry and Western blot analyses demonstrated that this transcript is expressed as a protein containing an expanded polyQ tract. Third, we used two antibodies relatively specific for polyQ to demonstrate the accumulation of NIs containing polyQ in two independent lines of BAC-HDL2 mice and found the spatial distribution of NI accumulation to be strikingly similar to that found in HDL2 patients. Fourth, we used two control BAC mouse lines with normal CTG/CAG repeats to demonstrate that disease pathogenesis in BAC-HDL2 mice, including NI formation, is dependent on the CTG/CAG repeat expansion. Finally, by genetic silencing of the JPH3 sense strand in BAC-HDL2-STOP mice, preventing expression of CUG-containing transcripts, we provided definitive genetic evidence that the expression of the HDL2-CAG transcript alone can lead to the formation of polyQ-containing NIs and manifestation of motor deficits. Taken together, our analysis of the series of HDL2 mouse models provide an important mechanistic insight that the expression of a novel expanded polyQ protein could play a critical role in HDL2 pathogenesis in vivo.
Figure 8.
A schematic model for novel pathogenic mechanisms revealed by HDL2 mouse model in which a novel antisense expanded CAG transcript is mediating polyQ pathogenesis in vivo. A novel expanded CAG repeat (HDL2-CAG) antisense to the JPH3 sense strand is transcribed from its own promoter immediately 5′ to the CAG repeat, and the transcript is then translated into a novel expanded polyQ protein termed HDL2-CAG. Mutant HDL2-CAG is translocated and accumulated in the nucleus to form prominent NIs consisting of HDL2-CAG protein, ubiquitin and at a later time point, CBP. Transcriptional dysregulation, in part due to interference of CBP-mediated activation of gene expression (e.g. BDNF), may be a shared molecular pathogenic mechanism between HD and HDL2.
Prior to this study, the expression of an expanded antisense CAG-containing transcript or a novel polyQ protein had not been demonstrated using postmortem patient brain tissues (Holmes et al., 2001; Margolis et al., 2005). There are several possible explanations to account for the more sensitive detection of the CAG transcript and polyQ expanded protein in HDL2 mice than in patients. First, unlike the postmortem brain tissues, the HDL2 mouse brains used for the studies do not exhibit robust neurodegeneration that may lead to the loss of neurons expressing mutant CAG transcripts at high levels. Second, the sequence difference between the human mutant JPH3 transgene and murine wildtype locus in the same HDL2 mouse permits the easier detection of the mutant antisense transcripts. Third, the much longer polyQ repeat in the BAC-HDL2 mouse model (120Q, as compared to 40–50Q in patients) also permits more sensitive detection of the small amount of soluble mutant polyQ protein using antibodies that provide signal strength in Western blots depending on the polyQ length (e.g. 3B5H10). Since the brain regional and subcellular distribution of the NIs are remarkably similar between HDL2 mice and patients, it would strongly argue that the molecular pathogenesis for such NIs in HDL2 mice and patients are also likely to be similar. Future studies using additional patient samples and more sensitive detection methods may be needed to demonstrate HDL2-CAG transcript and protein in the patient brains.
Although our study is focused on the mechanistic investigation of the novel antisense CAG repeat transcript and its polyQ containing protein product, our study does not exclude a contribution of other potential mechanisms to aspects of HDL2 pathogenesis, such as partial loss of JPH3 function or CUG repeat RNA gain-of-function toxicity (Rudnicki et al., 2007). Despite the fact that our study was not designed to address these alternative mechanisms, our models do support a potential role of CUG RNA in disease pathogenesis, since BAC-HDL2 mice accumulate CUG RNA foci that can sequester Mbnl1 hence mimicking those in the patients. Importantly, the CUG RNA foci and NIs in both HDL2 mice and patients appear to be distinct entities, consistent with the interpretation that independent pathogenic mechanisms lead to their formation (ibid; Supplemental Figure S2, Panel B). Since expanded CUG repeat RNA is clearly pathogenic in DM1 via an RNA gain-of-function mechanism (Mankodi et al., 2001), in part via sequestration and depletion of MBNL1 (Kanadia et al., 2003), future mouse genetic studies are needed to address whether the expression of expanded CUG repeat transcripts also could mediate CUG repeat RNA toxicities in vivo.
