Abstract
Chromosome 4 from Drosophila melanogaster has several unusual features that distinguish it from the other chromosomes. These include a diffuse appearance in salivary gland polytene chromosomes, an absence of recombination, and the variegated expression of P-element transgenes. As part of a larger project to understand these properties, we are assembling a physical map of this chromosome. Here we report the sequence of two cosmids representing ∼5% of the polytenized region. Both cosmid clones contain numerous repeated DNA sequences, as identified by cross hybridization with labeled genomic DNA, BLAST searches, and dot matrix analysis, which are positioned between and within the transcribed sequences. The repetitive sequences include three copies of the mobile element Hoppel, one copy of the mobile element HB, and 18 DINE repeats. DINE is a novel, short repeated sequence dispersed throughout both cosmid sequences. One cosmid includes the previously described cubitus interruptus (ci) gene and two new genes: that a gene with a predicted amino acid sequence similar to ribosomal protein S3a which is consistent with the Minute(4)101 locus thought to be in the region, and a novel member of the protein family that includes plexin and met–hepatocyte growth factor receptor. The other cosmid contains only the two short 5′-most exons from the zinc-finger-homolog-2 (zfh-2) gene. This is the first extensive sequence analysis of noncoding DNA from chromosome 4. The distribution of the various repeats suggests its organization is similar to the β-heterochromatic regions near the base of the major chromosome arms. Such a pattern may account for the diffuse banding of the polytene chromosome 4 and the variegation of many P-element transgenes on the chromosome.
Chromosome 4 of Drosophila melanogaster is one of the smallest chromosomes of a metazoan. Pulsed-field electrophoresis indicates that its overall length is between 4.5 and 5.2 Mb (Locke and McDermid 1993). Most of the chromosome, ∼3–4 Mb, is transcriptionally inactive, and comprised of simple satellite repeats (Lohe et al. 1993). The remaining ∼1.2Mb of chromosome 4 contains the genes and can be seen in salivary gland polytene chromosomes as the short-banded segment of cytogenetic region 101E–102F extending from the chromocenter. Although this region of chromosome 4 has polytene chromosome bands, it is unlike the typical banded euchromatic regions found on the other chromosomes in various respects.
First, the polytene region 101E–102F displays several features typical of β-heterochromatin. This term was first used by Heitz (1934) to describe the diffuse and poorly banded regions that comprise much of the chromocenter of Drosophila virilis polytene chromosome spreads. In this species, the chromocenter contains a very dark staining mass that he called the α-heterochromatin that is now known to form from the pericentric, satellite-rich DNA. Because this central dark mass is missing from the chromosome spreads of D. melanogaster, the utility of the two terms, α- and β-heterochromatin to describe the molecular basis of heterochromatin in Drosophila has been questioned (Lohe and Hilliker 1995). Clearly there are large blocks of satellite DNA distributed throughout the chromocenter in D. melanogaster that are conveniently referred to as α-heterochromatin even though it is not cytologically identifiable in this species.
The classical view of β-heterochromatin put forward by Heitz (1934) is that it represents the transition between the α-heterochromatin and the euchromatin at the base of the polytene chromosome arms. This transition view has to be re-examined in the light of several studies on D. melanogaster that show that within the extended blocks of pericentric, simple sequence DNA, are interspersed unique or low-copy sequences (Traverse and Pardue 1989; Zhang and Spradling 1995), which could well loop out to form the diffuse chromocenter (Traverse and Pardue 1989). The most satisfactory description of β-heterochromatin is espoused by Gatti and Pimpinelli (1992). They suggest that there are two distinct molecular organizations that lead to cytologically distinguishable β-heterochromatin. “Proximal” β-heterochromatin comprises the low-copy sequences embedded within the satellite arrays in the pericentric regions of each chromosome, whereas “distal” β-heterochromatin is comprised of the diffuse regions that lie at the base of most of the chromosome arms. This latter form is a mosaic of middle and low-copy repetitive DNA, often transposons, interspersed with unique DNA (Miklos et al. 1988; Vaury et al. 1989; Devlin et al. 1990).
Distal β-heterochromatin has been particularly well studied in polytene division 20, located on the X chromosome of D. melanogaster (Miklos et al. 1988). It does not appear well banded in polytene spreads, although it is known to contain 10 genes (Schalet and Lefevre 1976). The banded region of chromosome 4, throughout which ∼80 genes are distributed often shows a diffuse and poorly defined appearance similar to the base of the X chromosome. Chromosome 4 exhibits a further similarity to β-heterochromatin in that the chromosomal protein HP1, thought to be an important constituent of heterochromatin, binds to several sites along the chromosome (Eissenberg et al. 1992).
A further property that chromosome 4 shares with heterochromatin is that P-element transgenes inserted onto chromosome 4 often show variegated expression of the white+ marker gene (Wallrath and Elgin 1995; Wallrath et al. 1996), whereas P elements inserted in the banded regions of the other chromosomes rarely variegate. Nearly half of the variegating P-element transgenes recovered by Wallrath and Elgin (1995) were located on chromosome 4. P-element inserts distributed throughout the banded region of the chromosome show variegated expression (Wallrath et al. 1996), which suggests that a chromatin configuration leading to variegation is present at numerous sites along the whole chromosome.
The above discussion indicates that the banded region of chromosome 4 is different from the banded regions of the other chromosomes. An explanation of these unusual properties may reside in the atypical distribution of repetitive sequences along the chromosome. In situ hybridization to polytene chromosomes showed that long stretches of CA/TG repeats were present throughout the euchromatic regions but absent from the heterochromatic regions as well as from the banded region of chromosome 4 (Pardue et al. 1987). Secondly, Miklos et al. (1988) observed that repetitive DNA sequences normally confined to β-heterochromatic regions on chromosomes X, 2, and 3 were distributed along the banded portion of chromosome 4. These repeated sequences include transposable elements, which are also found at various euchromatic sites, and other repeats, such as the Dr. D element, which localize specifically to β-heterochromatic regions and chromosome 4. This great abundance of repeated sequences on chromosome 4 clearly differentiates it from normal euchromatin elsewhere in the Drosophila genome (Miklos and Cotsell 1990). On chromosomes X, 2, and 3, where most of the DNA is outside the pericentric regions and the polytene banding is normal, the repeat sequences are distributed in the long period interspersion pattern (Manning et al. 1975). Since this early work, a number of chromosomal walks in the euchromatic domain (Carramolino 1982; Bender et al. 1983a,b; Pirrotta et al. 1983) have confirmed the rare interspersion of repetitive DNA stretches (usually several kilobases) within long regions of single-copy sequences (∼35 kb). On chromosome 4, however, the abundance of repeats more closely resembles the short period interspersion pattern (short single-copy DNA interspersed with short repeats) found in the distal β-heterochromatin and throughout the genomes of many animals (Bonner et al. 1973; Davidson et al. 1973; Graham et al. 1974). As repeated DNA sequences are known to affect chromatin structure (Dorer and Henikoff 1994), a short period interspersion pattern of repeats may be responsible for the occasional variegation of transgenes in mammals (Martin and Whitelaw 1996) and the β-heterochromatin-like behavior of chromosome 4 in Drosophila.
The final unusual property of chromosome 4 is an absence of crossover during normal meiosis. Rare crossovers on chromosome 4 have been observed in three special situations: (1) a diplo-4 condition in a triploid female (Sturtevant 1951), (2) in the presence of the meiotic mutations, mei-41 and mei-218 (Sandler and Szauter 1978), and (3) following heat treatment of the oocyte (Grell 1971). Despite these instances, the absence of crossover in most situations has made conventional mapping difficult for the ∼80 genes on this chromosome. As a result, most gene locations have been only roughly determined by use of two deletions, Df(4)M and Df(4)G (Hochman 1976). To obtain a better understanding of how chromosome 4 differs from the other chromosomes at the molecular level, and to position the genes and repeated sequences, we are constructing and analyzing a physical map. As part of this project, we report here an analysis of the sequence of two chromosome 4 cosmid clones determined by a new, directed DNA sequencing strategy (Ahmed and Podemski 1997). Clone cosL7L, centered on the cubitus interruptus (ci) gene, includes part or all of two adjacent genes. The other clone, cos16J20, includes the 5′ end of the zinc-finger-homolog-2 (zfh-2) gene but appears devoid of other coding sequences. Both clones have a complex array of repeated DNA sequences dispersed throughout their length in a pattern that clearly distinguishes chromosome 4 from the euchromatic portions of the other Drosophila chromosomes.
