Abstract
The analysis of the sequence of ∼150 kb of a genomic region corresponding to the wnt1 gene of the Japanese pufferfish Fugu rubripes confirms the compact structure of the genome. Fifteen genes were found in this region, and 26.6% of the analyzed sequence is coding sequence. With an average intergenic distance of <5 kb, this gene density is comparable to that of Caenorhabditis elegans. The compactness of this region corresponds to the reduction of the overall size of the genome, consistent with the conclusion that the gene number in Fugu and human genomes is approximately the same. Eight of the genes have been mapped in the human genome and all of them are found in the chromosomal band 12q13, indicating a high degree of synteny in both species, Fugu and human. Comparative sequence analysis allows us to identify potential regulatory elements for wnt1 and ARF3, which are common to fish and mammals.
[The sequence data described in this paper have been submitted to GenBank under accession no. AF056116.]
With 400 Mb, the pufferfish Fugu rubripes (Fugu) has one of the smallest genomes of all vertebrates, but the number of genes is similar to that of mammals (Brenner et al. 1993; for review, see Angrist 1998). The size difference is attributable to a marked reduction in intron size and intergenic distances, as well as to the paucity of dispersed repetitive sequences. Sequencing of its compact genome has several uses. The high gene density facilitates gene discovery (Trower et al. 1996) and because there is some conservation of gene order between fish and man (Elgar et al. 1996), it can be used for the analysis of linkage in the more complex mammalian genomes. Information on the Fugu genome, however, is still sparse. Linkage data for genomic fragments >100 kb are available only for the Hox gene clusters (Aparicio et al. 1997). We therefore decided to analyze a larger genomic fragment for its gene content and synteny to human. We have chosen the region around wnt1, because human wnt1 maps to chromosome 12q13, where a large amount of mapping information is available.
In addition, the expression of wnt1 is highly regulated and very well studied in vertebrates (Echelard et al. 1994). Regulatory elements involved in these processes, however, are difficult to identify. An efficient approach is the comparative sequence analysis of noncoding regions with the compact Fugu genome that has been successfully used previously (Marshall et al. 1994; Aparicio et al. 1995; Poepperl et al. 1995). In this study we have extended this approach to the wnt1 region.
The 150 kb of sequence described in this paper contains 15 genes and, overall, 26.6% of the sequence is coding sequence, confirming the high gene density in the Fugu genome. For eight genes, the human counterparts have been mapped, and all are assigned to human chromosomal position 12q13. This supports the idea that the Fugu genome can in some instances serve as a good model system for human in genome research. We also could identify putative regulatory elements for the wnt1 and ARF3 genes. One of these elements has already been used in functional assays (Rowitch et al. 1998), which confirmed the importance of the element for the regulation of wnt1 expression.
RESULTS AND DISCUSSION
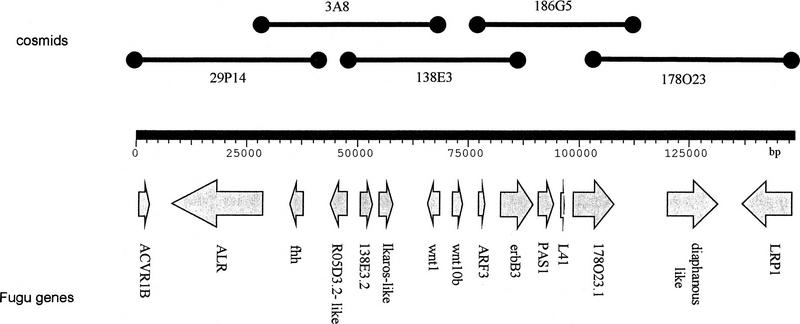
In an initial screen with a specific Fugu probe, we isolated a cosmid containing wnt1. By two steps of cosmid walking from each end of the initially isolated cosmid 138e3, we ended up with five overlapping cosmids (Fig.1), which were then fully sequenced. The entire Fugu genomic sequence of 148640 bp (GenBank accession no. AF056116) has been analyzed with a combination of different methods. All subclones were submitted to “gap-blast” homology searches (Altschul et al. 1997) to reveal any specific homologies with sequences in the databases. Additionally, after aligning all sequences to obtain one contig, the whole sequence was analyzed with the gene prediction program XGRAIL (Xu et al. 1994) to find potential exons. For some genes, a second gene prediction program, FGENES, was used (Solovyev et al. 1995). Exon–intron boundaries were also analyzed by NETGENE (Brunak et al. 1991).
The combination of these approaches led to the discovery of 15 genes within the 148-kb fragment (Fig. 1). These can be divided into three groups. Genes of the first group are highly homologous to their mammalian counterparts (Table 1A), genes of the second group have some homology to known genes and are most likely new members of the same family (Table 1B), whereas genes of group three do not show any striking homology to any known mammalian genes (Table 1C).
Figure 1.
Schematic representation of the genomic organization of the Fugu wnt1 locus and adjacent genes. Cosmids from the Elgar Fugu library that were fully sequenced are shown as dumbbells. Also cosmids 28L23, 34D20, and 133H20 cover this region and were sample sequenced (data not shown). Arrows indicate the direction of transcription of the identified genes. Gene names are shown below.
