Abstract
NF-κB (nuclear factor κB) has a pivotal role in many cellular processes, including the inflammatory and immune responses and, therefore, its activation is tightly regulated by the IKK (IκB kinase) complex and by IκBα degradation. When Shigella bacteria multiply within epithelial cells they release peptidoglycans, which are recognized by Nod1 and stimulate the NF-κB pathway, thus leading to a severe inflammatory response. Here, we show that IpaH9.8, a Shigella effector possessing E3 ligase activity, dampens the NF-κB-mediated inflammatory response to the bacterial infection in a unique way. IpaH9.8 interacts with NEMO/IKKγ and ABIN-1, a ubiquitin-binding adaptor protein, promoting ABIN-1-dependent polyubiquitylation of NEMO. Consequently, polyubiquitylated NEMO undergoes proteasome-dependent degradation, which perturbs NF-κB activation. As NEMO is essential for NF-κB activation, we propose that the polyubiquitylation and degradation of NEMO during Shigella infection is a new bacterial strategy to modulate host inflammatory responses.
The ability of the innate immune system to discriminate infecting bacteria as ‘non-self ’ is critical to limiting bacterial colonization. Major bacterial components, such as peptidoglycans (PGNs), lipopolysac-charides (LPSs) and nucleic acids, are recognized by Toll-like receptors (TLRs) and Nod-like receptors (NLRs), which in turn stimulate inflammatory signalling cascades and activate the host immune system1–3. As NF-κB has a central role in the regulation of numerous genes involved in stress responses, cell proliferation and the inflammatory responses4, NF-κB activation is tightly regulated by the multi-protein IKK complex and the NF-κB inhibitor IκB. Upon stimulation of the IKK complex, which is composed of IKKα, IKKβ, NEMO/IKKγ and ELKS, IκB is phosphorylated by IKKα and IKKβ, polyubiquitylated and degraded by the proteasome, thereby allowing NF-κB to translocate to the nucleus and activate transcription of proinflammatory cytokines and antimicrobial peptide genes. Importantly, activation of the IKK complex is primarily dependent on modification of NEMO by Lys 63-linked polyubiquitin chains5,6.
During multiplication, cytoplasm-invading bacteria, such as Shigella, release PGNs and LPSs, which induce a strong inflammatory response7. Nevertheless, such pathogens are able to efficiently colonize the intestinal epithelium because they possess highly evolved systems that modulate the host inflammatory and immune responses8,9. For example, Shigella deliver a number of effectors, including OspG, OspF and IpaH9.8 (a member of the IpaH family), into the host cell cytoplasm and nucleus through their type III secretion system (TTSS)10–14. The IpaH family is of great interest because bacterial effectors produced by mammal, fish and plant pathogens, including the Yersinia, Salmonella, Edwardsiella, Bradyrhizobium, Rhizobium and some Pseudomonas species, share several structural and functional characteristics with the members of the IpaH family, namely, their amino-terminal portions contain highly conserved leucine-rich repeats (LRR), while their conserved carboxy-terminal regions possess E3 ligase activity15. The crystal structure of IpaH proteins has recently been described, and based on this structure the IpaH family has been identified as a new class of E3 ubiquitin ligases found in pathogenic and symbiotic bacteria16,17. Although the host target molecule(s) of the IpaH family E3 ligase activity remains unknown, in a yeast-cell system, one of the IpaH family, IpaH9.8, was shown to have activity that interferes with the pheromone response by ubiquitylation and proteasome degradation of the MAPK kinase Ste7 (ref. 15). Furthermore, previous studies have suggested that Shigella IpaH9.8 affects NF-κB-dependent gene expression and the activity of U2AF35, an alternative splicing factor, and that IpaH9.8 has an important role in modulating the host inflammatory response12,18. Therefore, in this study, we attempted to identify the host target protein of IpaH9.8 E3 ligase activity and to elucidate its role in Shigella infection.
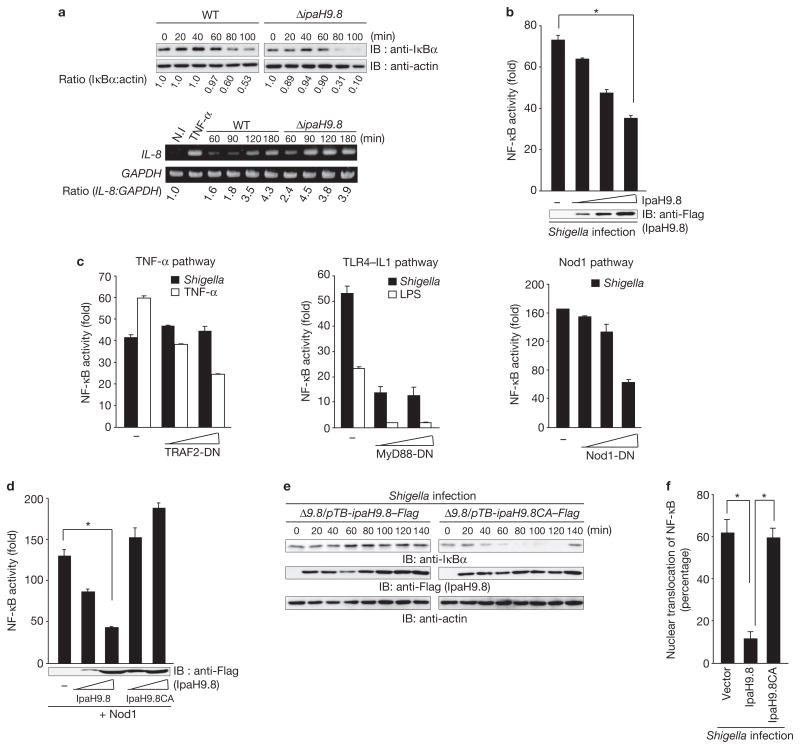