RESULTS
Cosmids from Chromosome 4 Contain Repeated DNA Sequences
To compare the distribution of repeated sequences on chromosome 4 with those elsewhere in the genome, we chose a random set of 15 cosmid clones, not on chromosome 4, from the same library that yielded the chromosome 4 clones below. In situ hybridization of five of these clones to polytene chromosomes was carried out to determine to what extent the set was representative of the genome as a whole. Four clones (3A3, 3A4, 27P2, and 27P3) were located at unique sites within the euchromatin, despite the fact that clones 27P2 and 27P3 contained some repetitive DNA (see below). Clone 3A1 was confined to a very localized region within the chromocenter. These results indicate that the set of random clones is representative of the different types of sequence organization found in the Drosophila genome. Each cosmid was restricted, fractionated on agarose (Fig. 1A), Southern blotted, and probed with labeled Drosophila total genomic DNA (Fig. 1B). The hybridization conditions were such that cosmid fragments containing repeated sequences would hybridize with the probe whereas those with only unique sequence would not. Furthermore, the relative intensity of the signal would reflect the abundance in the genome of the repeats within the fragment.
Figure 1.
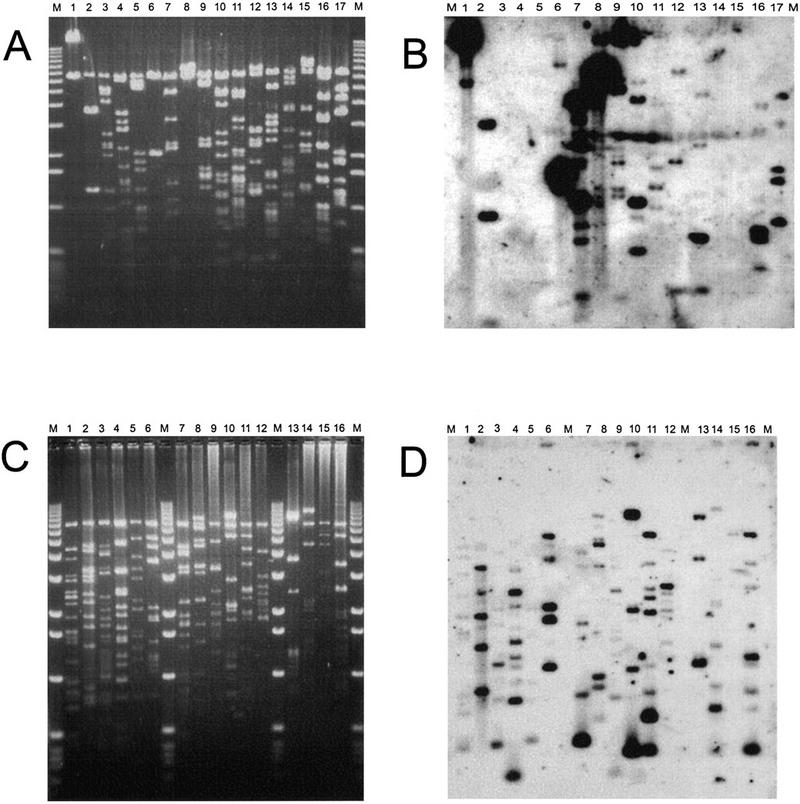
Cosmid clones hybridized with genomic DNA to identify fragments with repeated DNA sequences. Cosmid DNA samples were digested with HindIII and EcoRI (except ci no.1 in which the EcoRI was accidentally omitted) and separated on a 1% agarose gel (A, C). The fragments were Southern transferred to a membrane and probed with genomic DNA labeled by nick translation. The resulting autoradiographs are shown in B and D. DNA from clones 38P3 and 19O3 are in both sets and provide a basis for comparison between the two blots. (M)1-kb size markers (BRL). For randomly chosen cosmids (A, B): (lane 1)3A1; (lane 2)3A2; (lane 3)3A3; (lane 4)3A4; (lane 5)3A5; (lane 6)3A6; (lane 7)3A7; (lane 8)3A8; (lane 9)27P1; (lane 10)27P2; (lane 11)27P3; (lane 12)27P5; (lane 13)27P8; (lane 14)27P9; (lane 15)27P10; (lane 1619O3; (lane 1738P3. For cosmids located on chromosome 4 (C, D):(lane 1zfh2 #1; (lane 214B14; (lane 319F4; (lane 416J20; (lane 558i20; (lane 638P3; (lane 732D22; (lane 819O3; (lane 937L7; (lane 1019H1; (lane 1111H1(lane 1210M7(lane 13 ci #1; (lane 1454A21; (lane 1548F16; (lane 1612A5. Note that ci no.1 is very similar to cosL7L.
The autoradiograph (Fig. 1B) gave a typical pattern of hybridizing fragments with some containing highly repeated, some middle repeated, and others unique sequences. In this set, four clones show very strong hybridization (Fig. 1B, lanes 1,6,7,8), indicating the presence of fragments containing highly repeated sequences. From the insert sizes and paucity of restriction sites in these clones (Fig. 1A) we conclude that they are comprised of simple tandem repeats (satellites). This conclusion is strengthened by our observation that hybridization of one such clone (3A1, lane 1) to polytene chromosomes was confined to a highly localized region within the chromocenter (data not shown), suggesting the clone comprises part of the under-replicated α-heterochromatin. Of the remaining 12 clones, 5 show no hybridization indicating that they are devoid of repeated sequences. In situ analysis of two of these clones, 3A3 (lane 2) and 3A4 (lane 4), revealed unique sites of hybridization to polytene regions 86C and 60E respectively (data not shown). The final seven cosmids had some fragments that contained repeated sequences. Two of these clones, 27P2 and 27P3, localized to unique chromosomal locations (97C and 63F, respectively), which implies the repetitive sequences were too short to yield a hybridization signal. In total, only ∼30% of the HindIII–EcoRI fragments from these 15 random clones hybridized and thus contain repeated sequences.
As part of our chromosome 4 genome mapping project, we have used a variety of techniques to collect cosmids that are distributed along chromosome 4. These cosmids have been assembled into a small set of unambiguous contigs on which all of the known genes mapped to chromosome 4 have been located (Locke et al., in prep.). We have chosen for analysis in this work a subset of 16, which represents a total of 450–500 kb, or almost half of the polytene-banded region of chromosome 4. All of these cosmids are located within the banded region (data not shown), therefore, we feel that the set provides a diverse representation along the polytenized region of the chromosome. A stained agarose gel on which the digestion products of these cosmids were fractionated is shown in Figure 1C. When this gel was Southern blotted and probed with labeled Drosophila total genomic DNA, the resulting autoradiogram showed that all the clones contained repeated DNA sequences (Fig. 1D). Furthermore, about half of the HindIII–EcoRI fragments hybridized and these repeats appear relatively evenly distributed among the clones. From the intensity of the hybridization signal, we infer that these repeats are not highly repeated, as was seen in the four random cosmids described above.
From this comparison, we conclude that on balance, the clones from the polytenized region of chromosome 4 have a higher average frequency of repeated sequences and that repeated sequences are more evenly distributed along the chromosome than is found in typical Drosophila DNA. Furthermore, clones without repeats and clones with highly repeated tandem arrays are absent from our chromosome 4 set of cosmids.
With the distribution of repeated sequences in mind, we went on to explore two of these cosmid clones in more detail; cos16J20 contains the 5′ end of the zfh-2 gene and cosL7L contains the ci gene. These two clones were chosen because (1) they represented proximal (ci) and distal (zfh-2) sites on the chromosome; (2) both had numerous subfragments containing repeated DNA (Fig. 1D; lanes 4,13); and (3) they contained appropriate restriction sites for a novel, rapid DNA sequencing technique (Ahmed and Podemski 1997) that would allow an examination of the gene and repeat organization at the DNA sequence level.
