Abstract
Apolipoprotein E (ApoE) ε4 genotype is a strong risk factor for developing Alzheimer’s disease (AD). Conversely, the presence of the ε2 allele has been shown to mitigate cognitive decline. Tensor-based morphometry (TBM), a novel computational approach for visualizing longitudinal progression of brain atrophy, was used to determine whether cognitively intact elderly participants with the ε4 allele demonstrate greater volume reduction than those with the ε2 allele. Healthy “younger elderly” volunteers, aged 55–75, were recruited from the community and hospital staff. They were evaluated with a baseline and follow-up MRI scan (mean scan interval = 4.72 years, s.d. = 0.55) and completed ApoE genotyping. Twenty-seven participants were included in the study of which 16 had the ε4 allele (all heterozygous ε3ε4 genotype) and 11 had the ε2ε3 genotype. The two groups did not differ significantly on any demographic characteristics and all subjects were cognitively “normal” at both baseline and follow-up time points. TBM was used to create 3D maps of local brain tissue atrophy rates for individual participants; these spatially detailed 3D maps were compared between the two ApoE groups. Regional analyses were performed and the ε4 group demonstrated significantly greater annual atrophy rates in the temporal lobes (p = 0.048) and hippocampus (p = 0.016); greater volume loss was observed in the right hippocampus than the left. TBM appears to be useful in tracking longitudinal progression of brain atrophy in cognitively asymptomatic adults. Possession of the ε4 allele is associated with greater temporal and hippocampal volume reduction well before the onset of cognitive deficits.
Keywords: Aging, Alzheimer’s disease, Apolipoprotein E, asymmetry, healthy elderly, hippocampus, magnetic resonance imaging, tensor-based morphometry, temporal lobe, white matter
INTRODUCTION
Apolipoprotein E (ApoE) ε4 genotype, coded on chromosome 19, is a strong risk factor for developing Alzheimer’s disease (AD), second only to advancing age. The lifetime risk of AD for an individual without the ε4 allele is approximately 10%, whereas the lifetime risk for an individual carrying at least one ε4 allele is 30% [1]. Epidemiological studies have demonstrated that ApoE ε4 genotype shifts the age of onset for AD by more than a decade, thus accounting for the vast majority of observed cases of AD that occur before age 80 [2–5]. Conversely, presence of an ApoE ε2 allele has been shown to mitigate cognitive decline [6–8] and delay the onset age for dementia [2, 5, 9, 10].
Several magnetic resonance imaging (MRI) studies have attempted to map brain structural changes associated with the ApoE gene in non-demented elderly subjects. These investigations, focused primarily on the hippocampus, have yielded mixed results. Within cross-sectional designs, a handful of studies have reported reduced hippocampal volume in carriers of the ε4 allele relative to non-carriers [11–13], but many others have failed to detect an ApoE effect on hippocampal size [14–21]. Longitudinal reports were more consistent in finding an association between accelerated hippocampal atrophy and ApoE ε4 genotype [15–17] although not all investigators have confirmed these findings [18].
Whole brain volume has thus far not been shown to be sensitive to the effects of ApoE in multiple cross-sectional [11, 13, 14] and longitudinal [17] studies, as global brain atrophy likely occurs in later stages of AD. A recent longitudinal report did show greater annual rate of whole brain atrophy for homozygous but not heterozygous ε4 carriers when compared to non-carriers [22]. The effect of the ApoE gene on white matter integrity has also been investigated. Using diffusion tensor imaging (DTI), lower fractional anisotropy (FA) values were obtained for ε4 carriers, compared to non-carriers, in the posterior regions of the corpus callosum [23]. More recently, Honea et al. [24] and Nierenberg et al. [25] both reported decreased FA in the left parahippocampal gyrus white matter. We previously demonstrated, using transverse relaxation rate (R2), that ApoE genotype can affect the trajectory of age-related myelin breakdown in healthy, cognitively-intact older adults [26]. Specifically, individuals with the ε4 allele displayed a steeper slope of age-related decline in R2 of frontal white matter and genu of the corpus callosum than subjects with the ε2 genotype.
Tensor-based morphometry (TBM) is a novel computational approach that can compare longitudinally acquired images and visualize the spatial profile of brain atrophy over time, including estimates of tissue volume loss rates at each voxel in the brain (reviewed in [27]). This approach has been successfully used to study neurodegenerative and psychiatric disorders, as well as normal brain development [28–30]. We applied the TBM method to characterize and compare the pattern of brain atrophy in cognitively intact elderly adults who are either ε2 or ε4 carriers. We specifically chose to study these two groups because of the contrasting effects of these two genotypes on the development of incipient AD. We hypothesized that ε4 carriers would demonstrate greater rate of brain atrophy than ε2 carriers, specifically in the temporal lobes and hippocampus, regions associated with AD pathology.