Table 1.
Identified F. rubripes Genes on a Stretch of 148 kb of Genomic DNA
| A: Genes with vertebrate homologs | ||||
| Fugu and human name | Length of Fugu protein (amino acids) | Percent identity | Accession no. | Assigned to chromosome |
|---|---|---|---|---|
| ACVR1B | 506 | 83 human | P36896 | 12q13 |
| 88 zebrafish | X94119 | |||
| ALR | 4825 | 45 human | 2358285 | 12q13 |
| wnt1 | 370 | 76 human | P04628 | 12q13 |
| 92 zebrafish | P24257 | |||
| wnt10b | 390 | 60 human | 1932789 | 12q13 |
| 60 mouse | P48614 | |||
| ARF3 | 181 | 100 human | P16587 | 12q13 |
| erbB3 | 1328 | 51 human | P21860 | 12q13 |
| PAS1 | 392 | 79 human | 1216526 | 12q13 |
| 79 mouse | P50580 | |||
| L41 | 25 | 100 human | P28751 | N.A. |
| LRP1 | 1581 | 72 human | Q07954 | 12q13 |
| (partial) | 74 chicken | P98157 | ||
| B: New genes of a known gene family | ||||
| Fugu name | Length of Fugu protein (amino acids) | Closest homolog | Accession no. | Percent identity (amino acids) |
|---|---|---|---|---|
| Ikaros-like | 417 | Ikaros/Lyf-1 (human) | 1289371 | 85 (zinc finger domains) |
| 50 (full protein) | ||||
| fugu hedgehog (fhh) | 442 | shh (human) | 663157 | 48 |
| shh (mouse) | A49425 | 52 | ||
| ihh (mouse) | 1890097 | 52 | ||
| C. New vertebrate genes | |||||
| Fugu name | Length of Fugu protein (amino acids) | No. of exons | Significant hits with dbEST entries (acc. no.) | Exons | E-value |
|---|---|---|---|---|---|
| R05D3.2-like | 497 | 15 | AA307511 | 3–8 | 8e-58 |
| AA037301 | 12–15 | 1e-39 | |||
| AA304712 | 11–14 | 1e-27 | |||
| AA307337 | 8–13 | 4e-23 | |||
| 138e3.2 | 255 | 4 | HUMHBC4912 | 2 | 3e-15 |
| AA905621 | 4 | 2e-06 | |||
| AA170127 | 3–4 | 4e-11 | |||
| 178o23.1 | 966 | 23 | T27154 | 3–8 | 6e-54 |
| H82462 | 4–9 | 6e-35 | |||
| AA312185 | 15–17 | 4e-29 | |||
| AA378749 | 9–11 | 6e-18 | |||
| AA773555 | 21–23 | 5e-16 | |||
| H10578 | 13–14 | 7e-13 | |||
| R88176 | 1 | 4e-09 | |||
| diaphanous-like | 1037 | 24 | AA309980 | 17–19 | 4e-48 |
| AA224424 | 19–22 | 3e-46 | |||
| T07474 | 9–12 | 4e-39 | |||
| AA459557 | 5–7, 9–10 | 4e-39 | |||
| U69179 | 17–23 | 9e-32 | |||
| Z41620 | 15–16 | 9e-28 | |||
| AA421752 | 4–5 | 1e-14 | |||
| M78344 | 23–24 | 1e-09 | |||
Homology searches were made with the putative amino acid sequences of the identified Fugu genes using the GAP-BLAST program (Altschul et al. 1997). (A) Fugu genes with highly homologous human counterparts. Genomic location data for human and mouse were obtained from NCBI; http://www.ncbi.nlm.nih.gov. (N.A.) Data not available. (B) New Fugu genes belonging to a known vertebrate gene family. (C) Fugu genes with no or only weak homology to any characterized vertebrate gene. Strong homologies to human entries of the dbEST library can be found for all four Fugu genes.
Group 1: Highly Homologous Genes
The nine genes of the first group code for activin A receptor type IB (ACVR1B), ALL-1 related protein (ALR), wnt1, wnt10b, ADP-ribosylation factor 3 (ARF3), the receptor tyrosine kinase erbB3, proliferation associated protein 1 (PAS1), ribosomal protein L41, and LDL receptor related protein (LRP). All deduced protein sequences show 45% or more identity to their human counterparts. For the majority of the exons of the nine genes, prediction of coding sequences was consistent with both XGRAIL prediction and blast analysis. The first exons of some genes (e.g., the secreted protein wnt10b and part of the intracellular domain of erbB3), however, are not conserved and therefore are difficult to predict from the genomic sequence. In these cases, additional information was obtained using the program NETGENE (Brunak et al. 1991).