Cloning and Sequencing cosL7L and cos16J20
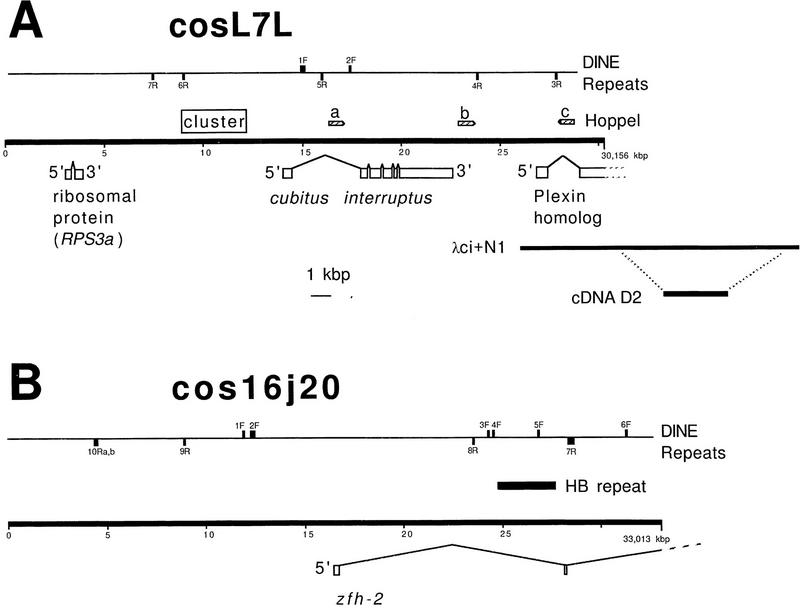
Previous work showed that the ci gene and adjacent sequences were contained on a single ∼30-kb BamHI fragment (Locke et al. 1996). This BamHI fragment was cloned into the cosmid vector pAL-L to give cosL7L. The sequence of the complete BamHI fragment was determined by use of a new method that reduces the number of templates from the normal 700–900 that would have been required in shot-gun methods to <300 (Ahmed and Podemski 1997). Each template’s sequence was determined on an automated DNA sequencer and the few regions not included in these original templates were completed by use of oligomers to direct sequence from known ends (GenBank accession no.U66884). The restriction map predicted from the 30,156-bp sequence is consistent with that determined from the cosmid DNA, λ clones (Locke et al. 1996), and Southern blots of genomic DNA. The only discordance observed is the absence in the sequence of an NsiI site at ∼16,600, which appears in the DNA of most stocks on genomic Southern analysis. This probably represents a polymorphism in the population of Oregon-R used to make the library. It is possible to create an appropriately placed NsiI site by a single-base substitution (16579 T → A or 16704 T → G). In addition to ci, this cosmid contains a complete ribosomal protein gene and the 5′ end of a plexin homolog (Fig. 2A).
Figure 2.
Diagram of the sequenced cosmids cosL7L (A) and cos16J20 (B). cosL7L contains the ci , plexin–homolog, and the ribosomal protein genes, whereas cos16J20 contains the 5′ end of the zfh-2 gene. The λ clone λci+N1 and the cDNA clone D2 are also shown relative to the cosL7L clone. Shown on each cosmid are the positions for the Hoppel and DINE repeats and the HB repeat described in the text. The DINE repeats are numbered and are shown above the line (F) for one orientation or below the line (R) for the other. The location of the clustered repeat is shown upstream from the ci transcript. Physical mapping shows that ci is transcribed in a distal to proximal direction along the chromosome, whereas the orientation of cos16J20 on the chromosome is such that zfh-2 is transcribed in a proximal to distal direction (data not shown).
The 33, 013-bp sequence of clone cos16J20 was determined by the improvement of the selected template system used for cosL7L described in Ahmed and Podemski (1997), (GenBank accession no.U87286). Within cos16J20, we were only able to identify two 5′exons of the zfh-2 gene (Fig. 2B) and the remainder of this gene is located in an adjacent overlapping cosmid. No other ORFs could be reasonably assembled into a putative gene.
Sequence Analysis of the Hoppel Repeats
cosL7L contains three copies of a mobile element, called Hoppel (Kurenova et al. 1990) as shown in Figure 2A. Two Hoppel elements, labeled a and b, are oriented in the same direction (with respect to each other) near the ci transcript with one of the elements present within the first intron of ci and the other ∼240 bp downstream from its 3′ end (Fig. 2A). The third Hoppel element, labeled c, is in the opposite orientation from the other two, and lies within the first intron of the plexin-homolog gene (see below). The cos16J20 clone lacks Hoppel elements but has one other previously described mobile element, an HB repeat, positioned within the first intron of the zfh-2 gene (Fig. 2B).
A sequence comparison of these three Hoppel elements with three previously described sequences shows many small deletions and insertions (Fig. 3). Hoppel-a is an almost complete element, being the most similar of these three to the previously published Hoppel4579 element. One region of dissimilarity is found at the right end and includes the inverted terminal repeat, whereas another dissimilarity is found in the central region in which the spacing for Hoppel-a and Hoppel4579 is similar despite a difference in sequence. Hoppel-b has lost ∼200 bp in the central region, in which Hoppel-a and Hoppel4579 differ. Hoppel-b retains both inverted terminal repeats and has a target site duplication of 7 bp. Hoppel-c has lost ∼100 bp near the central region, ∼150 bp near the right end (as they are shown in Fig. 3), and both terminal repeats.
Figure 3.

A DNA sequence comparison of three Hoppel elements (a, b, and c) within cosL7L with the previously described Hoppel sequences made with ClustalW (Thompson et al. 1994) on the default settings. The similarity in sequence is shown in uppercase letters, whereas differences from the most common base are shown as lowercase letters. The location of the inverted repeats is shown by the broken arrow above the sequences; while the location of the short tandem repeats are indicated by an underlined sequence. (Broken lines) Indicate gaps to facilitate alignment. The nucleotide count is given at right. The abbreviation hop4579 is Hoppel4579 (accession no. X78388), hop1360 is Hoppel1360 (accession nos. M55078 and M38659), hop-ste is a composite assembly of sequences from two adjacent elements (accession no. Z11734) in which the sequences 985-1 are joined to 2837–2660 at a common MluI site. The hopL7La, hopL7Lb, and hopL7Lc sequences are from cosmid L7L.
From this alignment and dot matrix comparisons (not shown), it is clear that the central region of Hoppel elements are quite variable. The regions near and at the ends, especially the first 450–600 bp, which are present in all six elements, show greatest similarity. The inverted terminal repeats, however, are frequently lost. Of the six Hoppel element sequences (assuming the partial sequences of Hoppel174 and Hoppel178 from Kurenova et al. 1990 are combined into one element), only four have retained both inverted repeats, one has one repeat, and one has neither.
A Novel Short Repeat (DINE) Is Interspersed throughout Both Clones
A series of short (∼100–300 bp) repeated sequences belonging to a family we have called DINE (Drosophila interspersed element) is also present in the cosmids (Fig. 2). These repeats are present in both orientations, F and R (forward and reverse), in both clones. Each appears to be a partial sequence from some larger, as yet unidentified, repeated element. The element 1F in cosL7L (Fig. 2A) appears to retain the most sequence and with the 1F sequence as the archetype, we found 17 other repeats, or partial repeats, within cosL7L and cos16J20 (Fig. 2). Although there are many deletions and insertions apparent, these sequences can be aligned as shown in Figure 4. Many of the apparent insertions are, in fact, duplications of the adjacent sequence. For example, the large insert in 1F is a duplication of its rightmost sequences (in Fig. 4 the comparison was made to the rightmost repeat to facilitate the alignment of the other more terminal elements).
Figure 4.
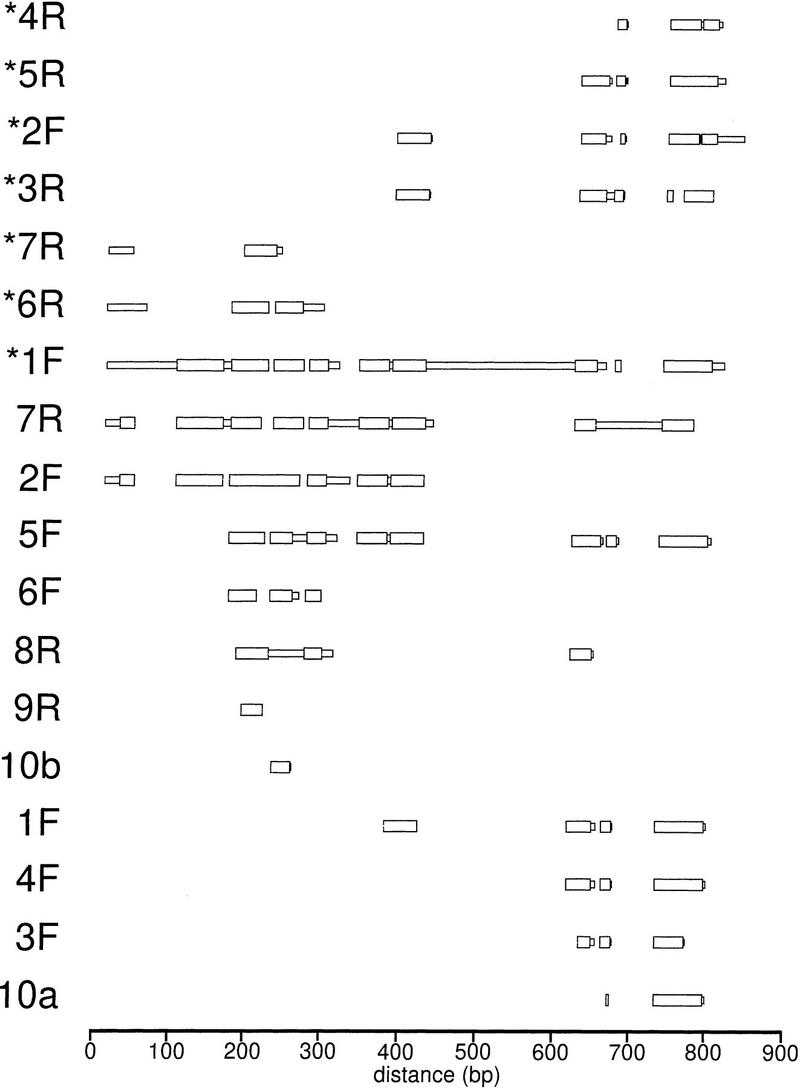
A diagram showing the alignment of blocks of similar sequence of the 18 DINE repeats from clones cosL7L and cos16J20 as shown in Fig. 2. Thick regions indicate blocks of sequence similar to the archetype 1F element; while thin regions indicate regions of sequence without similarity identified by the MACAW program.
