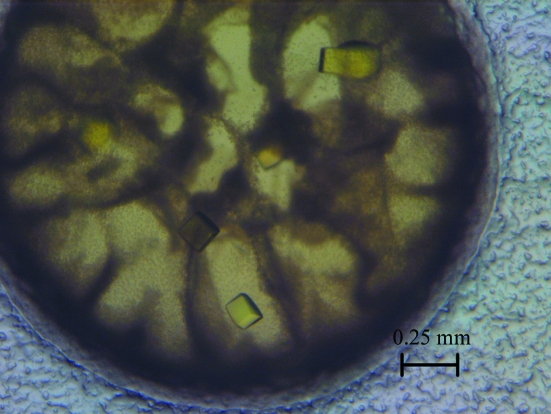
In this study, the catalytic subunit of E. coli AHAS II was cocrystallized with its cofactors Mg2+, FAD and ThDP using the sitting-drop vapour-diffusion method and the crystals diffracted to 2.80 Å resolution.
Keywords: acetohydroxyacid synthase, Escherichia coli, AHAS II, branched-chain amino-acid synthesis
Abstract
Acetohydroxyacid synthase (AHAS) is the first common enzyme in the branched-chain amino-acid biosynthesis pathway and is the target of several classes of commercial herbicides. In this study, the Escherichia coli ilvG gene that encodes the catalytic subunit of AHAS II was cloned into the pET28a vector and expressed in soluble form at high levels in E. coli strain BL21 (DE3) cells. The protein was purified using Ni2+-chelating chromatography followed by size-exclusion chromatography. The catalytic subunit of E. coli AHAS II was cocrystallized with its cofactors Mg2+, FAD and ThDP using the sitting-drop vapour-diffusion method and the crystals diffracted to 2.80 Å resolution.
1. Introduction
Bacteria, archaea, fungi, algae and plants all possess the branched-chain amino-acid (BCAA; valine, leucine and isoleucine) biosynthesis pathway. However, animals do not possess this pathway, acquiring BCAAs from their diet. Acetohydroxyacid synthase (AHAS; EC 2.2.1.6) is the first common enzyme in the BCAA biosynthesis pathway. AHAS catalyzes the synthesis of 2-acetolactate from two molecules of pyruvate and also the synthesis of 2-aceto-2-hydroxybutyrate from one molecule of pyruvate and one molecule of 2-ketobutyrate. Almost all of the AHASs from different species (except for those from archaea) contain two subunits: a catalytic subunit and a regulatory subunit. Flavin adenine dinucleotide (FAD), thiamine diphosphate (ThDP) and Mg2+ are essential cofactors for full activity of the enzyme and bind to the catalytic subunit. In general, the regulatory subunit can enhance the activity of the catalytic subunit and can bind feedback signals that down-regulate the activity. AHAS has thus become the target of several classes of commercial herbicides, including sulfonylureas, imidazolinones, triazolopyrimidines, sulfonylaminocarbonyltriazolinones and pyrimidinylthiobenzoates, and the characteristics of different AHASs have been extensively researched (McCourt & Duggleby, 2006 ▶). The crystal structure of the Saccharomyces cerevisiae AHAS catalytic subunit with the three cofactors was reported in 2002 (Pang et al., 2002 ▶) and structures with five sulfonylureas in the presence of the three cofactors were subsequently resolved (Pang et al., 2003 ▶; McCourt et al., 2005 ▶). Structures of the Arabidopsis thaliana AHAS catalytic subunit in complex with five sulfonylureas and one imidazolinone cocrystallized with all three cofactors were resolved soon after (Pang et al., 2004 ▶; McCourt et al., 2006 ▶). Furthermore, AHAS can also serve as a potential target for fungicides and antimicrobials (Zohar et al., 2003 ▶; Kingsbury et al., 2004 ▶).
Escherichia coli contains three AHAS isoenzymes: AHAS I, AHAS II and AHAS III (Umbarger, 1996 ▶). AHAS II has the smallest regulatory subunit of three isoenzymes and has the most specific activity; it is not sensitive to valine inhibition but is the most sensitive to herbicides. Therefore, E. coli AHAS II has been used to study mechanisms of herbicide resistance and to develop new herbicidal compounds based on activity assays (Li et al., 2006 ▶; Xi et al., 2006 ▶).
In this study, we obtained crystals of the catalytic subunit of E. coli AHAS II cocrystallized with FAD, ThDP and Mg2+ and collected a complete X-ray data set. The structural study will provide important insights into both substrate binding and the catalytic mechanism of AHAS.