An overarching goal of this study was to shed light on potential common pathogenic mechanisms shared between HD and HD-like disorders. By the development and analysis of the first BAC mouse genetic model for an HD-like disorder, we have already gained some initial insights toward this important objective. First, our cumulative analysis of these models suggests that expanded polyQ-mediated pathogenesis may be a shared pathogenic mechanism between HD and HDL2. The finding that a mutant polyQ protein may contribute to HDL2 pathogenesis in BAC-HDL2 mice is consistent with the presence of NIs that stain with both 1C2 and 3B5H10 antibodies in HDL2 brain, and the comparable pathogenic CAG repeat threshold for HDL2 and HD (about 40 triplets). It is striking that this threshold is similar to other polyglutamine diseases, but is much shorter than the threshold for most of the non-polyglutamine repeat expansion disorders, most prominently DM1. Thus, our study provides experimental evidence to suggest that therapeutics that could ameliorate expanded polyQ toxicity could benefit both HD and HDL2.
Another potentially shared pathogenic mechanism between HD and HDL2 is transcriptional dysregulation mediated by sequestration and/or functional interference of CBP. Prior studies have demonstrated that mutant huntingtin N-terminal fragments can bind to and/or sequester CBP into aggregates leading to changes in CBP mediated transcription (Kazantsev et al., 1999; Nucifora et al., 2001; Steffan et al., 2000). Although physical depletion of CBP is not consistently observed in all HD mouse models (Yu et al., 2002), functional interference of CBP has been observed in HD as well as other polyQ disorders. Indirect restoration of CBP function via histone deacetylase (HDAC) inhibition has been shown to be beneficial in several animal models of HD (Steffan et al., 2001) and in other polyQ disorders such as SBMA (McCampbell et al., 2000; Taylor et al., 2003). Our analysis of BAC-HDL2 mice provides evidence that expression of BNDF is significantly reduced in the mutant cortex at 15 months of age, and our in vivo ChIP analysis and primary neuron study reveal that mutant HDL2-CAG can selective interfere with CBP co-activator function at BDNF promoter IV (Hong et al., 2008). Together, our study supports the notion that mutant HDL2-CAG protein can perturb CBP-mediated transcription, in part but not necessarily exclusively, through the sequestration of CBP into NIs. Our current study does not rule out the possibility that mutant HDL2-CAG protein may also disrupt the function of other critical nuclear transcription factors such as TBP (Rudnicki et al., 2008). Additional gene expression and epigenetic profiling, along with functional manipulation of these molecular pathways in HDL2 mice, will be necessary to critically evaluate their contribution in disease pathogenesis.
Finally, our study reveals the complexity of disease pathogenesis mediated by trinucleotide repeat expansion. Together with SCA8 (Moseley et al., 2006), our models provide another compelling example in which bi-directional transcription across an expanded CTG/CAG repeat leads to the expression of an antisense CAG transcript and previously unrecognized polyQ protein toxicity. Since the predicted HDL2-CAG protein has no known homology to any other protein in the human proteome beyond the polyQ stretch (data not shown), the function of this novel transcript and the small protein it encodes remains to be explored. Given the recent discovery that antisense transcription is nearly ubiquitous throughout the mammalian genome (Katayama et al., 2005), our study highlights the importance of examining antisense repeat-containing transcripts and their ORFs in the pathogenesis of other brain disorders.