The small size and scattered distribution of these repeats suggested they might be similar to mammalian short interspersed elements (SINEs). Among our DINE elements, only three contain the left portion (as shown in Fig. 4) of the repeat, whereas 11 contain the right; a distribution consistent with mammalian SINEs. No long ORFs could be found among these elements. The frequency of this repeat on the two chromosome 4 clones is 18 repeats in ∼63 kb or one every 3.5 kb. Despite these three similarities with SINEs, no RNA polymerase III promoters have been identified, and the run of adenosines, which is characteristic of the 3′ end of a SINE, is not found at either end. The termini of the DINE elements differ among the repeats and no short, direct repeats were observed at the junction between element and adjacent genomic DNA.
BLAST searches identified a few other sequences with similarities to the 1F repeat in the databases of the D. melanogaster and other Drosophilid genomes (data not shown). Mostly, the DINE-like repeats were isolated copies and nowhere in the current database (which reflects predominately the euchromatic domain) was an interspersed pattern of DINEs found like that for the two chromosome 4 cosmids. The closest exception was one sequence, located near su(f) at the base of the X chromosome, with two repeats ∼9 kb apart. In situ hybridization of the 1F DINE to polytene chromosomes revealed its distribution throughout the banded region of chromosome 4, at the base of the major chromosome arms, at several telomeres and within the chromocenter (Locke et al., in prep.). This pattern of hybridization indicates that DINE elements are present in the distal β-heterochromatin and suggests a similarity between chromosome 4 and this chromatin domain. Hybridization to the chromocenter could indicate the presence of DINE elements in the proximal β-heterochromatin as well, but only very limited sequence of this region is available (Zhang and Spradling 1995) and a search of this DNA showed that DINE sequences were not present.
Noncoding Regions of Chromosome 4 Have an Abundance of 13/15 Repeats
The sequence of cosL7L was examined by dot matrix analysis by use of different conditions that varied the window lengths and identity requirements for matches. Selection of long window lengths (>30 bp) showed essentially a random distribution of sequence matches as might be expected to occur in any stretch of DNA of this length. However, when shorter lengths were tested, we observed a nonrandom distribution of sequence matches. For example, Figure 5 shows the pattern of matches when 13 bp of a 15-bp window needs to be identical (13/15). When this pattern is compared with the position of coding regions of the ci gene, it appears that the noncoding DNA contains a much higher repeat density than the ci coding region. The difference between coding and noncoding regions also appears true for the other two genes, although their smaller size makes it more difficult to resolve. An examination of cos16J20 also showed an abundance of 13/15 matches, similar to the noncoding regions of cosL7L, all along the sequence. However, when cosmid-sized sequences of typical euchromatic regions from the other five chromosome arms were examined under these conditions (see Materials and Methods), only a low frequency of 13/15 matches, similar to that of the ci coding region, was found throughout the sequences. Because the noncoding regions in chromosome 4 clones contain an abundance of 13/15 repeats, whereas sequences from other chromosomes do not, it appears this bias is a general property of the sequence organization of chromosome 4.
Figure 5.
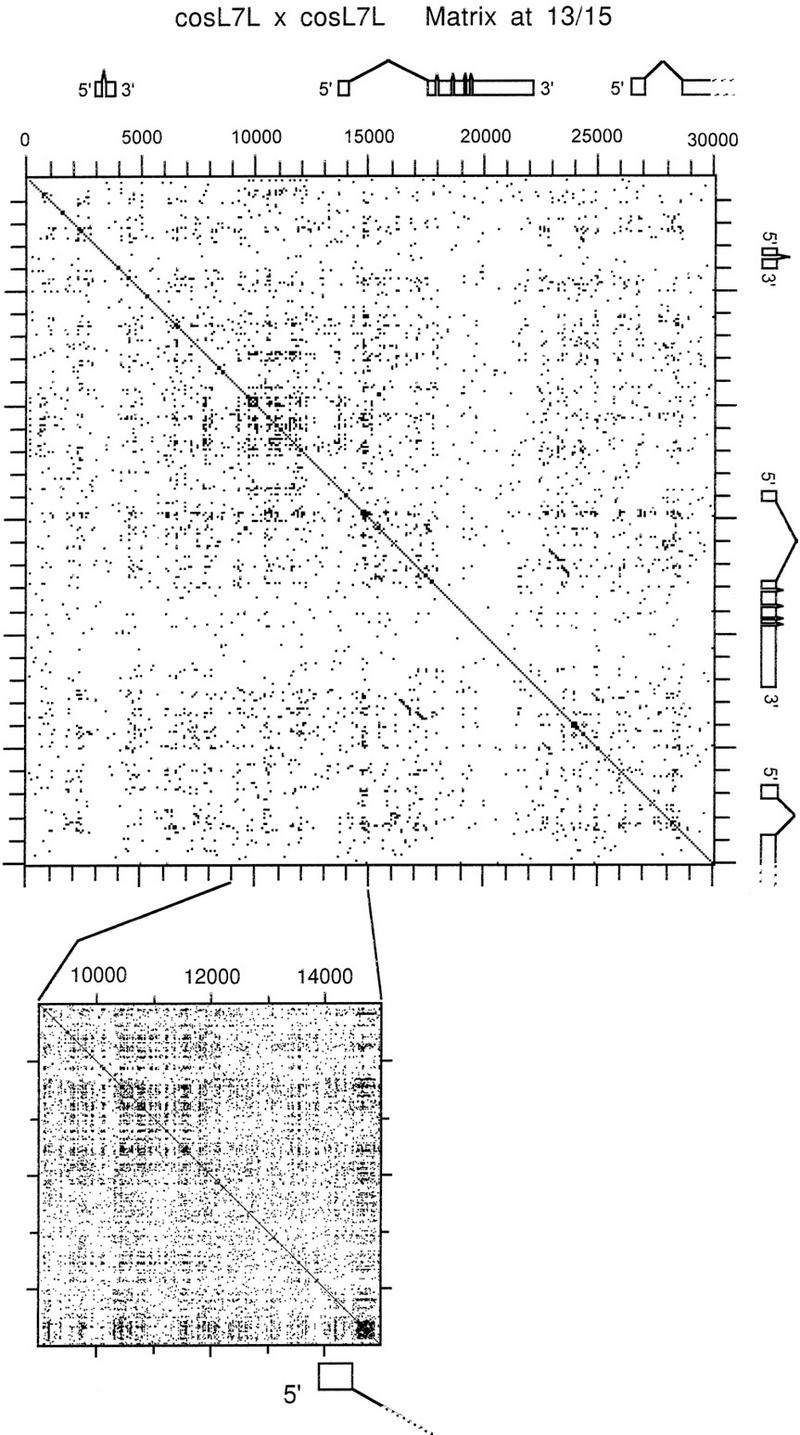
A dot matrix comparison of cosmid L7L with itself showing the generally high frequency of 13/15 repeats as well as the clustered repeat upstream from the ci transcript. The whole cosmid comparison is shown at top; while an enlarged matrix of the cluster is shown at bottom in expanded view. The transcription unit positions, as described in Fig. 1, are also shown so as to permit comparison of repeat frequency in coding and noncoding regions.
A Cluster of Repeats Upstream from the ci Gene
Over and above the generally increased frequency of short similar repeats in noncoding sequences, is an exceptionally dense region of repeats upstream from the ci gene. This cluster, located between position ∼9000 and ∼12,000 on cosL7L, appears as a box shape in the dot matrix comparison (Fig. 5), and overlaps the distal regulatory region of the ci gene defined by Schwartz et al. (1995) as shown in Figure 2A. A more detailed analysis of this region indicates a repeating substructure. There is a periodicity to the repeats in that a similar, nonidentical sequence is present approximately every 200 bp over the 3 kb of the cluster. It was not possible to derive a consensus sequence for the repeat because the repeating units are very degenerate, but they do appear to be AT- rich. Thus, we were able to detect a periodic fluctuation in the percentage of GC throughout this region of cosL7L. Although most of the divergent tandem repeats appear clustered, the dot matrix analysis also indicates the presence of similar sequences elsewhere in cosL7L.
cosL7L Has Three Genes
The ci Gene
From our complete genomic sequence of cosL7L, we confirmed the limited upstream sequences of ci determined by Schwartz et al. (1995) and noted some differences with a previously published cDNA sequence (Orenic et al. 1990). Table 1 shows the differences found within the putative transcription unit and relates the consequence of the DNA change to that of the putative amino acid sequence of the CI protein. Of the 13 alterations (involving 20 nucleotides), only 9 alter the putative amino acid sequence. The most significant change (at position 8264) involves a shift in the position of the termination codon such that an additional 19 amino acids are added to the polypeptide’s carboxyl terminus. The new DNA and protein sequences have GenBank accession number U66884.