Eight of the nine genes have been mapped to a definite chromosomal position in the human genome, and all are assigned to 12q13 (Arheden et al. 1988; Kraus et al. 1989; Forus and Myklebost 1992; Hirai et al. 1996; Lamartine et al. 1997; Prasad et al. 1997; Roijer et al. 1998). In addition, murine wnt1 and wnt10b (previously also called wnt12) are tightly linked within 2.3 cM on murine chromosome 15 (Adamson et al. 1994). The linkage between the two wnt genes may be an evolutionary conserved pattern, because also the Drosophila wnt1 homolog, wingless, and Dwnt4, are closely linked (Graba et al. 1995). Furthermore, human wnt3 and wnt15 are found in the chromosome band 17q21 within a distance of 125 kb (Bergstein et al. 1997). It would be most logical to assume that the linkage of two wnt genes occurred once in evolution, and that in the evolution of the vertebrates this pattern may have duplicated on several chromosomes (Ruddle et al. 1994).
For L41, the precise location has not been determined in human or mouse. The L41 gene encodes a ribosomal protein that is only 25 amino acids long and is interrupted by two introns, with the middle exon only 23 bp in length. Because in other eukaryotes there are several unclustered copies of genes for ribosomal proteins (Woolford et al. 1979; D’Eustachio et al. 1981), some of which are pseudogenes, we cannot state if the Fugu gene is normally expressed.
Conserved synteny between Fugu and human was found previously for the c-fos region (Trower et al. 1996) and the PDGF-receptor region (How et al. 1996). It may therefore be possible in many cases to use the Fugu genome for positional cloning of genes important in human diseases, an efficient process because of the smaller size of introns and intergenic regions in Fugu. The sequence information of the Fugu gene can then be used to isolate the human gene of interest. We do not have sufficient data as yet to define the average length of conserved synteny between Fugu and human. In addition to the genes discussed in this paper, however, Fugu homologs have been isolated for both the HoxC cluster and the genes for adenylate cyclase VI and L-type calcium channel polypeptide, which also map to human chromosome 12q13 or 12q12–q13 (Elgar et al. 1996; Aparicio et al. 1997). Further genome walking experiments could extend the present region of synteny.
Group 2: Fugu Genes with Some Homology to Known Vertebrate Genes
Of the 15 genes defined within the 148 kb contig, 2 show some homology to human genes but are classified as new members of a known gene family. The first gene shows the highest homology to the zinc finger containing transcription factor Lyf-1, encoded by the gene Ikaros (Molnar and Georgopoulos 1994). Conserved sequence is almost exclusively found at the six zinc finger domains (exons 3–5 and end of exon 8 in Fugu, exon 7 in mouse), where 145 of 171 amino acids are identical. The remainder of the sequence is completely different, except for two stretches comprising 40 amino acids with 75% identities between Fugu and human. Human and mouse Ikaros, on the other hand, share a high homology of 95% throughout the whole amino acid sequence (Molnar et al. 1996). Therefore, we believe that this Fugu gene is only a relative of Ikaros and call it Ikaros-like.
Evidence for another human family member of Lyf/Ikaros exists in the dbEST database of expressed sequence tags (Boguski et al. 1993). Like Fugu Ikaros-like, the partially sequenced clone 249118 (GenBank accession nos. H81649 and H81650) shows high homology only to the zinc finger domains of Ikaros. Fugu Ikaros-like and dbEST clone 249118 do not share any homology outside the zinc finger domains either, indicating that the clone 249118 might be a third member of the Lyf/Ikaros gene family.
Interestingly, Ikaros has been mapped previously to human chromosome 7p13–p11 and mouse chromosome 11 (Molnar et al. 1996) close to Egfr, a member of the erbB gene family. It is known that paralogous genes of different families are linked to all four Hox clusters (Ruddle et al. 1994). For instance, HoxC is linked to wnt1 and erbB3, whereas HoxB is linked to wnt3 and erbB2. Genes from additional families, such as aquaporins, integrins, keratins, and collagens are located around the four Hox clusters. Therefore, it is possible that the Lyf/Ikaros family is another example of such families, with Ikaros and Ikaros-like linked to HoxA and HoxC, respectively.
The second gene, fugu hedgehog (fhh), shows, like all hedgehog genes (Zardoya et al. 1996), about 75% identity to human sonic hedgehog in the amino-terminal half of the protein, but less conservation in the carboxy-terminal half. In Fugu, three more hedgehog genes have been isolated (K. Gellner and S. Brenner, unpubl.), one is clearly sonic hedgehog (shh), the others are homologs to desert hedgehog (dhh) and indian hedgehog (ihh). Therefore, it seems that fhh is a fourth member of the hedgehog gene family.