The levels of IκBα and IL-8 in HeLa cells infected with Shigella strain YSH6000 (wild type) or a Shigella ΔipaH9.8 mutant were measured. IκBα levels in cells infected with ΔipaH9.8 were found to be lower than in cells infected with the wild-type strain, while IL-8 levels in cells infected with ΔipaH9.8 were higher than in cells infected with the wild-type strain (Fig. 1a). To confirm the effect of IpaH9.8, cells carrying pNF-κB–Luc (an NF-κB-luciferase reporter plasmid) plus an IpaH9.8-expressing plasmid (pIpaH9.8) or a mock plasmid were infected with Shigella; the NF-κB activity in the presence of IpaH9.8 was found to decrease in a dose-dependent manner (Fig. 1b). We then investigated whether the effect of IpaH9.8 was specific to the NF-κB pathway by measuring the levels of IκBα, phosphorylated-Erk and phosphorylated-JNK in HeLa/IpaH9.8–Flag cells. The results showed that the IκBα signalling pathway, but not other signalling pathways, such as the MAPK pathway, is involved in the IpaH9.8-dependent downregulation (Supplementary Information, Fig. S1). When dominant-negative (DN) forms of TRAF2, MyD88 or Nod1 were expressed in cells, MyD88-DN and Nod1-DN, but not TRAF2-DN, reduced NF-κB activity (Fig. 1c), suggesting that IpaH9.8 preferentially targets Nod1 or TLR4 signalling. When pNF-κB with or without pIpaH9.8 was transfected with pNod1, pRICK or pIKKβ, NF-κB activation by Nod1 and RICK, but not by IKKβ, was inhibited by IpaH9.8 (Supplementary Information, Fig. S2). These results suggested that IpaH9.8 targets the Nod1–RICK–NF-κB pathway.
Figure 1.
IpaH9.8 inhibits NF-κB activity. (a) HeLa cells were infected with Shigella wild-type or ΔipaH9.8 mutant bacteria harbouring an afimbrial adhesin (Afa) expression plasmid (multiplicity of infection, MOI = 10). After Shigella invasion, cell lysates were prepared at the indicated time points and subjected to immunoblotting (upper panel) or semi-quantitative reverse transcription-PCR (RT-PCR) analysis (lower panel). The blots were quantified by measuring relative intensity, and values are indicated below the blots. (b) Luciferase assays were performed after Shigella infection (for 3 h) of 293T cells transiently transfected with an NF-κB reporter plasmid (50 ng) and an empty vector or a Flag–IpaH9.8-expressing plasmid (0, 50, 100 or 200 ng). Results are presented as fold relative to the activity of non-infected cells. Data are mean ± s.e.m., n = 4. (c) Luciferase assays of 293T cells transiently transfected with an NF-κB reporter plasmid and an empty vector or increasing amounts of dominant-negative forms of TRAF2 (TRAF2-DN), MyD88 (MyD88-DN) or Nod1 (Nod1-DN). After 24 h, cells were infected with Shigella or treated with TNF-α (10 ng ml−1) or LPS (100 ng ml−1) for 3 h and luciferase activity was measured. Results are presented as fold relative to the activity of non-infected or non-stimulated cells. Data are mean ± s.e.m., n = 3. (d) Luciferase assay of 293T cells transfected with an NF-κB reporter plasmid (50 ng) and empty vector, Flag–IpaH9.8 or Flag–IpaH9.8CA expressing plasmid (50 or 200 ng), plus plasmids expressing Nod1 as an NF-κB activator. Results are presented as fold relative to the activity of non-stimulated cells. Data are mean ± s.e.m., n = 4. (e) HeLa cells were infected with Shigella ΔipaH9.8 harbouring pTB-ipaH9.8–Flag or pTB-ipaH9.8CA–Flag (pTB is an IPTG-inducible vector). The cell lysates prepared at the indicated time points were subjected to immunoblotting. (f) The percentage of nuclear translocation of NF-κB (p65) after Shigella infection for 1 h was determined by counting the number of nuclear translocated p65 in at least 300 GFP-, GFP–IpaH9.8- or GFP–IpaH9.8CA-expressing cells using fluorescence microscopy. Data are mean ± s.e.m., n = 3. *P < 0.001. WT, wild-type. IB, immunoblot. N.I, non-infected. For full scans of blots in a, b, d and e, see Supplementary Information, Fig. S7.
As the Cys 337 residue of the IpaH family proteins, including IpaH9.8, is critical to their E3 ligase activity15–17, we investigated the involvement of Cys 337 in the suppression of NF-κB activation using a NF-κB reporter assay. NF-κB activation by Nod1 was reduced by IpaH9.8 in a dose-dependent manner, but not by IpaH9.8CA (E3 ligase-deficient mutant with Ala substituted for Cys 337; Fig. 1d). HeLa cells infected with Shigella expressing IpaH9.8, but not IpaH9.8CA, showed inhibited IκBα degradation (Fig. 1e). Similarly, the nuclear translocation of NF-κB stimulated by Shigella infection was greatly reduced in the cells expressing IpaH9.8 but not IpaH9.8CA (Fig. 1f). These results clearly indicate that the E3 ligase activity of IpaH9.8 is critical to dampen NF-κB activation during Shigella infection.