2. Materials and methods
2.1. Cloning, expression and purification
The ilvG gene of E. coli was amplified by PCR from the plasmid pQE-GMwt containing the ilvGM gene (encoding the holoenzyme) using the primers 5′-CGGGATCCATGAATGGCGCACAG-3′ and 5′-CCGCTCGAGTCAGGCGCGGATTTG-3′. The amplified fragment was inserted into the expression vector pET-28a(+) (Novagen) between BamHI and XhoI restriction sites to yield pNXH201a. This construct expressed the ilvG gene with a 34-residue (3.56 kDa) vector-encoded N-terminal fusion protein containing a hexahistidine tag and was verified by DNA sequencing.
The expression plasmid pNXH201a was transformed into E. coli strain BL21 (DE3) cells. A single colony was inoculated into 20 ml Luria–Bertani (LB) broth containing 50 µg ml−1 kanamycin. After overnight incubation at 310 K and 200 rev min−1, the culture was scaled up to 1 l containing 50 µg ml−1 kanamycin and incubated until the OD600 reached about 0.6. The culture was cooled to 291 K, 0.5 ml 1 M isopropyl β-d-1-thiogalactopyranoside was added and the culture was incubated for 16 h at 291 K and 200 rev min−1. The cells were harvested by centrifugation at 3000g for 10 min at 277 K and the cell pellet was resuspended in buffer A (20 mM Tris–HCl, 500 mM NaCl, 10 µM FAD pH 7.5) and stored at 253 K.
A two-step purification was performed to obtain the hexahistidine-tagged target protein: immobilized metal-affinity chromatography followed by size-exclusion chromatography. The frozen suspension was thawed and the cells were disrupted by sonication in an ice–water bath. Insoluble material was removed by centrifugation twice at 58 000g for 20 min at 277 K. The supernatant was passed through a 0.2 µm filter, loaded onto a 5 ml Ni2+-chelating affinity column (HiTrap Chelating HP 5 ml, GE Healthcare, USA) previously charged with 100 mM nickel sulfate and equilibrated with buffer A using a peristaltic pump. All subsequent chromatography steps were performed using an ÄKTA Explorer 100 (GE Healthcare, USA) in a cold room.
Unbound proteins were washed out with 30 ml buffer A and nonspecifically bound proteins were washed out with 30 ml 10% buffer B (20 mM Tris–HCl, 500 mM NaCl, 10 µM FAD, 500 mM imidazole pH 7.5) in buffer A. The hexahistidine-tagged protein was eluted with 15 ml 100% buffer B. Fractions containing the desired protein were pooled in the presence of 2 mM dithiothreitol (DTT). The protein solution was concentrated to a volume of 1 ml by ultrafiltration (Ultra-15, 30 kDa cutoff, Millipore Amicon, centrifugation at 3000g). Denatured and precipitated protein was eliminated by centrifugation at 16 000g for 10 min at 277 K and the supernatant containing the target protein was further purified using a HiLoad Superdex 75 column (GE Healthcare, USA) with buffer C (20 mM Tris–HCl, 200 mM NaCl, 10 µM FAD, 2 mM DTT pH 7.5). Fractions containing the AHAS II catalytic subunit were collected and concentrated by ultrafiltration to 40 mg ml−1 using the Bio-Rad Protein Assay (Bio-Rad Laboratories Inc., USA; bovine serum albumin as standard). The purified protein was stored in small aliquots at 193 K. The purity of the target protein was checked by SDS–PAGE during each step. Only one main band with the expected molecular weight (∼60 kDa) was visible on the SDS–PAGE assay after size-exclusion purification (Fig. 1 ▶).
Figure 1.
SDS–PAGE of the purified catalytic subunit of E. coli AHAS II. Lane M contains molecular-weight markers (labelled in kDa) and lane P contains a sample of the purified protein.
2.2. Crystallization and data collection
Crystallization trials were performed at 293 K using the sitting-drop vapour-diffusion method. An initial screen for suitable crystallization conditions was carried out using Crystal Screen, Crystal Screen 2 and Index Screen (Hampton Research, California, USA). Before each experiment, the cofactors were added to a freshly thawed protein aliquot to final concentrations of 1 mM FAD, 1 mM ThDP and 1 mM MgCl2. For the sitting drop, 1 µl reservoir solution was added to 1 µl protein solution. The droplets were equilibrated against 0.1 ml reservoir solution.
Crystals with different shapes were observed in 5–10 d in a number of conditions. Optimal crystal-growth conditions were obtained by decreasing the protein concentration to about 15 mg ml−1 and using a reservoir solution consisting of 0.8 M potassium/sodium tartrate, 0.1 M Na HEPES pH 7.5.