Group 3: New Vertebrate Genes
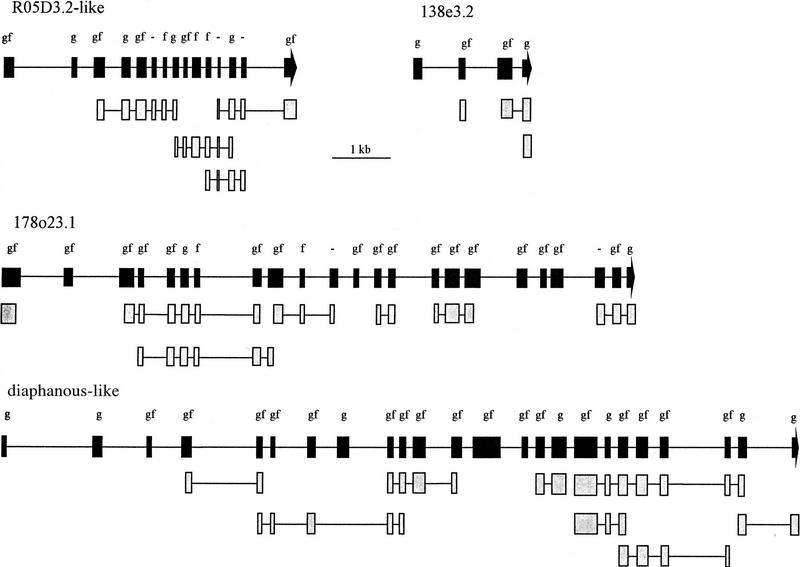
We have defined four genes that have no or only weak homology to characterized mammalian genes (Table 1C). One of these is similar to the Caenorhabditis elegans gene R05D3.2. In the Fugu gene 178o23.1 we could identify sequences similar to the C2 domain, found in protein kinase C and Synaptotagmin. The Fugu gene diaphanous-like, shows similarities in the WW domain to the Drosophila gene diaphanous. All four Fugu genes show strong homology to human cDNA fragments from the dbEST database (Table 1C), indicating that these genes also have as yet uncharacterized human counterparts. Defining the exact coding sequence, however, can be difficult. Comparing the predicted Fugu protein sequences against the predicted protein sequences of all dbEST database entries with TBLAST gave us some additional clues. Fifty-two of the 66 exons of the four new genes are based on homology with dbEST entries (Fig. 2). In many cases, the homology extends over several exons. Of these 52 exons, 42 are confirmed by XGRAI, and 37 by FGENES. The 14 remaining exons are based only on the two gene prediction programs, and nine of these are predicted by both programs.
Figure 2.
Predicted exon–intron structure of the four new vertebrate genes of group 3. Exons are indicated by solid boxes or solid arrows (last exon). Shaded boxes indicate regions with significant homology to entries of the dbEST library. Letters above solid boxes show whether exons have been predicted by XGRAIL (g), FGENES (f), by both (gf), or by neither (−) of these two gene prediction programs.
Xu et al. (1994) have stated that GRAIL II predicts 93% of the exons in a human genomic fragment, with at least one edge correctly matching. By using the human dataset for the XGRAIL analysis of Fugu fragments, we expected a slightly lower accuracy. If we analyze the accuracy of XGRAIL for eight Fugu genes classified in this report to group 1, we find that only 80% of the exons are defined by XGRAIL. The exons that GRAIL failed to predict, but that were found by homology searches, are small exons surrounded by small introns. Only one false-positive exon has been predicted. Therefore, the predicted exons of the four genes of group 3 are most likely correct, but we assume that we may have overlooked some small exons, rather than added unreal ones.
Genomic Organization
The overall genomic structure of the analyzed fragment is very compact (Fig. 1). The predicted 15 genes are separated by intergenic regions <5 kb on average, counting from the end of one coding sequence to the beginning (or end) of the next. Only 45% of the sequence is intergenic sequence. Of the analyzed sequence, 26.6% is predicted to be coding. Eighty-four percent of the introns of the genes classified in group 1 and 2 are <200 bp, the majority are <100 bp. Genes of group 3 have larger introns, but we assume that, because of the lack of homology data, some exons have not been predicted and therefore the size of some introns may appear larger than they really are. The only noticable repetitive areas are some GT or AT dinucleotide repeats, but they never extend >60 bp.
In terms of gene density, the Fugu genome resembles strongly that of C. elegans. In a 2.2-Mb C. elegans genomic fragment of Chromosome III, 29% of the sequence is presumed coding and 52% is intergenic (Wilson et al. 1994). For human it is estimated that only 1%–3% of the genome codes for protein (Jones 1995). If the above predicted numbers were true for the whole Fugu genome, we can calculate that genes are packed eight times tighter in Fugu compared to human and, because of the difference in the genome sizes, we can confirm the original estimate (Brenner et al. 1993) that Fugu and human have about the same number of genes in their genomes. The size difference between the two genomes is only caused by the expansion of intergenic regions and introns in human.
Comparing Noncoding Sequences
The detection of elements involved in gene regulation is the most challenging area of genome research. It is possible that many processes common to fish and man, such as early development, involve common sets of regulatory elements which may show complete conservation. Comparative sequencing of noncoding regions of two species, which are widely separated in evolution, can uncover these. Fugu is very well suited for this approach for three reasons. Fugu has small noncoding regions, putative regulatory elements between different vertebrate species are highly conserved in many cases, and the evolutionary distance between teleost fish and mammals offers maximum divergence for mutational changes in nonfunctional sequences, highlighting any conserved sequences.