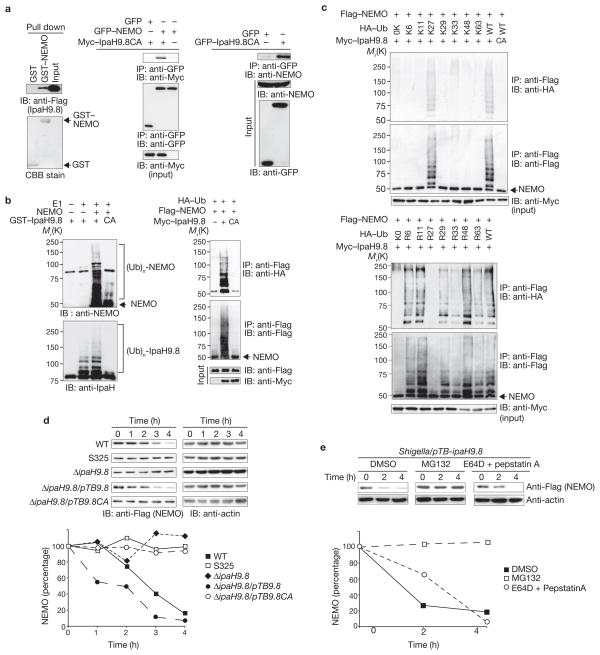
Yeast two-hybrid screening of HeLa cell and human brain cDNA libraries with IpaH9.8 as bait identified NEMO/IKKγ and ABIN-1 (A20-binding inhibitor of NF-κB) as IpaH9.8 binding partners. ABIN-1 binds to NEMO, facilitating A20-mediated removal of polyubiquitin chains from NEMO, and preventing NF-κB activation19–21. To confirm the interactions observed in the yeast two-hybrid screening, we performed a GST pulldown and immunoprecipitation assays (Fig. 2a, 4a). NEMO contains two coiled-coil domains (CC1 and C2), a leucine zipper (LZ) and a C-terminal zinc finger (ZF) domain, required for correct assembly of the IKK complex. The use of NEMO truncations enabled the identification of a region (residues 347–396) between the LZ and ZF domain that is responsible for the interaction with IpaH9.8, and IpaH9.8 truncations allowed us to identify the IpaH9.8 LRR region that is required for the interaction with NEMO (Supplementary Information, Fig. S3a, b).
Figure 2.
IpaH9.8 promotes NEMO ubiquitylation and degradation. (a) IpaH9.8–Flag bound to NEMO, as found in GST pulldown assays (left panel) and immunoprecipitation analysis of 293T cell lysates (center and right panel). (b) IpaH9.8 targets NEMO, and promotes its ubiquitylation. In vitro ubiquitylation assay with NEMO and a mixture of E1, UbcH5b, ATP and ubiquitin in the presence or absence of GST–IpaH9.8 or GST–IpaH9.8CA (left panel). Cells expressing Flag–NEMO, HA–Ub and Myc6–IpaH9.8 or Myc6–IpaH9.8CA were immunoprecipitated using an anti-Flag antibody and analysed for ubiquitylation by immunoblotting using anti-HA and anti-Flag antibodies. Whole cell lysates were immunoblotted with anti-Flag and anti-Myc antibodies (right panel). (c) Cells expressing Flag–NEMO, Myc6-IpaH9.8 and each of the HA-ubiquitin mutants as indicated were lysed and immunoprecipitated using an anti-Flag antibody. Ubiquitylated NEMO was analyzed by immunoblotting. (d) HeLa cells were transfected with Flag–NEMO and infected with Shigella strains. After infection, cell extracts were prepared at the indicated time points and subjected to immunoblotting. The remaining NEMO was quantified (graph). (e) HeLa cells were transfected with Flag–NEMO and infected with Shigella/pTBipaH9.8. After infection, cells were treated with DMSO, MG132 (40 μM) or E64D + pepstatin A, and cell extracts were prepared at the indicated time points. Samples were subjected to immunoblotting. The remaining NEMO was quantified (graph). WT, wild-type. IP, immunoprecipitate. IB, immunoblot. Ub, ubiquitin. For full scans of blots in a–d, see Supplementary Information, Fig. S7.
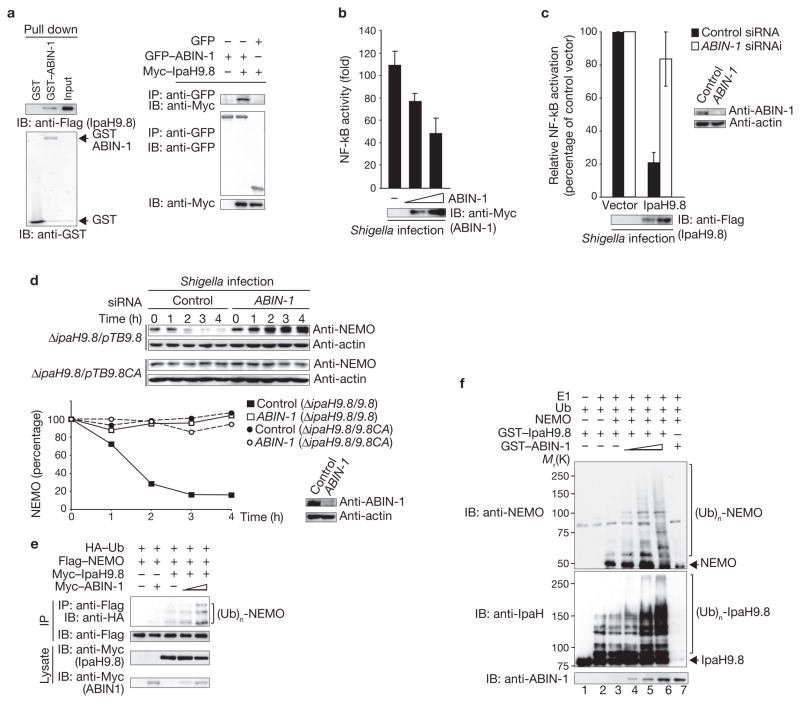
Figure 4.