Table 1.
Adjustments to the DNA and Putative Protein Sequence of the cubitus interruptus gene
| DNA base position(s)a | DNA alterationb | Alteration to the putative amino acid sequence |
|---|---|---|
| 4594 | T → C | |
| 4906 | G → A | |
| 5115 | T → C | |
| 5222 | C → A | Thr → Lys |
| 5385 | G → C | Arg → Ser |
| 6540 | TATAT → ATATA | Ile Ser → Tyr Thr |
| 7033 | T → A | Ser → Thr |
| 7176 | T → C | |
| 7258 | CT → TC | Leu → Ser |
| 7585 | insert AAG | add Lys |
| 7594 | G → T | Val → Leu |
| 8264 | TATA → ATAT | Val Amb → Asp Met + 19 |
| more amino acids | ||
| 8346 | T → A | in the UTR |
Base number 1 is the A in the AGT transciption start site proposed by Schwartz et al. (1995).
Changes are from those from Orenic et al. (1990) to those in this report.
A Ribosomal Protein S3a Gene
A BLASTN DNA sequence search of the whole cosmid cosL7L identified sequences with similarity to several ribosomal protein S3a genes. An ORF, interrupted by one intron, that encodes a putative protein of 168 amino acids, showed similarity to the ribosomal protein S3a of several species of which four are shown in Figure 6. The alignments suggest that this gene encodes Drosophila ribosomal protein S3a. Additional comparison of this genomic sequence with that from a previously reported cDNA clone encoding a putative RPS3a protein from D. melanogaster (accession no. 1752661) shows several amino acid differences but is not sufficient to indicate that more than one gene is present in the Drosophila genome. The cDNA sequence confirms our positioning of an intron based on the consensus splice site sequences (Padgett et al. 1986) and from the reading frame change in the genomic DNA sequence. Our sequence is consistent with that of van Beest et al. (1998), with the exception of 11-bp differences at seven sites in AT-rich stretches located outside the protein coding region and one silent GC → CG transition within the coding region. No consensus poly(A) signal (AATAAA) is present 3′ to the TAA stop but the sequence ATTAAA is present immediately after the stop codon and likely serves as the signal.
Figure 6.
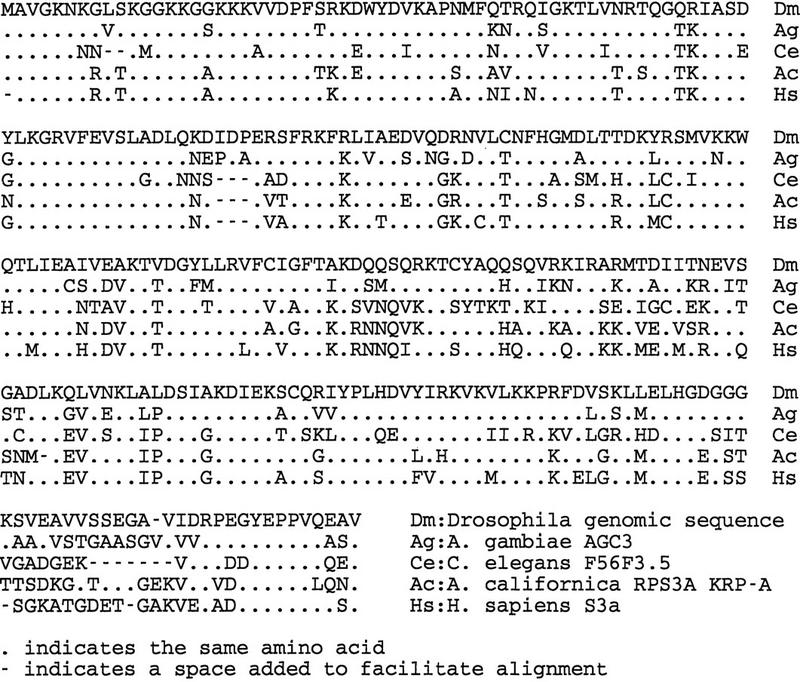
Alignment to compare the deduced amino acid sequence of the Drosophila ribosomal protein gene homolog with the sequences derived from the RPS3a genes from Anopheles gambiae (Ag; accession no. X98186), C. elegans (Ce; accession no. Z32681), Aplysia californica (Ac; accession no. X68555) (Auclair et al. 1994), and Homo sapiens (Hs; Metspalu et al. 1992). Dots indicate the same amino acids as those found in Drosophila; Dashes indicate spaces added to facilitate alignment.
Previous associations between ribosomal protein genes and Minute loci led us to suspect that this gene encodes the Minute(4)101 locus located near ci on chromosome 4. Using PCR primers, which span the RPS3a gene’s coding region, we examined the three pre-existing M(4)101 mutations for the presence and integrity of this gene. We confirmed the presence of suitable template DNA in each sample by obtaining a product using a primer pair from the vestigial locus.
Despite Farnsworth (1951) indicating M(4)101 mutations are embryonic lethals, Hochman (1973) found that among the progeny of M/+ heterozygote parents, M(4)10157g and Df(4)M(4)101-63a homozygotes died, not as embryos but as larvae. Therefore, to examine M(4)101 homozygotes, we collected eggs from M/+ parents and assayed them with the expectation that, on average, 25% would be of the genotype M/M. For both M(4)10157g and Df(4)M(4)101-63a progeny, no alteration of the PCR product was observed in >14 randomly chosen embryos indicating the RPS3a coding sequence is intact in both alleles. Interestingly, van Beest et al. (1998) located a Doc retroposon within the promoter region of the M(4)10157g allele (∼100 bp upstream from our primer) indicating that it is responsible for this Minute allele.
No RPS3a gene PCR product could be amplified from homozygotes of Df(4)M(4)101-62f, showing this region, or at least one primer site, is absent. Such homozygotes could be identified unambiguously and recovered as dead embryos from crosses of heterozygous parents. They die as embryos because of the absence of other essential genes within the deletion that are embryonic lethals (e.g., l(4)13 and l(4)17).
A New Drosophila Plexin Homolog
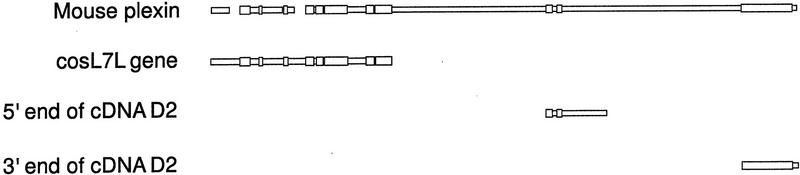
Downstream from the ci gene and proximal to it on the chromosome, we have identified sequences similar to the 5′ end of a family of genes encoding neuronal cell surface proteins called plexins. Because the sequences in cosL7L contained only the 5′ end for this gene, we used a fragment from an adjacent λ clone, called λci+N1 (Locke and Tartof 1994; Locke et al. 1996), which overlaps cosL7L and extends downstream (Fig. 2A) to generate a probe with which we screened a cDNA library derived from third instar imaginal disc mRNA. This probe identified a clone, called D2 (Fig. 2), that contained an ∼3.0-kb insert and shows that this putative gene is expressed. The first 500–600 bases of both ends of this cDNA clone were determined. When compared with the full-length mouse plexin protein sequence, both ends of D2 showed nucleotide and conceptual amino acid sequence similarity to the plexin family of related genes (Fig. 7). The similarities of D2 are in the center and at the 3′ end of the plexin, whereas the genomic sequence from cosL7L shows nucleotide and conceptual amino acid sequence similarity at the 5′ end. Although D2 hybridizes with other fragments from the λci+N1 genomic clone, it does not extend 5′ far enough to hybridize to any cosL7L sequences. When similar comparisons of the Drosophila amino acid sequences were made with the plexins from Xenopus and Caenorhabditis elegans, similarities corresponding to the 5′, central, and 3′ regions were seen. From this observation, we conclude that D2 is a partial cDNA clone containing only the central and 3′ end portion of the full transcript that begins within the cosL7L clone.
Figure 7.