We carried out a comparison of noncoding flanking sequences for wnt1 and ARF3, for which vertebrate genomic noncoding sequence information is available. The Fugu genomic sequence was specifically screened for a homology to the murine putative regulatory sequence in the 3′ flanking region of wnt1 (Echelard et al. 1994). This led to the detection of a highly homologous sequence in Fugu (Fig. 3A), ∼200 bp in length and located ∼1800 bp downstream of the stop codon. In mouse, it is as much as 5–6 kb downstream of the stop codon and the orientation is reversed. Transgenic studies with mice showed that 110 bp of that homologous region work as an enhancer element and trigger the full wnt1 expression in the early murine embryo (Rowitch et al. 1998). Furthermore, by DNase I footprinting, Iler et al. (1995) identified some AT-rich elements called HBS1 and HBS2 as binding sites for homeodomain transcription factors that have a role in restricting the expression of wnt1 to the developing forebrain. Identical elements were found in the Fugu sequence within the 200-bp stretch of high homology at the 3′ end of wnt1 (Fig. 3A).
Figure 3.
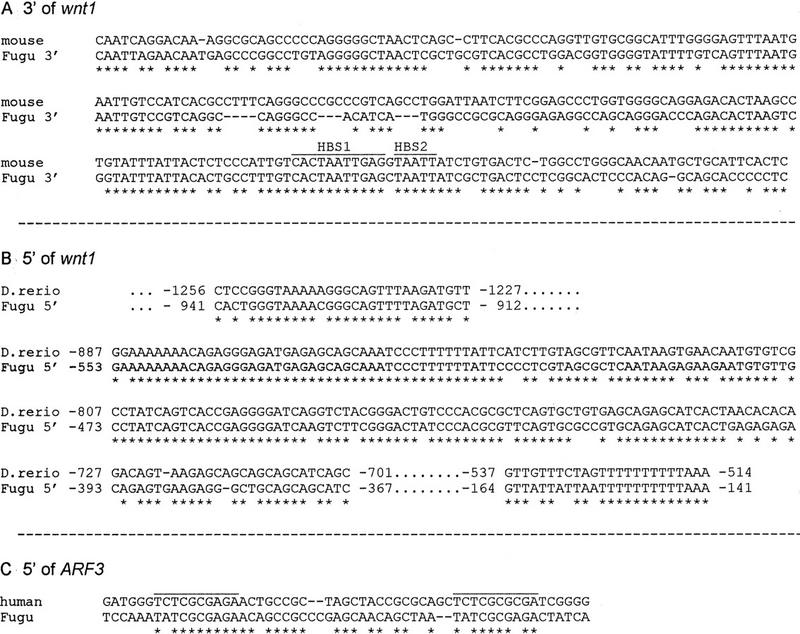
Highly conserved noncoding vertebrate sequences. Stars indicate identical nucleotides. (A) A stretch of ∼240 bp at the 3′ end of wnt1 is 68% homologous between mouse and Fugu. Homeodomain binding sites HBS1 and HBS2 (Iler et al. 1995) are labeled. (B) Extremely high conservation of the 5′ wnt1 noncoding region of Fugu and zebrafish (Danio rerio). The core sequence of 187 bp shows 85% identity in both species. Base pairs are counted from the translation start sites. (C) Region of homology 5′ of human and Fugu ARF3.
Screening of the 5′ noncoding sequence of Fugu wnt1 in a BLAST search revealed another highly conserved region, in this case with the zebrafish 5′ wnt1 region (GenBank accession no. X58880). The conservation covers 800 bp with a core sequence of ∼200 bp, which is 85% identical in both species (Fig. 3B). In zebrafish, several putative TATA boxes were found just downstream of this sequence (Molven et al. 1991). This might indicate that this region serves as the promoter of wnt1. In both species several potential engrailed binding sites can be defined in similar positions 1070, 1560, and 2010 bp upstream of the Fugu and 1420, 1670, and 1900 bp upstream of the zebrafish wnt1 start codon, respectively (data not shown). No homology could be found between the Fugu 5′ wnt1 flanking region and the putative promoter sequence of any other vertebrate wnt1 gene. The murine and human wnt1 promoter share a sequence of 83% identity over a length of 270 bp. This conserved sequence can not be found in the 5′ flanking region of the Fugu wnt1 gene, which could indicate that much of the common mammalian sequence is not important for the regulation of wnt1 expression.
About 1300 bp of human 5′ untranscribed sequence of the ARF3 gene has been published (Haun et al. 1993). To identify homologous regions between Fugu and human 5′ ARF3, both sequences were aligned using the program “bestfit” of the GCG package. Only one short stretch showed significant homology (Fig. 3C), just upstream of the human transcription start site. In both species it consists of the 10-bp nonperfect double repeat T(C/A)TCGCG(C/A)GA. For human ARF3, it has been shown by extensive expression studies with deletion constructs that this specific region is crucial for proper expression (Haun et al. 1993), and this may therefore be another control element uncovered by comparative sequencing analysis.