ABIN-1 promotes IpaH9.8-mediated NEMO ubiquitylation. (a) IpaH9.8–Flag bound to GST–ABIN-1 in a GST-pulldown assay. Proteins pulled down were immunoblotted with an anti-Flag antibody (left panel), and 293T cell lysates were subjected to imunoprecipitation analysis (right panel). (b) Shigella induced luciferase activity in 293T cells transfected with a plasmid encoding NF-κB luciferase reporter and a plasmid encoding Myc-ABIN-1 (0, 50 or 100 ng). Results are presented as fold relative to the activity of non-infected cells. Data are mean ± s.e.m., n = 3. (c) 293T cells transfected with siRNA targeting ABIN-1 or GFP (control) were co-transfected with a NF-κB reporter plasmid and empty vector or Flag–IpaH9.8, and then infected with Shigella. NF-κB activity is presented as a percentage of cells expressing empty vector (set to 100%). Data are mean ± s.e.m., n = 3. (d) HeLa cells transfected with siRNA targeting ABIN-1 or GFP (control) were infected with ΔipaH9.8/pTB-ipaH9.8 or ΔipaH9.8/pTB-ipaH9.8CA bacteria. Quantified NEMO levels are shown (graph). (e) 293T cells were transfected with Flag–NEMO, HA–Ub, a semi-optimal amount of Myc–IpaH9.8 and increasing amounts of Myc–ABIN-1. Cell extracts were immunoprecipitated with an anti-Flag antibody and analysed by immunoblotting with an anti-HA antibody. (f) In vitro ubiquitylation assay with NEMO and a mixture of E1, UbcH5b, ATP, ubiquitin and GST–IpaH9.8 in the presence or absence of increasing amounts of GST–ABIN-1. Samples were subjected to immunoblotting with anti-NEMO, anti-IpaH and anti-ABIN-1 antibodies. IP, immunoprecipitate. IB, immunoblot. Ub, ubiquitin. For full scans of blots in a, d, e and f see Supplementary Information, Fig. S7.
As polyubiquitylation of NEMO, (Ub)n-NEMO, is required for the activation of the IKK complex and NF-κB in response to various stimuli5,6, recombinant NEMO was incubated with E1, UbcH5b (as E2), ubiquitin (Ub) and GST–IpaH9.8 or GST–IpaH9.8CA, and NEMO was analysed for polyubiquitylation. IpaH9.8, but not IpaH9.8CA, was auto-ubiquit-ylated, and NEMO was ubiquitylated in the presence of IpaH9.8, but not IpaH9.8CA (Fig. 2b, left panel). NEMO was also shown to be ubiq-uitylated in an IpaH9.8-dependent manner in 293T cells (Fig. 2b, right panel). Lys 63-linked ubiquitylation is sometimes required for signal transduction processes, including NF-κB activation, while Lys 48-linked ubiquitylation typically leads to proteasome degradation22. We therefore created a series of ubiquitin mutants possessing single lysine residues to investigate IpaH9.8-mediated NEMO ubiquitylation. NEMO was ubiq-uitylated by the Lys 27-Ub (K27-Ub) mutant to a similar extent as by wild-type ubiquitin (Fig. 2c, upper panel). We then examined ubiquitin in which only one of each lysine residue had been replaced by arginine and confirmed that Lys 27 has a major role in the IpaH9.8-mediated ubiquitylation of NEMO (Fig. 2c, lower panel). These findings suggest that IpaH9.8 is a bacterial ubiquitin E3 ligase that is capable of catalysing Lys 27-linked polyubiquitylation of NEMO.
To investigate the fate of ubiquitylated NEMO, we measured the half-life of NEMO in cells expressing, or not expressing, pFlag–IpaH9.8 or pFlag–IpaH9.8CA. NEMO levels in cells expressing IpaH9.8 decreased 3 h after cycloheximide treatment in comparison with cells expressing IpaH9.8CA. Intriguingly, the rate of IpaH9.8-mediated NEMO degradation was higher in Nod1-stimulated cells than in non-stimulated cells (Supplementary Information, Fig. S4). Analysis of NEMO degradation kinetics in HeLa/pFlag–NEMO cells infected with Shigella (wild-type), S325 (TTSS-deficient mutant), ΔipaH9.8, ΔipaH9.8/pTB-ipaH9.8 (IpaH9.8 overproduction) or ΔipaH9.8/pTB-ipaH9.8CA (IpaH9.8CA overproduction) bacteria showed that NEMO levels had decreased to less than 20% of their original level by 4 h after infection with wild-type bacteria, but not after infection with S325 or ΔipaH9.8 bacteria. The effect on NEMO stability was greater in cells infected with ΔipaH9.8/pTB-ipaH9.8 bacteria. NEMO levels were unchanged following infection with ΔipaH9.8/pTB-ipaH9.8CA bacteria (Fig. 2d). As MG132 (a proteasome inhibitor), but not E64D plus pepstatin A (a lysosome inhibitor), prevented NEMO degradation in cells infected with Shigella/pTB-ipaH9.8 (Fig. 2e) bacteria, we concluded that NEMO ubiquitylated by IpaH9.8 undergoes proteasomal degradation.