EXPERIMENTAL PROCEDURES
Generation of BAC Transgenic Mouse Models of HDL2
Human BAC (RP11-33A21) containing the JPH3 genomic locus from BACPAC Resource Center (Oakland Children’s Hospital, Oakland, CA) was engineered using homologous recombination and microinjected into FvB/N embryos to generate the BAC transgenic mouse lines, BAC-HDL2, BAC-HDL2-STOP and BAC-JPH3 (Yang et al., 1997; Gong et al., 2002). These mouse lines were maintained in FvB/NJ inbred background. A second BAC control mouse was generated using the wildtype JPH3 BAC (CTD-2195P9), and was created and maintained in the C57/BL6 background (BAC-JPH3b6). More details about the transgene constructs and initial characterization of the mouse lines are in Supplemental Experimental Procedures.
Reverse Transcriptase PCR Analyses of Strand-specific RNA Expression
For RT-PCR analyses of JPH3 sense strand and antisense HDL2-CAG transcripts, total RNA was extracted using the RNeasy Lipid mini-kit (Qiagen, Valencia, CA). Synthesis of cDNA was primed using either oligo(dT)20 (Invitrogen, Carlsbad, CA) or strand-specific oligonucleotide primers (see Supplemental Table S1 for primers). Both 5′ and 3′ RACE analyses were performed using FirstChoice® RLM-RACE kit (ABI) using the manufacturer’s instructions. A random-primed reverse transcription reaction and nested PCR was used to amplify 5′ and 3′ ends of the transcript (see Supplemental Table S1). Quantitative RT-PCR analyses of BDNF transcripts in BAC-HDL2 and control cortices were preformed using published protocol (Gray et al., 2008).
Accelerating Rotarod, Brain Weights, and Stereological Measurement of Brain Volumes
See published protocols for the rotarod, forebrain and cerebellar weights, and stereological measurement of cortical and striatal volumes (Gray et al., 2008; Gu et al., 2009; Supplemental Experimental Procedures).
Immunohistochemistry
Mouse brains were perfused, sectioned and immunostained using established protocols (Gray et al., 2008; Gu et al., 2009; Supplemental Experimental Procedures). HDL2 brain samples used in the study were described in detail before (Rudnicki et al. 2008). The following antibodies were used to stain NIs in HDL2 models: 3B5H10 (1:1000) (Sigma, St. Louis, MO), 1C2 (1:3000) (Chemicon, Billerica, MA), CBP (1:3000) (A-22 & sc-583, Santa Cruz, Santa Cruz, CA), Ubiquitin (1:1000) (DakoCytomation, Carpinteria, CA), 3B5H10 (1:2000), MBNL1 antibody (A2764, 1:10000 dilution; Lin et al., 2006). Antigen retrieval for polyQ NI detection using 3B5H10 was performed according to published protocols (Osmand et al., 2006). More details on immunohistochemical methods and reagents, and quantitation of NI sizes can be found in Supplemental Experimental Procedures.
Western blotting
Brain extracts and nuclear/cytoplasmic fractionations were preformed using established methods (Gray et al., 2008; Gu et al., 2009; see Supplemental Experimental Procedures). Antibodies for western blot included: JPH3exon4 (1:1000) (Takeshima H., Tohoku University, Japan), M2-Flag (1:500), α-tubulin (1:2000), 3B5H10 (1:1000) (Sigma, St. Louis, MO), and 1C2 (1:2000) (Chemicon, Billerica, MA).
Chromatin Immunoprecipitation (ChIP)
ChIP analyses were performed using our established method (Martinowich et al., 2003; Tao et al., 1998; see Supplemental Experimental Procedures) using the following antibodies: anti-CBP (sc-583, Santa Cruz), anti-IgG (sc-66931, Santa Cruz). Real-time quantitative PCR was performed using iQ SYBR Green Supermix (Bio-Rad). For quantification of relative level of CBP occupancy, we calculated the percentage of immunoprecipitated DNA over whole cell extract. Primer sequences used in ChIP-qPCR are listed in Supplemental Experimental Procedures.
DNA constructs, Cortical Primary Neuron Culture, Transfection and Luciferase Reporter Assays
See details in Supplemental Experimental Procedures.