A diagram showing the alignment of blocks of similar deduced amino acid sequence between the mouse plexin (Kameyama et al. 1996) and the sequences from clones cosL7L and the 5′ and 3′ ends of cDNA clone D2 (as shown in Fig. 1). Thick regions indicate blocks of similar sequence aligned among the sequences; whereas thin regions indicate regions without similarity.
DISCUSSION
Repeated DNA Sequences on Chromosome 4
Various repeated DNA sequences have been localized to chromosome 4 by in situ hybridization (Miklos et al. 1988). Although this study provided a rough positioning of repeated sequences along the chromosome, we still know little about their distribution at the molecular level and their locations relative to genes on chromosome 4.
This is the first examination of the DNA sequence structure of chromosome 4 at the nucleotide level. The cosmids, cosL7L and cos16J20, come from the proximal (101EF) and middle positions (102C), respectively, of the polytene-banded region of chromosome 4. Their sequences total ∼63 kb, >5% of the polytenized region, and should be sufficient to offer a generalized insight into the DNA sequence structure of this unusual chromosome.
We can now see that chromosome 4 has an abundance of repeated sequences at several levels of DNA organization that makes it distinctly different from the euchromatic regions of other chromosomes. In most of the Drosophila genome, the distribution of repeated sequences is, for the most part, of the long-period pattern (also called the Drosophila pattern), in which relatively long repeat sequences of several kilobases are interposed between even longer stretches (∼35 kb) of unique DNA (Manning et al. 1975; Carramolino 1982; Bender et al. 1983a,b; Pirrotta et al. 1983). This generalized long-period pattern of the Drosophila genome is not applicable to chromosome 4. Our hybridization results in Figure 1, the distribution of repeated DNA elements represented in Figure 2, and the greater frequency of short repeats revealed in the dot matrix analysis (Fig. 5), together, show that the pattern of repeats on chromosome 4 is reminiscent of the short-period interspersion (also called the Xenopus pattern) found in many vertebrates, including humans. The short-period pattern is characterized by short repeats of several hundred basepairs interposed between short stretches (several kilobases) of unique DNA (Davidson et al. 1973). On chromosome 4, the short repeats could be the DINE elements that are similar in size and distribution to the SINE elements of mammals (e.g. Alu elements). A similar mosaic of repeats and unique DNA is found in the distal β-heterochromatin at the base of the major chromosome arms (Miklos et al. 1988; Vaury et al. 1989; Devlin et al. 1990). BLAST searches of the database reveals that two copies of the DINE element are located in close proximity to su(f), a gene that is embedded within the β-heterochromatin at the base of the X chromosome (Schalet and Lefevre 1976). The prevalence of the DINE element throughout the distal β-heterochromatin has been confirmed by an in situ hybridization to polytene chromosomes (Locke et al., in prep.). This similarity in the organization of repeated sequences suggests that chromosome 4 chromatin closely resembles that of the heterochromatic regions at the base of the major chromosome arms.
To what extent is the short period interspersion pattern of chromosome 4 responsible for its unusual properties? Recently, it has been shown that the absence of recombination on chromosome 4 is due to the proximity of its genes to the centromere and not to the sequence organization per se (Osborne 1998). However, the diffuse, heterochromatic-like appearance of the banded part of the chromosome in polytene spreads could well be caused by the high density of repeats, as it has been suggested that secondary structures between repetitive sequences could act as nucleation sites for the assembly of heterochromatin-specific proteins (Dorer and Henikoff 1994). Finally, the variegation of many P-element borne transgenes on chromosome 4 could also stem from its DNA sequence organization. In particular, the proximity to multiple DINE elements might cause transgene variegation because the genomic distribution of the DINE (see Results) corresponds exactly with regions in which P elements variegate.
Our DNA sequence analysis shows that chromosome 4 genes are often interrupted by repeated sequences, some of which are transposable elements. The insertion of a transposable element within or near gene transcription units normally would be expected to have a substantial deleterious effect on the expression of those genes. The insertion of mobile elements into introns or protein coding sequences is the basis for many of the spontaneous mutations found in laboratory and wild populations of Drosophila (ten Have et al. 1995). However, it seems that such insertions are tolerated and may even be the normal situation on chromosome 4 as both the ci and plexin-homolog genes have Hoppel elements within their first intron and the zfh-2 gene has an HB repeat within its first intron. Although the presence of the Hoppel or HB element within these transcription units appears not to be detrimental, not all such transposable inserts on chromosome 4 are benign. Several spontaneous mutant alleles of ci are due to the insertion of transposable elements (Locke and Tartof 1994) and M(4)101, a mutation in the gene encoding dRPS3a, is apparently caused by the insertion of the Doc transposon (van Beest et al. 1998). What makes some inserts innocuous and others detrimental is not clear.
Gene Identification by DNA Sequence Analysis
M(4)101 Locus
Over 50 Minute loci are dispersed throughout the Drosophila genome. The Minute locus M(3)99D was the first to be shown to encode a ribosomal protein, rp49 (Kongsuwan et al. 1985). Of the remaining loci, another eight have been shown to be genes encoding ribosomal proteins and the others are tightly linked to such genes (see Saebøe-Larssen et al. 1997, 1998). Our data and a recent study by van Beest et al. (1998) indicate a clear similarity in predicted protein sequence (Fig. 6) between the Drosophila RPS3a protein and other S3a ribosomal protein genes. Furthermore, Reynaud et al. (1997) have shown that dRPS3a is a ubiqitous protein, localized in the cytoplasm and associated with the 40S ribosome subunit. It is encoded by the gene dRps3a, located on cosL7L, and van Beest et al. (1998) have shown that M(4)101 is an allele. We are puzzled by our PCR results that show that dRps3a is intact in Df(4)M63a, a deletion that reportedly removes the M(4)101 locus (Hochman 1976). The presence of a PCR product can be explained if Df(4)M63a has sustained a second site point mutation in the M(4)101 locus in addition to the visible deletion. Alternatively, Df(4)M63a could be due to a loss of regulatory elements located outside of the PCR primers sites or to a position effect as described for the adjacent ci locus (Locke and Tartof 1994).
Plexin-Homolog Gene
The plexin proteins are neuronal cell surface molecules that have extracellular regions that resemble the cysteine-rich domains of the c-met proto-oncogene protein product (Ohta et al. 1995). The c-Met family of genes (MET/RON/SEA) encodes transmembrane proteins that have a similar extracellular domain that binds ligands and signals intracellularly via a large cytoplasmic domain. The sequence similarity between our Drosophila sequences, both genomic and cDNA, and the reported vertebrate plexin genes indicates that this gene is most likely the Drosophila plexin homolog.
These two cosmid sequences have provided an initial look into the organization of the DNA on chromosome 4. The gene density on chromosome 4 is similar to that found on other chromosomes; however, it is clear that the distribution of repeated sequences is different from that found in the euchromatic portions of the other chromosomes. Chromosome 4 appears very similar in its sequence organization to the regions of β-heterochromatin found at the base of polytene chromosome arms of the major chromosomes. Completion of the physical map and a determination of the DNA sequence of chromosome 4, therefore, should provide important insights into the unusual properties of this component of the Drosophila genome.
METHODS
Drosophila Stocks and Maintenance
D. melanogaster mutations used in this study are described in Lindsley and Grell (1968) or Lindsley and Zimm (1992), unless otherwise cited. Cultures were grown at 21°C on standard yeast/sucrose medium (Nash and Bell 1968).
Three existing alleles of M(4)101 were examined. M(4)10157g is an X-ray-induced mutation with no cytologically visible alteration to the polytene banding pattern. Df(4)M(4)101-62f is a deletion that spans cytological regions 101E to 102B10-17 and fails to complement alleles of M(4)101, ci, l(4)13, and l(4)17. Df(4)M(4)101-63a is a smaller deletion that spans the cytological region 101F-102A1 to 102A2-5 and fails to complement M(4)101 and ci only (Hochman 1976).
Southern Transfer
Cosmid DNA was digested with HindIII and EcoRI in a common buffer and the products were separated on a 1.2% agarose gel. After staining with ethidium bromide and photo documentation, the DNA was transferred to a GeneScreen membrane (NEN Life Science Products, Boston, MA) and then probed with D. melanogaster total genomic DNA labeled with 32P by nick translation (Rigby et al. 1977). The probe was hybridized to the blot at 65°C overnight and washed twice in 2× SSC, 0.1% SDS and once in 0.2× SSC, 0.1% SDS at 60°C before exposure to X-ray film.