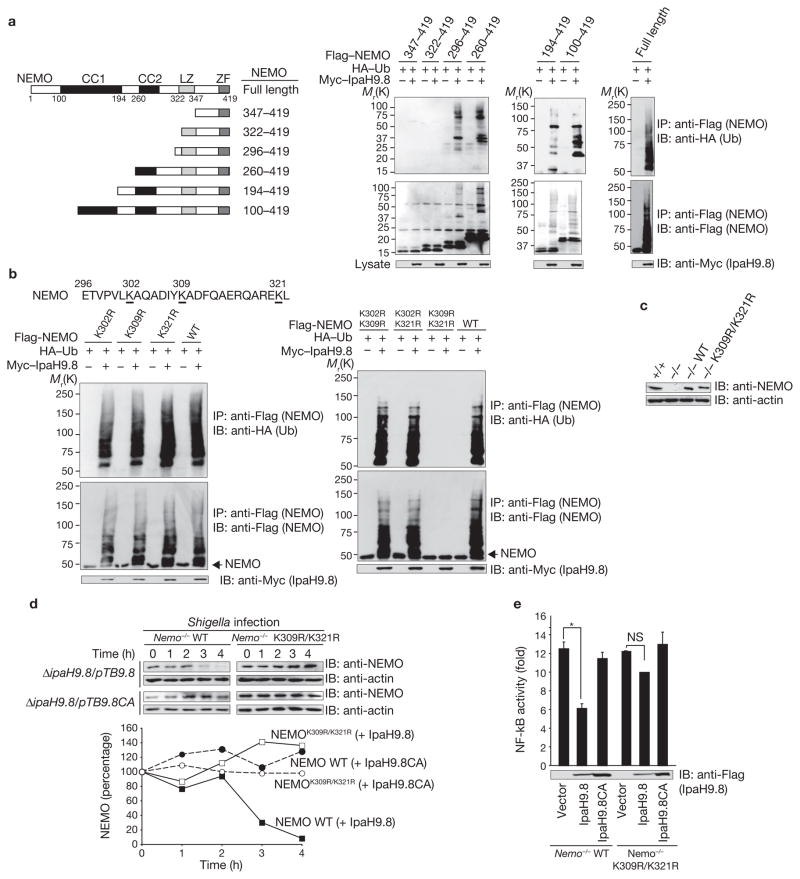
Previous studies found that the particular NEMO lysine residue that accepts ubiquitin depends on the stimulus; for example, T-cell receptor signalling triggers Lys 63-linked ubiquitylation of NEMO at Lys 399 (refs 23, 24). MDP-induced Nod2–RICK stimulation triggers Lys 63-linked polyubiquitylation of NEMO at Lys 285, whereas LPS-induced TLR stimulation triggers Lys 285 and Lys 399 for ubiquitylation25,26. LUBAC, a newly identified E3 ligase complex, triggers linear polyubiquitylation of NEMO at both Lys 285 and Lys 309 (ref. 27). We therefore tested a series of Flag-tagged truncations of NEMO for IpaH9.8-mediated ubiquitylation, by immunoprecipitation. The C-terminal portion (residues 296–322) was ubiquitylated (Fig. 3a), and as three lysine residues are present in this part of the protein (Fig. 3b), we investigated ubiquitylation of Flag–NEMOK302R, Flag–NEMOK309R and Flag–NEMOK321R, by immunoprecipitation. Because all three mutants were polyubiquitylated (Fig. 3b left), we analysed NEMO double mutants for ubiquitylation. The results showed that NEMOK302R/K309R and NEMOK302R/K321R were polyubiquitylated, but that NEMOK309R/K321R was not (Fig. 3b, right; Supplementary Information, Fig. S5a), suggesting that Lys 309 and Lys 321 are involved in IpaH9.8-mediated polyubiquitylation. It is notable that the binding capability of IpaH9.8 to NEMO wild-type and to NEMOK309R/K321R was similar (Supplementary Information, Fig. S5b), suggesting that the lower extent of NEMOK309R/321R ubiqitination by IpaH9.8 was not related to binding. To confirm this, we retrovirally transduced Nemo-deficient mouse embryonic fibroblasts (Nemo−/−)28 with NEMO wild type (Nemo−/− NEMO-wild type) and NEMOK309R/K321R (Nemo−/− NEMOK309R/K321R; Fig. 3c). The level of NEMOK309R/K321R, but not of NEMO-wild type, was unaffected in Nemo−/− NEMO-wild-type cells or Nemo−/− NEMOK309R/K321R cells infected with ΔipaH9.8/pTB-ipaH9.8. Indeed, the levels of both NEMO-wild type and NEMOK309R/K321R was unaffected in cells infected with ΔipaH9.8/pTB-ipaH9.8CA (Fig. 3d). As expected, the activity of Shigella-induced NF-κB was reduced by IpaH9.8 E3 ligase activity in Nemo−/− NEMO-wild-type cells but not in Nemo−/− NEMOK309R/K321R cells (Fig. 3e). These results indicate that IpaH9.8 predominantly targets both the Lys 309 and Lys 321 residues of NEMO for ubiquitylation.
Figure 3.
IpaH9.8 targets NEMO Lys 309 and Lys 321 residues for ubiquitylation. (a) Schematic representation of NEMO truncations used for ubiquitylation assays (left panel). Cells expressing each of various truncated versions of Flag–NEMO, HA–Ub and Myc6–IpaH9.8 were immunoprecipitated using an anti-Flag antibody and analysed by immunoblotting using anti-HA and -Flag antibodies. Whole cell lysates were immunoblotted with an anti-Myc antibody (right panel). (b) Cells expressing Flag–NEMO wild type or Flag–NEMO point mutants, HA–Ub and Myc6–IpaH9.8 were immunoprecipitated with an anti-Flag antibody and analysed by immunoblotting with anti-HA and anti-Flag antibodies. Whole cell lysates were immunoblotted with an anti-Myc antibody. (c) Nemo+/+, Nemo−/− or Nemo−/− cells stably expressing NEMO-wild type or the NEMOK309R/K321R mutant were immunoblotted with anti-NEMO and anti-actin antibodies. d) Nemo−/− cells stably expressing NEMO-wild type or NEMOK309R/K321R were infected with ΔipaH9.8/pTB-ipaH9.8 or ΔipaH9.8/pTB-ipaH9.8CA bacteria. Cell lysates prepared at each time point were subjected to immunoblotting with anti-NEMO and anti-actin antibodies. The levels of NEMO were quantified (graph). (e) Nemo−/− cells stably expressing NEMO-wild type or NEMOK309R/K321R were transiently co-transfected with an NF-κB reporter plasmid and empty vector, Flag–IpaH9.8- or Flag–IpaH9.8CA-expressing plasmid. After 3 h infection with Shigella at multiplicity of infection (MOI) =100, Shigella-induced NF-κB luciferase activity was measured. Results are presented as fold relative to the activity of non-infected cells. Data are mean ± s.e.m., n = 4. *P < 0.001. NS, not significant. WT, wild-type. IP, immunoprecipitate. IB, immunoblot. Ub, ubiquitin. For full scans of blots in a, b and d see Supplementary Information, Fig. S7.