Our findings extend previous work that found conservation of enhancer elements for Hoxb4 (Aparicio et al. 1995) and Hoxb1 (Marshall et al. 1994, Poepperl et al. 1995), both identified using Fugu genome sequences. The high sequence conservation of these noncoding regions corresponds to a conserved regulatory function. Therefore, it seems that a comparative genomic sequencing approach, using the pufferfish genome is likely to reveal many potential regulatory elements common to all vertebrates. The large evolutionary distance separating the pufferfish and mammals, about 350 million years, ensures that most sequences that can diverge will have diverged and that only sequences of functional importance, such as coding sequences and regulatory elements, will have been preserved.
METHODS
Isolation of Cosmids Containing wnt1
Degenerate PCR primers wnt1f (CAA/GGAA/GTGT/CAAA/GTGT/CCAT/CGG) and wnt1r (ACA/GTGA/GCAA/GCACCAA/GTGA/GAA) (Sidow 1992) were used to amplify and clone several fragments of Fugu wnt exon 4 from Fugu genomic DNA, including wnt1, wnt3a, wnt3b, wnt4, wnt5a, wnt5b, wnt6, wnt7a, wnt7b, wnt8, wnt10a, and wnt10b. The 400-bp wnt1 fragment was used to screen a gridded Fugu genomic cosmid library (Baxendale et al. 1995; Elgar et al. 1995). Four positive cosmids were found and confirmed by Southern hybridization with the Fugu wnt1 fragment. DNA of cosmid 138E3 was isolated and fragmented by sonication. Fragments of an average size of 1000 bp were subcloned into pBluescript SK (Stratagene) and picked into 96-well microtiter plates.
Cosmid Walking
Cosmid DNA was digested with EcoRI or SacI, ethanol-precipitated, and ligated with EcoRI- or SacI-digested pBluescript SK (Stratagene). One microliter of the ligation mix was used in a 50-μl PCR reaction with one pBluescript primer and one lawrist primer in all four combinations, expecting two amplification products, one for each end of the cosmid. The PCR fragments then were used as probes to screen the Fugu cosmid library. Positive clones 29P14, 3A8, 186G5, and 178O23 were processed as cosmid 138E3.
DNA Sequencing
Subclones were amplified by PCR, DNA was precipitated with PEG (Rosenthal and Charnock-Jones 1992), and then sequenced with KS and SK primers (Stratagene Inc.) and ABI dye terminator chemistry on an ABI 373 or 377 automated DNA sequencer. Sequences were assembled with the program SEQMAN from the laser-gene package (DNASTAR, Inc.). Sequencing with custom primers was used for gap closure and for completion of both strands. For each cosmid, on average 550 sequences were aligned to obtain one contig. The absence of coligation events in regions covered by only a single cosmid was confirmed by long PCR on Fugu genomic DNA with custom primers, spanning up to 13 kb (data not shown). PCR was performed with Bio-X-act (Bioline, UK) under the supplier’s suggested conditions. After denaturing for 2 min at 94°C, the following cycle was used 25 times: 10 sec at 94°C, 30 sec at 60°C, 12 min at 68°C.
Sequence Analysis
All subclone sequences were searched with BLAST (Altschul et al. 1997). Prediction of coding sequences was based on the blast results and on the analysis of the entire sequence with the gene prediction programs XGRAIL_1.3c (Xu et al. 1994) and FGENES of the BCM Gene Finder package (Solovyev et al. 1995). The XGRAIL_1.3c client server has been downloaded via ftp from arthur.epm.ornl.gov and installed on a Solaris 2.5 Sun station. The analysis was made with the human sublibrary on version GRAIL 2. Highly conserved noncoding regions were found by blast searches or with the alignment programs “bestfit” of the GCG package and CLUSTALW (Thompson et al. 1994). Linkage data were obtained from the Online Mendelian Inheritance in Man (OMIM) database for human and “mousedb” for mouse, respectively. All programs and databases were accessed through the UK Medical Research Council Human Genome Mapping Project Resource Center (http://www.hgmp.mrc.ac.uk) or the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov).
Acknowledgments
We thank David Rowitch, Paul Danielian, and Andy McMahon for sharing unpublished results, and Greg Elgar for the Fugu cosmids. We thank Judy Huynh for help with sequencing. Part of this work was supported by the Louis Jeantet Research Foundation. The Molecular Sciences Institute was founded by a donation from Philip Morris Companies Inc.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kgellner@pan.zoo.uni-jena.de; FAX 493641 949162.