Statistical Analysis
All data is shown as the mean ± SEM. SPSS 14.0 statistics software (SSPS, Chicago, IL) was used to perform all statistical analyses. The significance level was set at 0.05. See more details in Supplemental Experimental Procedures and in our published methods (Gray et al., 2008; Gu et al., 2009).
Supplementary Material
Acknowledgments
This work was generously supported by independent research grants from the Hereditary Disease Foundation (HDF) to X.W. Yang, to R.L. Margolis, to A. Osmand, and to D.D. Rudnicki. X.W.Y. is also supported by NIH/NINDS R01NS049501, and by the David Weil Fund to Semel Institute at UCLA. R.L.M. and D.D.R. by NIH/NINDS R21NS057516 and R01NS064138. We would like to thank N. S. Wexler and C. Johnson for their tremendous support of this project. We would also like to thank members of Yang lab for discussions and reading of the manuscript. B. Wilburn and S.P. Chang were supported by predoctoral fellowships from the UCLA Center for Neurobehavioral Genetics Predoctoral Training Program funded by NIH/NIMH (5T32MH073526).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brooks E, Arrasate M, Cheung K, Finkbeiner SM. Using antibodies to analyze polyglutamine stretches. Methods Mol Biol. 2004;277:103–128. doi: 10.1385/1-59259-804-8:103. [DOI] [PubMed] [Google Scholar]
- Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- Dorsman JC, Pepers B, Langenberg D, Kerkdijk H, Ijszenga M, den Dunnen JT, Roos RA, van Ommen GJ. Strong aggregation and increased toxicity of polyleucine over polyglutamine stretches in mammalian cells. Hum Mol Genet. 2002;11:1487–1496. doi: 10.1093/hmg/11.13.1487. [DOI] [PubMed] [Google Scholar]
- Gambazzi L, Gokce O, Seredenina T, Katsyuba E, Runne H, Markram H, Giugliano M, Luthi-Carter R. Diminished activity-dependent brain-derived neurotrophic factor expression underlies cortical neuron microcircuit hypoconnectivity resulting from exposure to mutant huntingtin fragments. J Pharmacol Exp Ther. 2010;335:13–22. doi: 10.1124/jpet.110.167551. [DOI] [PubMed] [Google Scholar]
- Gong S, Yang XW, Li C, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res. 2002;12:1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, Andre VM, Wilburn B, Lu XH, Tao J, Yamazaki I, Li SH, Sun YE, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein PE, Vonsattel JP, Margolis RL, Joseph JT. Huntington’s disease like-2 neuropathology. Mov Disord. 2007;22:1416–1423. doi: 10.1002/mds.21417. [DOI] [PubMed] [Google Scholar]
- Gu X, Greiner ER, Mishra R, Kodali R, Osmand A, Finkbeiner S, Steffan JS, Thompson LM, Wetzel R, Yang XW. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes SE, O’Hearn E, Rosenblatt A, Callahan C, Hwang HS, Ingersoll-Ashworth RG, Fleisher A, Stevanin G, Brice A, Potter NT, et al. A repeat expansion in the gene encoding junctophilin-3 is associated with Huntington disease-like 2. Nat Genet. 2001;29:377–378. doi: 10.1038/ng760. [DOI] [PubMed] [Google Scholar]
- Hong EJ, McCord AE, Greenberg ME. A biological function for the neuronal activity-dependent component of Bdnf transcription in the development of cortical inhibition. Neuron. 2008;60:610–24. doi: 10.1016/j.neuron.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc Natl Acad Sci U S A. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- Margolis RL, Holmes SE, Rosenblatt A, Gourley L, O’Hearn E, Ross CA, Seltzer WK, Walker RH, Ashizawa T, Rasmussen A, et al. Huntington’s Disease-like 2 (HDL2) in North America and Japan. Ann Neurol. 2004;56:670–674. doi: 10.1002/ana.20248. [DOI] [PubMed] [Google Scholar]