As ABIN-1 binds to NEMO and inhibits NF-κB activation by cooperating with A20, which mediates de-ubiquitylation of NEMO19, we performed a NF-κB reporter assay to determine the effect of ABIN-1 on Shigella-induced NF-κB activity. NF-κB activity in cells decreased in an ABIN-1 dose-dependent manner (Fig. 4b), and IpaH9.8 strongly inhibited Shigella-induced NF-κB activity in control cells, but only weakly inhibited Shigella-induced NF-κB activity in ABIN-1 knockdown cells (Fig. 4c). Furthermore, NEMO levels were reduced in control cells, but unaffected in ABIN-1 knockdown cells during ΔipaH9.8/pTB-ipaH9.8 infection, whereas NEMO levels were unaffected in both control and ABIN-1 knockdown cells during ΔipaH9.8/pTB-ipaH9.8CA infection (Fig. 4d). These results indicate that the interaction between IpaH9.8 and ABIN-1 is involved in inhibition of NF-κB activity. To determine the role of ABIN-1 in IpaH9.8-mediated NF-κB inhibition, we investigated the ability of a series of truncations to interact with IpaH9.8. The results showed that ABIN-1 residues 351–367 were involved in interacting with IpaH9.8, and by using various IpaH9.8 truncations we were able to determine that the IpaH9.8 carboxy-terminal region is involved in the interaction with ABIN-1 (Supplementary Information, Fig. S3c, d).
To confirm ABIN-1 involvement in IpaH9.8-mediated NEMO ubiquitylation, we analysed the levels of ubiquitylated NEMO by immunoprecipitation. ABIN-1 promoted IpaH9.8-mediated NEMO ubiquitylation in a dose-dependent manner; however, ABIN-1 failed to induce NEMO ubiquitylation in the absence of IpaH9.8 (Fig. 4e). Next, we assessed NEMO ubiquitylation in vitro (Fig. 4f). NEMO was ubiquitylated in the presence of ubiquitin and GST–IpaH9.8 (lane 3), and when ABIN-1 was added to the reaction the extent of NEMO ubiquitylation increased in a dose-dependent manner (lanes 4–6), strongly suggesting that ABIN-1 does not possess E3 ubiquitin ligase activity, but rather acts as an adaptor for IpaH9.8-mediated ubiquitylation of NEMO.
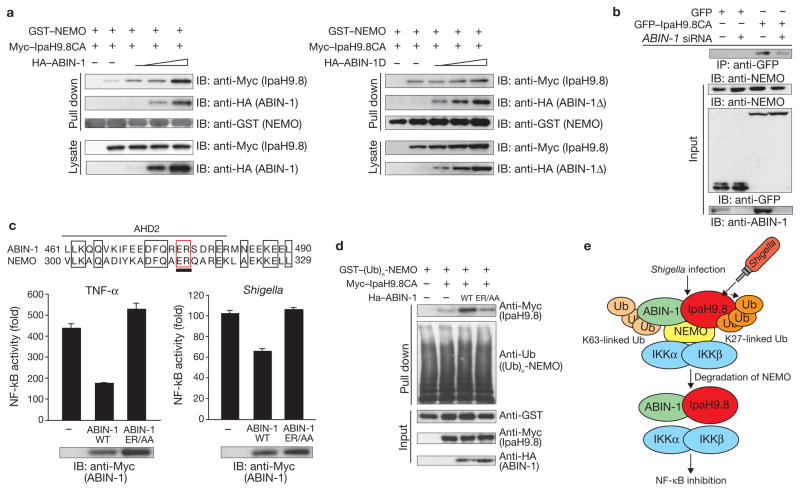
In the A20–NEMO–ABIN-1 complex, ABIN-1 promotes the association between A20 and NEMO, thereby increasing A20-mediated NEMO deubiquitylation and inhibiting NF-κB activation19. Because this raises the possibility that ABIN-1 contributes to the association between IpaH9.8 and NEMO, we measured the amounts of ABIN-1 and IpaH9.8 that interacted with NEMO in lysates of cells expressing pGST–NEMO, pMyc–IpaH9.8CA and pHA–ABIN-1 or pHA-ABIN-1Δ, which lacks the IpaH9.8-binding domain but retains the NEMO-binding domain. Increased ABIN-1 levels enhanced the extent of interaction between IpaH9.8 and NEMO (Fig. 5a, left panel), but increased ABIN-1Δ levels had little effect on the IpaH9.8–NEMO interaction (Fig. 5a, right panel). Furthermore, an RNA interference (RNAi) experiment showed that the amount of NEMO protein bound to IpaH9.8 was lower in ABIN-1 knockdown cells (Fig. 5b). These findings further support our proposal that ABIN-1 promotes IpaH9.8 binding to, and ubiquitylation of, NEMO.
Figure 5.