PCR Reactions
Primers were synthesized by BioServe Biotechnologies, Ltd. (Laurel, MD). Primers RP5F (5′-GTCAACGTGATTCGACCTTTTCCG) and RP3R (5′-AATTTAAACAGCTTCCTGTACTGGG) were used as the forward and reverse primers in the detection of the putative ribosomal protein gene sequence and give an amplified fragment of 939 bp from wild-type genomic DNA. RP5F is positioned at −42 to −16 upstream from the putative ATG start, whereas RP3R overlaps the last six codons and the TAA stop codon of the ribosomal protein S3a gene. Reactions were amplified on a Stratagene Robocycler model 40 (1 cycle at 95°C/2 min; 54°C/1.5 min; 73°C/3 min and 29 cycles at 94°C/1 min; 54°C/1.25 min; 73°C/2 min). PCR amplifications were performed on single embryos by the method of Garozzo and Christensen (1994).
The control primers 1 (5′-ATCCCGCGCGGCGGTGAGAG) and 6 (5′-AATCAAGTGGGCGGTGCTTG), from the vestigial locus on chromosome 2 (Heslip and Hodgetts 1994), were used to confirm the presence of amplifiable DNA in each sample by the following conditions: 1 cycle at 95°C/3 min; 60°C/1 min; 73°C/2 min and 29 cycles at 92°C/1 min; 60°C/1 min; 73°C/2 min.
DNA Manipulations
Drosophila genomic DNA was prepared as described in Locke and Tartof (1994). The ci cosmid, cosL7L, was recovered, with a 3.6-kb XhoI fragment from within the ci gene as a probe, from a genomic library containing total BamHI-digested Oregon-R strain genomic DNA inserted into pAL-L and recloned into pAL-R, which are new vectors designed for rapid sequencing (Ahmed and Podemski 1997). The zfh-2 cosmid (16J20) was derived from Oregon-R strain genomic DNA partially digested with Sau3A and size selected for 30–45 kb fragments that were inserted into the cosDO cosmid vector (Ahmed 1994). This cosmid was identified in the library with a terminal probe from an adjacent cosmid that was part of a growing contig initiated at the zfh-2 gene in cytogenetic region 102C on chromosome 4. The sequencing details are described in Ahmed and Podemski (1997).
A cDNA clone (D2), containing a single ∼3.0-kb EcoRI fragment, was recovered from a λgt10 cDNA library of third instar imaginal disc poly(A)+ RNA by use of a 2.3-kb fragment extending ∼1.7–4 kb downstream from the BamHI site that defined the end of the sequenced cosL7L.
Sequence Analysis
Finished DNA sequence was analyzed with DNA Strider, GENEFINDER, and BCM GENEFIND for ORFs and repeated sequences within each cosmid. BLASTN and BLASTX were used to find similarities to sequences in public databases. DNA and protein sequences predicted from the genomic and cDNA sequences were aligned by the MACAW program (Mac68K, version 2.0.5; Schuler et al. 1991) for multiple sequence alignment. RNA polymerase III promoters were sought by use of Pol3scan (Pavesi et al. 1994).
Dot matrix analysis for 13/15 repeats used the following nonchromosome 4 sequences for comparison: accession number AL031883 (cosmid DMC11F6, distal on the X chromosome), AC005125 (P1 DS06478 at cytogenetic positon 30A7–8), AC004433 (P1 DS03659 at cytogenetic positon 57B1–6), AC005557 (P1 DS02777 at cytogenetic positon 62A10–B5), and AC001653 (P1 DS07876 at cytogenetic positon 84B1–2). In each case, the DNA Strider program accepted only the first 32,500 bp for the dot matrix analysis.
Acknowledgments
We thank Barb Hepperle, Hilary Kemp, Greg Rairdan, Brandy McGee, Scott Hanna, and Denise Adams for technical assistance. This work was funded by grants from the Canadian Genome Analysis and Technology Program (R.H. and J.L.), the Natural Sciences and Engineering Research Council of Canada (J.L.) and the Medical Research Council of Canada (J.L. and R.H.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL john.locke@ualberta.ca; FAX (403) 492-9234.
REFERENCES
- Ahmed A. Double-origin vectors for isolating bidirectional deletions useful in DNA sequence analysis. Gene. 1994;141:71–73. doi: 10.1016/0378-1119(94)90129-5. [DOI] [PubMed] [Google Scholar]
- Ahmed A, Podemski L. Use of ordered deletions in genome sequencing. Gene. 1997;197:367–373. doi: 10.1016/s0378-1119(97)00285-0. [DOI] [PubMed] [Google Scholar]
- Auclair D, Lang BF, Forest P, Desgroseillers L. Analysis of genes encoding highly conserved lysine-rich proteins in Aplysia californica and Saccharomyces cerevisiae. Eur J Biochem. 1994;220:997–1003. doi: 10.1111/j.1432-1033.1994.tb18704.x. [DOI] [PubMed] [Google Scholar]
- Bender W, Akam M, Karch F, Beachy PA, Peifer M, Spierer P, Lewis EB, Hogness DS. Molecular genetics of the Bithorax complex in Drosophila melanogaster. Science. 1983a;221:23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Bender W, Spierer P, Hogness DS. Chromosomal walking and jumping to isolate DNA from the Ace and rosy loci and the bithorax complex in Drosophila melanogaster. J Mol Biol. 1983b;168:17–33. doi: 10.1016/s0022-2836(83)80320-9. [DOI] [PubMed] [Google Scholar]
- Bonner J, Garrrard WT, Gottesfeld J, Holmes DS, Sevall JS, Wilkes M. Functional organization of the mammalian genome. Cold Spring Harbor Symp Quant Biol. 1973;38:303–310. doi: 10.1101/sqb.1974.038.01.034. [DOI] [PubMed] [Google Scholar]
- Carramolino L, Ruiz-Gomez M, del Carmen Guerrero M, Campuzano S, Modolell J. DNA map of mutations at the scute locus of Drosophila melanogaster. EMBO J. 1982;1:1191–1982. doi: 10.1002/j.1460-2075.1982.tb00011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Hough BR, Amenson CS, Britten RJ. General interspersion of repetitive with non-repetitive sequence elements in the DNA of Xenopus. J Mol Biol. 1973;77:1–23. doi: 10.1016/0022-2836(73)90359-8. [DOI] [PubMed] [Google Scholar]
- Devlin RH, Bingham B, Wakimoto BT. The organization and expression of the light gene, a heterochromatic gene of Drosophila melanogaster. Genetics. 1990;125:129–140. doi: 10.1093/genetics/125.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer D, Henikoff S. Expansions of transgenic repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Morris GD, Reuter G, Hartnett T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position effect variegation. Genetics. 1992;131:345–352. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth MW. Effects of the homozygous Minute-IV deficiency on the development of Drosophila melanogaster. Genetics. 1951;36:550. doi: 10.1093/genetics/42.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garozzo, M. and A.C. Christensen. 1994. Preparation of DNA from single embryos for PCR. Technical notes. Dros. Info. Newsl. 13.