- Margolis RL, O’Hearn E, Rosenblatt A, Willour V, Holmes SE, Franz ML, Callahan C, Hwang HS, Troncoso JC, Ross CA. A disorder similar to Huntington’s disease is associated with a novel CAG repeat expansion. Ann Neurol. 2001;50:373–380. doi: 10.1002/ana.1312. [DOI] [PubMed] [Google Scholar]
- Margolis RL, Rudnicki DD, Holmes SE. Huntington’s disease like-2: review and update. Acta Neurol Taiwan. 2005;14:1–8. [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- McCampbell A, Taylor JP, Taye AA, Robitschek J, Li M, Walcott J, Merry D, Chai Y, Paulson H, Sobue G, et al. CREB-binding protein sequestration by expanded polyglutamine. Hum Mol Genet. 2000;9:2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- Nishi M, Hashimoto K, Kuriyama K, Komazaki S, Kano M, Shibata S, Takeshima H. Motor discoordination in mutant mice lacking junctophilin type 3. Biochem Biophys Res Commun. 2002;292:318–324. doi: 10.1006/bbrc.2002.6649. [DOI] [PubMed] [Google Scholar]
- Nucifora FC, Jr, Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson VL, et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Osmand AP, Berthelier V, Wetzel R. Imaging polyglutamine deposits in brain tissue. Methods Enzymol. 2006;412:106–122. doi: 10.1016/S0076-6879(06)12008-X. [DOI] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- Rudnicki DD, Holmes SE, Lin MW, Thornton CA, Ross CA, Margolis RL. Huntington’s disease--like 2 is associated with CUG repeat-containing RNA foci. Ann Neurol. 2007;61:272–282. doi: 10.1002/ana.21081. [DOI] [PubMed] [Google Scholar]
- Rudnicki DD, Pletnikova O, Vonsattel JP, Ross CA, Margolis RL. A comparison of huntington disease and huntington disease-like 2 neuropathology. J Neuropathol Exp Neurol. 2008;67:366–374. doi: 10.1097/NEN.0b013e31816b4aee. [DOI] [PubMed] [Google Scholar]
- Schneider SA, Walker RH, Bhatia KP. The Huntington’s disease-like syndromes: what to consider in patients with a negative Huntington’s disease gene test. Nat Clin Pract Neurol. 2007;3:517–525. doi: 10.1038/ncpneuro0606. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, Wanker EE, Bates GP, Housman DE, Thompson LM. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci U S A. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H. Junctophilins: molecular components contributing junctional membrane complexes between the cell-surface membrane and endoplasmic/sarcoplasmic reticulum] Clin Calcium. 2001;11:758–762. [PubMed] [Google Scholar]
- Taylor JP, Taye AA, Campbell C, Kazemi-Esfarjani P, Fischbeck KH, Min KT. Aberrant histone acetylation, altered transcription, and retinal degeneration in a Drosophila model of polyglutamine disease are rescued by CREB-binding protein. Genes Dev. 2003;17:1463–1468. doi: 10.1101/gad.1087503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier Y, Lutz Y, Stevanin G, Imbert G, Devys D, Cancel G, Saudou F, Weber C, David G, Tora L, et al. Polyglutamine expansion as a pathological epitope in Huntington’s disease and four dominant cerebellar ataxias. Nature. 1995;378:403–406. doi: 10.1038/378403a0. [DOI] [PubMed] [Google Scholar]
- Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- Yu ZX, Li SH, Nguyen HP, Li XJ. Huntingtin inclusions do not deplete polyglutamine-containing transcription factors in HD mice. Hum Mol Genet. 2002;11:905–914. doi: 10.1093/hmg/11.8.905. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Botas J. Mouse and fly models of neurodegeneration. Trends Genet. 2002;18:463–471. doi: 10.1016/s0168-9525(02)02729-4. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, Timmusk T, Sipione S, Cattaneo E. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.