ABIN-1 acts as adaptor protein between IpaH9.8 and NEMO. (a) 293T cells were transfected with plasmids encoding GST–NEMO, Myc6–IpaH9.8CA and increasing amounts of HA–ABIN-1 (left panel) or HA-ABIN-1Δ (ABIN-1 mutant lacking the IpaH9.8-interacting domain, right panel). Cell extracts were bound to GST beads and subjected to immunoblotting with anti-Myc, anti-HA and anti-GST antibodies. (b) 293T cells treated with control (luciferase) siRNA or ABIN-1 siRNA were subjected to an immunoprecipitation assay. Cell extracts were immunoprecipitated with anti-GFP-conjugated beads and analysed by immunoblotting with an anti-NEMO antibody. (c) Sequence alignment of the ubiquitin-binding domain of ABIN-1 and NEMO (upper panel). Luciferase assays were performed after Shigella infection or TNF-α stimulation of 293T cells transfected with an NF-κB reporter plasmid and empty vector, Myc–ABIN-1- or Myc–ABIN-1-ER/AA-expressing plasmid (lower panels). Results are presented as fold relative to the activity of non-stimulated or non-infected cells. Data are mean ± s.e.m., n = 3. (d) 293T cells were transfected with GST–NEMO, Flag–Nod1 and HA–ubiquitin expression plasmids, and treated with iE-DAP to induce Nod1-stimulated ubiquitylation of NEMO. Cell extracts were bound to GST beads. The beads were subjected to GST pulldown assays with cell lysates expressing Myc–IpaH9.8CA with or without HA–ABIN-1 or HA–ABIN-1ER/AA and immunoblotted with anti-Myc, anti-Ub, anti-GST and anti-HA antibodies. (e) A proposed model for IpaH9.8-mediated NF-κB inhibition. WT, wild-type. IP, immunoprecipitate. Ub, ubiquitin. For full scans of blots in a, b and d see Supplementary Information, Fig. S7.
Recent reports have indicated that ABIN-1 acts as a ubiquitin-binding protein29–32. ABIN-1 interacts with NEMO through two domains. The C-terminal domain binds to NEMO, and the central AHD2 (ABIN homology domain 2) region is a ubiquitin-binding domain that interacts with ubiquitin moieties conjugated to NEMO29. We therefore investigated whether the ubiquitin-binding capacity of ABIN-1 that is required for interaction with NEMO is functionally involved in IpaH9.8-mediated down-regulation of NF-κB activation, by creating an ABIN-1-ER/AA mutant (a ubiquitin-binding-deficient mutant) and testing its effect on NF-κB activation by TNF-α or Shigella. The results showed that ABIN-1 had an inhibitory effect on NF-κB activation by both TNF-α and Shigella, but that ABIN-1-ER/AA had no inhibitory effect on activation by either (Fig. 5c), strongly suggesting that the interaction between ABIN-1 and NEMO through the polyubiquitin chain is pivotal to inhibition of NF-κB activation. Next, we investigated whether the ubiquitin-binding domain of ABIN-1 was involved in promoting the interaction between IpaH9.8 and polyubiquitnated-NEMO, which is stimulated through the Nod1 pathway. The results showed that ABIN-1-ER/AA interacted less strongly with IpaH9.8 and polyubiquitylated NEMO than ABIN-1-wild-type did (Fig. 5d), suggesting that IpaH9.8 requires ABIN-1 as a ubiquitin-binding adaptor to promote its interaction with and ubiquitylation of NEMO.
To establish the role of IpaH9.8 E3 ligase activity in Shigella infection, we infected mice intranasally with a sublethal dose of wild-type, ΔipaH9.8, ΔipaH9.8/ipaH9.8 (ipaH9.8 complementation strain) or ΔipaH9.8/ipaH9.8CA (ipaH9.8CA complementation strain) bacteria and analysed bacterial load, lung tissue inflammation and levels of myeloperoxidase and cytokines for 48 h. The mean numbers of wild-type and ΔipaH9.8/ipaH9.8 bacteria in the lung tissue were significantly higher than the mean numbers of ΔipaH9.8 and ΔipaH9.8/ipaH9.8CA bacteria (Supplementary Information, Fig. S6a). Although evidence of a strong inflammatory response, such as suppurative masses, neutrophil infiltration and macrophage infiltration, was observed in the lungs infected with all of the bacterial strains, the inflammatory response to wild-type and ΔipaH9.8/ipaH9.8 strains eventually declined to a level similar to that in uninfected control lungs (saline), whereas the inflammatory response to ΔipaH9.8 and ΔipaH9.8/ipaH9.8CA strains persisted for 48 h (Supplementary Information, Fig. S6b). Consistent with these observations, the levels of tissue myeloperoxidase, MIP-2, IL-6 and IL-1β induced by ΔipaH9.8 and ΔipaH9.8/ipaH9.8CA strains were higher than the levels induced by wild-type and ΔipaH9.8/ipaH9.8 strains (Supplementary Information, Fig. S6c, d).
Because numerous host sensors for bacterial components such as the TLR and NLR family, including the downstream signal pathways, are engaged in inducing and controlling the inflammatory response to bacterial infection, it is likely that large numbers of Shigella effectors, including IpaH9.8, are needed to modulate the various inflammatory signal pathways at different levels and times during bacterial infection of various host cells. In this study we showed that Shigella IpaH9.8 targets NEMO and ABIN-1, which results in the induction of Lys 27-mediated polyubiquitylation of NEMO, thereby facilitating the proteasome-dependent degradation of NEMO and interfering with NF-κB activation during bacterial infection (Fig. 5e). Although the reason is still unclear, the extent of NEMO degradation mediated by IpaH9.8 is more profound when Nod1 signalling is stimulated than TNF-α. Indeed, IpaH9.8-mediated inhibition of NF-κB activity is weaker in response to TNF-α than to stimulation by the Nod1 pathway (data not shown). Future studies will need to investigate the preferable NF-κB inhibition by IpaH9.8. The results of this study also demonstrate that the E3 ligase activity of IpaH9.8 contributes to promoting bacterial infection.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturecellbiology/.