- Gatti M, Pimpinelli S. Functional elements in Drosophila melanogaster heterochromatin. Annu Rev Genet. 1992;26:239–275. doi: 10.1146/annurev.ge.26.120192.001323. [DOI] [PubMed] [Google Scholar]
- Graham DE, Neufeld BR, Davidson EH, Britten RJ. Interspersion of repetitive and non-repetitive DNA sequences in the sea urchin genome. Cell. 1974;1:127–137. [Google Scholar]
- Grell R. Heat induced exchange in the fourth chromosome of diploid female of Drosophila melanogaster. Genetics. 1971;69:523–527. doi: 10.1093/genetics/69.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz E. Über α- und β-heterochromatin sowie konstanz und bau der chromomeren bei Drosophila. Biol Zentbl. 1934;54:588–609. [Google Scholar]
- Heslip TR, Hodgetts RB. Targetted transposition at the vestigial locus of Drosophila melanogaster. Genetics. 1994;138:1127–1135. doi: 10.1093/genetics/138.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochman B. Analysis of a whole chromosome in Drosophila. Cold Spring Harbor Symp Quant Biol. 1973;38:581–589. doi: 10.1101/sqb.1974.038.01.062. [DOI] [PubMed] [Google Scholar]
- ————— . The fourth chromosome of Drosophila melanogaster. In: Ashburner M, Novitski E, editors. The genetics and biology of Drosophila. 1b. London, UK: Academic Press; 1976. pp. 903–928. [Google Scholar]
- Kameyama T, Murakami Y, Suto F, Kawakami A, Takagi S, Hirata T, Fujisawa H. Identification of a neuronal cell surface molecule, plexin, in mice. Biochem Biophys Res Commun. 1996;226:524–529. doi: 10.1006/bbrc.1996.1388. [DOI] [PubMed] [Google Scholar]
- Kongsuwan K, Qiang Y, Vincent A, Frisardi M, Rosbash M. A Drosophila Minute gene encodes a ribosomal protein. Nature. 1985;317:555–558. doi: 10.1038/317555a0. [DOI] [PubMed] [Google Scholar]
- Kurenova EV, Leibovich BA, Bass IA, Bebikhov DV, Pavlova MN, Danilevskaya ON. Hoppel family of Drosophila melanogaster mobile elements flanked by short inverted repeats and possessing a preferential localization in the heterochromatic regions of the genome. Genetika. 1990;26:1701–1712. [PubMed] [Google Scholar]
- Lindsley DL, Grell EH. Genetic variations of Drosophila melanogaster. Washington, DC.: Carnegie Institution of Washington; 1968. [Google Scholar]
- Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. San Diego, CA: Academic Press; 1992. [Google Scholar]
- Locke J, McDermid H. Analysis of Drosophila chromosome 4 using pulsed field gel electrophoresis. Chromosoma. 1993;102:718–723. doi: 10.1007/BF00650898. [DOI] [PubMed] [Google Scholar]
- Locke J, Tartof KD. Molecular analysis of cubitus interruptus (ci) mutations suggest an explanation for the unusual ci-position effects. Mol & Gen Genet. 1994;243:234–243. doi: 10.1007/BF00280321. [DOI] [PubMed] [Google Scholar]
- Locke J, Rairdan G, McDermid H, Nash D, Pilgrim D, Bell J, Roy K, Hodgetts R. Cross-screening: A new method to assemble clones rapidly and unambiguously into contigs. Genome Res. 1996;6:155–165. doi: 10.1101/gr.6.2.155. [DOI] [PubMed] [Google Scholar]
- Lohe AR, Hilliker AJ. Return of the H-word (heterochromatin) Curr Opin Genet Dev. 1995;5:746–755. doi: 10.1016/0959-437x(95)80007-r. [DOI] [PubMed] [Google Scholar]
- Lohe AR, Hilliker AJ, Roberts PA. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics. 1993;134:1149–1174. doi: 10.1093/genetics/134.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning J, Schmid C, Davidson N. Interspersion of repetitive and nonrepetitive DNA sequences in the Drosophila melanogaster genome. Cell. 1975;4:141–155. doi: 10.1016/0092-8674(75)90121-x. [DOI] [PubMed] [Google Scholar]
- Martin DIK, Whitelaw E. The vagaries of variegating transgenes. BioEssays. 1996;18:919–923. doi: 10.1002/bies.950181111. [DOI] [PubMed] [Google Scholar]
- Metspalu A, Rebane A, Hoth S, Pooga M, Stahl J, Kruppa L. Human ribosomal protein S3a: Cloning of the cDNA and primary structure of the protein. Gene. 1992;119:316–319. doi: 10.1016/0378-1119(92)90289-2. [DOI] [PubMed] [Google Scholar]
- Miklos GLG, Cotsell JN. Chromosome structure at interfaces between major chromatin types: Alpha- and beta-heterochromatin. BioEssays. 1990;12:1–6. doi: 10.1002/bies.950120102. [DOI] [PubMed] [Google Scholar]
- Miklos GLG, Yamamoto M-T, Davies J, Pirrotta V. Microcloning reveals a high frequency of repetitive sequences characteristic of chromosome 4 and the β-heterochromatin of Drosophila melanogaster. Proc Natl Acad Sci. 1988;85:2051–2055. doi: 10.1073/pnas.85.7.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash D, Bell J. Larval age and the pattern of DNA synthesis in polytene chromosomes. Can J Genet Cytol. 1968;10:82–90. doi: 10.1139/g68-011. [DOI] [PubMed] [Google Scholar]
- Ohta K, Mizutani A, Kawakami A, Murakami Y, Kasuya Y, Takagi S, Tanaka H, Fujisawa H. Plexin: A novel neuronal cell surface molecule that mediates cell adhesion via a homophilic binding mechanism in the presence of calcium ions. Neuron. 1995;14:1189–1199. doi: 10.1016/0896-6273(95)90266-x. [DOI] [PubMed] [Google Scholar]
- Orenic TV, Slusarski DC, Kroll KL, Holmgren RA. Cloning and characterization of the segment polarity gene cubitus interruptus Dominant of Drosophila. Genes & Dev. 1990;4:1053–1067. doi: 10.1101/gad.4.6.1053. [DOI] [PubMed] [Google Scholar]
- Osborne JD. “Crossing over in a T(1,4) translocation in Drosophila melanogster.” MSc. thesis. Edmonton, Canada: University of Alberta; 1998. [Google Scholar]
- Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Pardue ML, Lowenhaupt K, Rich A, Nordheim A. (dC-dA)n (dG-dT)n sequences have evolutionary conserved locations in Drosophila with implications for roles in chromosome structure and function. EMBO J. 1987;6:1781–1789. doi: 10.1002/j.1460-2075.1987.tb02431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A, Conterio F, Bolchi AG, Ottonello S. Identification of new eucaryotic tRNA genes in genomic DNA databases by a multistep weight matix analysis of transcriptional control regions. Nucleic Acids Res. 1994;22:1247–1256. doi: 10.1093/nar/22.7.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V, Hadfield C, Pretorius GHJ. Microdissection and cloning of the white locus and the 3B1-3C2 region of the Drosophila X chromosome. EMBO J. 1983;2:927–934. doi: 10.1002/j.1460-2075.1983.tb01523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud E, Bolshakov VN, Barajas V, Kafatos FC, Zurta M. Antisense suppression of the putative ribosomal protein S3A gene disrupts ovarian development in Drosophila melanogaster. Mol & Gen Genet. 1997;256:462–467. doi: 10.1007/s004380050590. [DOI] [PubMed] [Google Scholar]
- Rigby PW, Dieckmann M, Rhodes C, Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977;113:237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sandler L, Szauter P. The effect of recombination-defective meiotic mutants on fourth-chromosome crossing over in Drosophila melanogaster. Genetics. 1978;90:699–712. doi: 10.1093/genetics/90.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalet A, Lefevre G. The proximal region of the X chromosome. In: Ashburner M, Novitski E, editors. The genetics and biology of Drosophila. 1b. London, UK: Academic Press; 1976. pp. 847–902. [Google Scholar]
- Schuler GD, Atschul SF, Lipman DJ. A workbench for multiple alignment construction and analysis. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Schwartz C, Locke J, Nishida C, Kornberg TB. Analysis of cubitus interruptus regulation in Drosophila embryos and imaginal disks. Development. 1995;121:1625–1635. doi: 10.1242/dev.121.6.1625. [DOI] [PubMed] [Google Scholar]
- Saebøe-Larrsen S, Urbanczyk Mohebi B, Lambertsson A. The Drosophila ribosomal protein L14-encoded gene, identified by a novel Minute mutation in a dense cluster of previously undescribed genes in cytogenetic region 66D. Mol Gen Genet. 1997;255:141–151. doi: 10.1007/s004380050482. [DOI] [PubMed] [Google Scholar]
- Saebøe-Larrsen S, Lyamouri M, Merriam J, Oksvold MP, Lambertsson A. Ribosomal protein insufficiency and the Minute syndrome in Drosophila: A dose-response relationship. Genetics. 1998;148:1215–1224. doi: 10.1093/genetics/148.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. A map of the fourth chromosome of Drosophila melanogaster based on crossing over in triploid females. Proc Natl Acad Sci. 1951;37:405–407. doi: 10.1073/pnas.37.7.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Have JF, Green MM, Howells AJ. Molecular characterization of spontaneous mutations at the scarlet locus of Drosophila melanogaster. Mol & Gen Genet. 1995;249:673–681. doi: 10.1007/BF00418037. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverse K, Pardue ML. Studies of the He-T DNA sequences in the pericentric regions of Drosophila chromosomes. Chromosoma. 1989;97:261–271. doi: 10.1007/BF00371965. [DOI] [PubMed] [Google Scholar]
- van Beest M, Morin M, Clevers H. Drosophila RpS3a, a novel Minute gene situated between the segment polarity genes cubitus interruptus and dTCF. Nucleic Acids Res. 1998;26:4471–4475. doi: 10.1093/nar/26.19.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaury C, Bucheton A, Pelisson A. The β heterochromatic sequences flanking the I elements are themselves defective transposable elements. Chromosoma. 1989;98:215–224. doi: 10.1007/BF00329686. [DOI] [PubMed] [Google Scholar]
- Wallrath LL, Elgin SCR. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes & Dev. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- Wallrath LL, Guntur VP, Rosman LE, Elgin SCR. DNA representation of variegating heterochromatic P-element inserts in diploid and polytene tissues of Drosophila melanogaster. Chromosoma. 1996;104:519–527. doi: 10.1007/BF00352116. [DOI] [PubMed] [Google Scholar]
- Zhang P, Spradling AC. The Drosophila salivary gland chromocenter contains highly polytenized subdomains of mitotic heterochromatin. Genetics. 1995;139:659–670. doi: 10.1093/genetics/139.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]