REFERENCES
- Adamson MC, Dennis C, Delaney S, Christiansen J, Monkley S, Kozak CA, Wainwright B. Isolation and genetic mapping of two novel members of the murine Wnt gene family, Wnt11 and Wnt12, and the mapping of Wnt5a and Wnt7a. Genomics. 1994;24:9–13. doi: 10.1006/geno.1994.1575. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrist M. Less is more: Compact genomes pay dividends. Genome Res. 1998;8:683–685. doi: 10.1101/gr.8.7.683. [DOI] [PubMed] [Google Scholar]
- Aparicio S, Morrison A, Gould A, Gilthorpe J, Chaudhuri C, Rigby P, Krumlauf R, Brenner S. Detecting conserved regulatory elements with the model genome of the Japanese puffer fish, Fugu rubripes. Proc Natl Acad Sci. 1995;92:1684–1688. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, Hawker K, Cottage A, Mikawa Y, Zuo L, Venkatesh B, Chen E, Krumlauf R, Brenner S. Organization of the Fugu rubripes HOX clusters: Evidence for continuing evolution of vertebrate Hox complexes. Nat Genet. 1997;16:79–83. doi: 10.1038/ng0597-79. [DOI] [PubMed] [Google Scholar]
- Arheden K, Mandahl N, Strombeck B, Isaksson M, Mitelman F. Chromosome localization of the human oncogene INT1 to 12q13 by in situ hybridization. Cytogenet Cell Genet. 1988;47:86–87. doi: 10.1159/000132513. [DOI] [PubMed] [Google Scholar]
- Baxendale S, Abdulla S, Elgar G, Buck D, Berks M, Micklem G, Durbin R, Bates G, Brenner S, Beck S, Lehrach H. Comparative sequence analysis of the human and pufferfish Huntington’s disease genes. Nat Genet. 1995;10:67–76. doi: 10.1038/ng0595-67. [DOI] [PubMed] [Google Scholar]
- Bergstein I, Eisenberg LM, Bhalerao J, Jenkins NA, Copeland NG, Osborne MP, Bowcock AM, Brown AM. Isolation of two novel WNT genes, WNT14 and WNT15, one of which (WNT15) is closely linked to WNT3 on human chromosome 17q21. Genomics. 1997;46:450–458. doi: 10.1006/geno.1997.5041. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST—database for “expressed sequence tags”. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Brenner S, Elgar G, Sandford R, Macrae A, Venkatesh B, Aparicio S. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature. 1993;366:265–268. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]
- Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- Bui TD, Rankin J, Smith K, Huguet EL, Ruben S, Strachan T, Harris AL, Lindsay S. A novel human Wnt gene, WNT10B, maps to 12q13 and is expressed in human breast carcinomas. Oncogene. 1997;14:1249–1253. doi: 10.1038/sj.onc.1200936. [DOI] [PubMed] [Google Scholar]
- D’Eustachio PD, Meyuhas O, Ruddle F, Perry RP. Chromosomal distribution of ribosomal protein genes in the mouse. Cell. 1981;24:307–312. doi: 10.1016/0092-8674(81)90320-2. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Vassileva G, McMahon AP. Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS. Development. 1994;120:2213–2224. doi: 10.1242/dev.120.8.2213. [DOI] [PubMed] [Google Scholar]
- Elgar G, Rattray F, Greystrong J, Brenner S. Genomic structure and nucleotide sequence of the p55 gene of the pufferfish Fugu rubripes. Genomics. 1995;27:442–446. doi: 10.1006/geno.1995.1075. [DOI] [PubMed] [Google Scholar]
- Elgar G, Sandford R, Aparicio S, Macrae A, Venkatesh B, Brenner S. Small is beautiful: Comparative genomics with the pufferfish (Fugu rubripes) Trends Genet. 1996;12:145–150. doi: 10.1016/0168-9525(96)10018-4. [DOI] [PubMed] [Google Scholar]
- Forus A, Myklebost O. A physical map of a 1.3-Mb region on the long arm of chromosome 12, spanning GLI and LRP loci. Genomics. 1992;14:117–120. doi: 10.1016/s0888-7543(05)80292-7. [DOI] [PubMed] [Google Scholar]
- Graba Y, Gieseler K, Aragnol D, Laurenti P, Mariol MC, Berenger H, Sagnier T, Pradel J. DWnt-4, a novel Drosophila Wnt gene acts downstream of homeotic complex genes in the visceral mesoderm. Development. 1995;121:209–218. doi: 10.1242/dev.121.1.209. [DOI] [PubMed] [Google Scholar]
- Haun RS, Moss J, Vaughan M. Characterization of the human ADP-ribosylation factor 3 promoter. J Biol Chem. 1993;268:8793–8800. [PubMed] [Google Scholar]
- Hirai M, Kusuda J, Hashimoto K. Assignment of human ADP ribosylation factor (ARF) genes ARF1 and ARF3 to chromosomes 1q42 and 12q13, respectively. Genomics. 1996;34:263–265. doi: 10.1006/geno.1996.0283. [DOI] [PubMed] [Google Scholar]
- How GF, Venkatesh B, Brenner S. Conserved linkage between the pufferfish (Fugu rubripes) and human genes for platelet-derived growth factor receptor and macrophage colony-stimulating factor receptor. Genome Res. 1996;6:1185–1191. doi: 10.1101/gr.6.12.1185. [DOI] [PubMed] [Google Scholar]
- Iler N, Rowitch DH, Echelard Y, McMahon AP, Abate-Shen C. A single homeodomain binding site restricts spatial expression of Wnt-1 in the developing brain. Mech Dev. 1995;53:87–96. doi: 10.1016/0925-4773(95)00427-0. [DOI] [PubMed] [Google Scholar]