Supplementary Material
Acknowledgments
We thank the members of the Sasakawa laboratory for their advice. This work was supported by Grand-in-Aid for Scientific Research (S) (20229006); a Grand-in-Aid for Exploratory Research (20659067); a Grant-in-Aid for Scientific Research on Priority Areas (18073003); the Strategic Cooperation to Control Emerging and Reemerging Infections Funded by The Special Coordination Funds for Promoting Science and Technology; and a Contract Research Fund for the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and the Core Research for Evolutional Science and Technology (CREST) from the Japan Science and Technology Agency (JST).
Footnotes
AUTHOR CONTRIBUTIONS
H.A. designed and performed the experiments. M.K. gave advice and designed the experiments. M.S.-S. provided NEMO materials. A.M. provided ABIN-1 materials. M.O. gave advice. C.S. conceived and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Published online at http://www.nature.com/naturecellbiology/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
Note: Supplementary Information is available on the Nature Cell Biology website.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Martnon F, Mayor A, Tchopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 3.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li QT, Verma LM. NF-κB regulation in the immune system. Nature Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 5.Sebban H, Yamaoka S, Courtois G. Posttranslational modifications of NEMO and its partners in NF-κB signaling. Trends in Cell Biol. 2006;16:569–577. doi: 10.1016/j.tcb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Chen ZJ. Ubiquitin signalling in the NF-κB pathway. Nature Cell Biol. 2005;7:758–766. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fritz JH, Ferrero RL, Philipott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nature Immunol. 2006;7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- 8.Sansonetti PJ, Di Santo JP. Debugging how bacteria manipulate the immune response. Immunity. 2007;26:149–161. doi: 10.1016/j.immuni.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Bhavsar AP, Guttman JA, Finlay BB. Manipulation of host-cell pathways by bacterial pathogens. Nature. 2007;449:827–834. doi: 10.1038/nature06247. [DOI] [PubMed] [Google Scholar]
- 10.Kim DW, et al. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci USA. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arbibe L, et al. An injected bacterial effector targets chromatin access for transcription factor NF-κB to alter transcription of host genes involved in immune responses. Nature Immunol. 2007;8:47–56. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- 12.Okuda J, et al. Shigella effector IpaH9.8 binds to a splicing factor U2AF35 to modulate host immune responses. Biochem Biophys Res Commun. 2005;333:531–539. doi: 10.1016/j.bbrc.2005.05.145. [DOI] [PubMed] [Google Scholar]
- 13.Ogawa M, Handa Y, Ashida H, Suzuki M, Sawakawa C. The versatility of Shigella effectors. Nature Rev Micro. 2008;6:1–7. doi: 10.1038/nrmicro1814. [DOI] [PubMed] [Google Scholar]
- 14.Ashida H, Toyotome T, Nagai T, Sasakawa C. Shigella chromosomal IpaH proteins are secreted via the type III secretion system and act as effectors. Mol Microbiol. 2007;63:680–693. doi: 10.1111/j.1365-2958.2006.05547.x. [DOI] [PubMed] [Google Scholar]
- 15.Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C. Type III secretion effectors of the IpaH family are E3 ubiquitin ligase. Cell Host Microbe. 2007;1:77–83. doi: 10.1016/j.chom.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Singer AU, et al. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nature Struct Mol Biol. 2008;15:1293–1301. doi: 10.1038/nsmb.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, et al. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nature Struct Mol Biol. 2008;15:1302–1308. doi: 10.1038/nsmb.1517. [DOI] [PubMed] [Google Scholar]
- 18.Haraga A, Miller SIA. Salmonella enterica serovar typhimurium translocated leucine-rich repeat effector protein inhibits NF-κB-dependent gene expression. Infect Immun. 2003;71:4052–4058. doi: 10.1128/IAI.71.7.4052-4058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mauro C, et al. ABIN-1 binds to NEMO/IKKγand co-operates with A20 in inhibiting NF-κB. J Biol Chem. 2006;281:18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 20.Heyninck K, et al. The zinc finger protein A20 inhibits TNF-induced NF-κB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-κB-inhibiting protein ABIN. J Cell Biol. 1999;145:1471–1482. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heyninck K, Kreike MM, Beyaert R. Structure-function analysis of the A20-binding inhibitor of NF-kappa B activation, ABIN-1. FEBS Lett. 2003;536:135–140. doi: 10.1016/s0014-5793(03)00041-3. [DOI] [PubMed] [Google Scholar]
- 22.Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou H, et al. Bcl10 activates the NF-κB pathway through ubiquitination of NEMO. Nature. 2004;427:167–171. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
- 24.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 25.Abbott DW, Wilkins A, Asara J, Cantley LC. The Crohn’s disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr Biol. 2004;14:2217–2227. doi: 10.1016/j.cub.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 26.Abbott DW, et al. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol Cell Biol. 2007;27:6012–6025. doi: 10.1128/MCB.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nature Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt-Supprian M, et al. NEMO/IKKγ-deficient mice model incontinemtia pigmenti. Mol Cell. 2000;5:981–992. doi: 10.1016/s1097-2765(00)80263-4. [DOI] [PubMed] [Google Scholar]
- 29.Wagner S, et al. Ubiquitin binding mediates the NF-κB inhibitory potential of ABIN proteins. Oncogene. 2008;21:1–7. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- 30.Ohshima S, et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustain embryonic development. Nature. 2009;457:906–909. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komander D, et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahighi S, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.