A complete diffraction data set was collected from a crystal of the catalytic subunit of E. coli AHAS II using a MAR 165 CCD detector on beamline 3W1A at Beijing Synchrotron Radiation Facility (BSRF), Beijing, People’s Republic of China. The crystal was flash-frozen in liquid nitrogen and maintained at 100 K using nitrogen gas (Oxford Instruments, UK). For cryoprotection, the crystals were soaked in reservoir solution containing 10% glycerol. The crystals were stable during soaking, showing no signs of cracking or dissolving for at least 5 min. The data were processed using the program XDS (Kabsch, 2010 ▶).
3. Results
The catalytic subunit of E. coli AHAS II with its cofactors (FAD, ThDP and Mg2+) was successfully crystallized in 0.8 M potassium/sodium tartrate, 0.1 M Na HEPES pH 7.5 (Fig. 2 ▶). Crystals were first observed after 2 d and took about 7 d to reach maximum size. The crystals were yellow, with a more intense colour than that of the mother liquor (as shown in Fig. 1 ▶), indicating incorporation of the cofactors into the crystals. The crystals diffracted to a resolution of 2.80 Å and belonged to space group P21212, with unit-cell parameters a = 139.1, b = 149.5, c = 59.0 Å. Assuming two molecules per asymmetric unit, the Matthews coefficient was 2.57 Å3 Da−1 and the solvent content was 52.11% (Matthews, 1968 ▶). The data-collection statistics are listed in Table 1 ▶.
Figure 2.
Crystals of the catalytic subunit of E. coli AHAS II cocrystallized with its cofactors.
Table 1. Data-collection statistics for E. coli AHAS II crystals.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.000 |
| Resolution range (Å) | 50.00–2.80 (2.90–2.80) |
| Space group | P21212 |
| Unit-cell parameters (Å) | a = 139.1, b = 149.5, c = 59.0 |
| No. of observed reflections | 153991 |
| No. of unique reflections | 30257 |
| Completeness (%) | 97.1 (99.9) |
| Rmerge† (%) | 6.4 (47.6) |
| Average I/σ(I) | 18.66 (3.31) |
| Molecules per asymmetric unit | 2 |
| VM (Å3 Da−1) | 2.57 |
| Solvent content (%) | 52.11 |
R
merge = 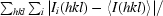
 .
.
Acknowledgments
We thank the beamline staff (Drs Yu-Hui Dong and Peng Liu) at BSRF for their kind help with data collection. We are also grateful to Dr Hong-fei Wang for reading the manuscript and making crucial comments. This work was supported by Peking University’s 985 and 211 grants.
References
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kingsbury, J. M., Yang, Z., Ganous, T. M., Cox, G. M. & McCusker, J. H. (2004). Microbiology, 150, 1547–1558. [DOI] [PubMed]
- Li, Y.-X., Luo, Y.-P., Xi, Z., Niu, C., He, Y.-Z. & Yang, G.-F. (2006). J. Agric. Food Chem. 54, 9135–9139. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCourt, J. A. & Duggleby, R. G. (2006). Amino Acids, 31, 173–210. [DOI] [PubMed]
- McCourt, J. A., Pang, S. S., Guddat, L. W. & Duggleby, R. G. (2005). Biochemistry, 44, 2330–2338. [DOI] [PubMed]
- McCourt, J. A., Pang, S. S., King-Scott, J., Guddat, L. W. & Duggleby, R. G. (2006). Proc. Natl Acad. Sci. USA, 103, 569–573. [DOI] [PMC free article] [PubMed]
- Pang, S. S., Duggleby, R. G. & Guddat, L. W. (2002). J. Mol. Biol. 317, 249–262. [DOI] [PubMed]
- Pang, S. S., Guddat, L. W. & Duggleby, R. G. (2003). J. Biol. Chem. 278, 7639–7644. [DOI] [PubMed]
- Pang, S. S., Guddat, L. W. & Duggleby, R. G. (2004). Acta Cryst. D60, 153–155. [DOI] [PubMed]
- Umbarger, H. (1996). Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd ed., edited by F. C. Neidhardt, Vol. I, pp. 352–367. Washington DC: American Society for Microbiology.
- Xi, Z., Yu, Z., Niu, C., Ban, S. & Yang, G. (2006). J. Comput. Chem. 27, 1571–1576. [DOI] [PubMed]
- Zohar, Y., Einav, M., Chipman, D. M. & Barak, Z. (2003). Biochim. Biophys. Acta, 1649, 97–105. [DOI] [PubMed]