- Jones SJM. An update and lesson from whole-genome sequencing projects. Curr Biol. 1995;5:349–353. doi: 10.1016/0959-437x(95)80050-6. [DOI] [PubMed] [Google Scholar]
- Kraus MH, Issing W, Miki T, Popescu NC, Aaronson SA. Isolation and characterization of ERBB3, a third member of the ERBB/epidermal growth factor receptor family: Evidence for overexpression in a subset of human mammary tumors. Proc Natl Acad Sci. 1989;86:9193–9197. doi: 10.1073/pnas.86.23.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamartine J, Seri M, Cinti R, Heitzmann F, Creaven M, Radomski N, Jost E, Lenoir GM, Romeo G, Sylla BS. Molecular cloning and mapping of a human cDNA (PA2G4) that encodes a protein highly homologous to the mouse cell cycle protein p38-2G4. Cytogenet Cell Genet. 1997;78:31–35. doi: 10.1159/000134621. [DOI] [PubMed] [Google Scholar]
- Marshall H, Studer M, Poepperl H, Aparicio S, Kuroiwa A, Brenner S, Krumlauf R. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994;370:567–571. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- Molnar A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol. 1994;14:8292–8303. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Wu P, Largespada DA, Vortkamp A, Scherer S, Copeland NG, Jenkins NA, Bruns G, Georgopoulos K. The Ikaros gene encodes a family of lymphocyte-restricted zinc finger DNA binding proteins, highly conserved in human and mouse. J Immunol. 1996;156:585–592. [PubMed] [Google Scholar]
- Molven A, Njolstad PR, Fjose A. Genomic structure and restricted neural expression of the zebrafish wnt-1 (int-1) gene. EMBO J. 1991;10:799–807. doi: 10.1002/j.1460-2075.1991.tb08012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poepperl H, Bienz M, Studer M, Chan S, Aparicio S, Brenner S, Mann RS, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- Prasad R, Zhadanov AB, Sedkov Y, Bullrich F, Druck T, Rallapalli R, Yano T, Alder H, Croce CM, Huebner K, Mazo A, Canaani E. Structure and expression pattern of human ALR, a novel gene with strong homology to ALL-1 involved in acute leukemia and to Drosophila trithorax. Oncogene. 1997;15:549–560. doi: 10.1038/sj.onc.1201211. [DOI] [PubMed] [Google Scholar]
- Roijer E, Miyazono K, Astrom AK, Geurts van Kessel A, ten Dijke P, Stenman G. Chromosomal localization of three human genes encoding members of the TGF-beta superfamily of type I serine/threonine kinase receptors. Mamm Genome. 1998;9:266–268. doi: 10.1007/s003359900745. [DOI] [PubMed] [Google Scholar]
- Rosenthal AR, Charnock-Jones DS. New protocols for DNA sequencing with dye terminators. DNA Sequence. 1992;3:61–64. doi: 10.3109/10425179209039697. [DOI] [PubMed] [Google Scholar]
- Rowitch DH, Echelard Y, Danielian PS, Gellner K, Brenner S, McMahon AP. Identification of an evolutionary conserved 110 base-pair cis-acting regulatory sequence which governs Wnt-1 expression in the murine neural plate. Development. 1998;125:2735–2746. doi: 10.1242/dev.125.14.2735. [DOI] [PubMed] [Google Scholar]
- Ruddle, F.H., K.L. Bentley, M.T. Murtha, and N. Risch. 1994. Gene loss and gain in the evolution of the vertebrates. Development (Suppl.) 155–161. [PubMed]
- Sidow A. Diversification of the Wnt gene family on the ancestral lineage of vertebrates. Proc Natl Acad Sci. 1992;89:5098–5102. doi: 10.1073/pnas.89.11.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev VV, Salamov AA, Lawrence CB. Identification of human gene structure using linear discriminant functions and dynamic programming. In: Rawling C, Clark D, Altman R, Hunter L, Lengauer T, Wodak S, editors. Proceedings of the third international conference on intelligent systems for molecular biology. Cambridge, UK: AAAI Press; 1995. pp. 367–375. [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trower MK, Orton SM, Purvis IJ, Sanseau P, Riley J, Christodoulou C, Burt D, See CG, Elgar G, Sherrington R, et al. Conservation of synteny between the genome of the pufferfish (Fugu rubripes) and the region on human chromosome 14 (14q24.3) associated with familial Alzheimer disease (AD3 locus) Proc Natl Acad Sci. 1996;93:1366–1369. doi: 10.1073/pnas.93.4.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- Woolford JL, Hereford LM, Rosbash M. Isolation of cloned DNA sequences containing ribosomal protein genes from Saccharomyces cerevisiae. Cell. 1979;18:1247–1259. doi: 10.1016/0092-8674(79)90236-8. [DOI] [PubMed] [Google Scholar]
- Xu Y, Mural R, Shah M, Uberbacher E. Recognizing exons in genomic sequence using GRAIL II. Genet Eng. 1994;16:241–253. [PubMed] [Google Scholar]
- Zardoya R, Abouheif E, Meyer A. Evolution and orthology of hedgehog genes. Trends Genet. 1996;12:496–497. doi: 10.1016/s0168-9525(96)20014-9. [DOI] [PubMed] [Google Scholar]