MATERIALS AND METHODS
Participants
Normal adult volunteers between the ages of 55 and 75 were recruited from the community and hospital staff for a study on healthy aging. All subjects received written and oral information about the study and signed written informed consents approved by the local institutional review board prior to study participation. Potential subjects were excluded if they had a history of neurological disorder or a family history of AD or other neurodegenerative disorder, psychiatric illness (including drug or alcohol abuse), or head injury resulting in loss of consciousness for more than 10 minutes. The subjects were physically very healthy and were excluded if they were obese (defined as body mass index [BMI] of >30 kg/m2), or if they had a history of diabetes, cardiovascular disease, or difficult to control hypertension. Determination of normal cognition and independent functioning was based on semi-structured clinical interview that included questions directed to subjects regarding performance of basic and instrumental activities of daily living, neurological examination by the study physician (GB), and formal neuropsychological testing. None of the participants met the diagnosis of dementia by Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV) criteria [31] or mild cognitive impairment based on Petersen criteria [32]. Specifically, none of the subjects had any subjective cognitive complaints or performed ≥1.5 s.d. below published normative means [33] on objective memory testing (California Verbal Learning Test; CVLT); all had normal global cognitive ability (≥27) on the Mini-Mental State Examination [34] and intact activities of daily living.
At baseline and at the 5-year follow-up visit, the subjects underwent MRI scan and a battery of neuropsychological tests; all subjects were genotyped at baseline. Eleven subjects with the ApoE ε2 allele (all with the ε2ε3 genotype) and 16 heterozygous ε4 carriers (all with ε3ε4 genotype) completed the 5-year follow-up visit. The demographic characteristics and cognitive test data are summarized in Table 1, indicating that the two Apoe groups were well-matched on demographic variables and exhibited intact memory functioning across time. There were 15 women and 12 men in the sample, and the racial composition was comprised of 22 (82%) Caucasians, 2 (7%) Asians, 1 (4%) African–American, and 2 (7%) Hispanics.
Table 1.
Demographic and cognitive variables by ApoE group
| Demographic variables | ApoE 2/3 (n = 11) | ApoE 3/4 (n = 16) | t or χ2 | p |
|---|---|---|---|---|
| Mean (s.d.) | Mean (s.d.) | |||
| Age, y | 67.0 (5.2) | 65.0 (4.5) | 1.06 | 0.301 |
| Education, y | 14.2 (1.9) | 15.6 (2.4) | −1.65 | 0.112 |
| Gender (m/f) | 5m/6f | 7m/9f | 0.008 | 0.930 |
| % white | 64% | 94% | 3.92 | 0.048 |
| Time between scans, y | 4.8 (0.5) | 4.8 (0.5) | 0.30 | 0.770 |
|
| ||||
| Cognitive variables | Mean ± s.d. (range) | Mean ± s.d. (range) | t | p |
|
| ||||
| MMSE | 28.2 ± 1.1 (27–30) | 28.7 ± 0.9 (27–30) | −0.92 | 0.376 |
| CVLT-long delay-baseline | ||||
| Raw scores | 11.5 ± 1.5 (9–14) | 11.6 ± 2.0 (8–15) | 0.05 | 0.960 |
| z-scores | 0.36 ± 0.50 (0.0 to +1.0) | 0.31 ± 0.79 (−1.0 to +1.0) | 0.19 | 0.852 |
| CVLT-long delay-follow-up | ||||
| Raw scores | 11.3 ± 3.1 (7–16) | 11.2 ± 1.9 (9–15) | −0.19 | 0.854 |
| z-scores | 0.60 ± 1.07 (−1.0 to +2.0) | 0.56 ± 0.81 (−1.0 to +2.0) | 0.10 | 0.920 |
MRI acquisition
At baseline, all the subjects were scanned using a single 1.5 Tesla (T) MR instrument with a T1-weighted spoiled gradient recalled (SPGR) 3D MRI sequence. The following scanning parameters were used: repetition time (TR) of 24 ms, echo time (TE) of 7 ms, flip angle of 35°, 25 cm field of view (FOV), 208 × 208 acquisition matrix with slice thickness of 1.5 mm, and acquired resolution of 1.2 × 1.2 × 1.5 mm. All follow-up scans were conducted on a different 1.5 T MRI scanner using T1-weighted spoiled GRASS (SPGR) sequences. A sagittal plane image acquisition protocol was used with TR = 25 ms; TE = 5 ms; flip angle of 35°; acquisition matrix = 256 × 192 × 25 with slice thickness of 1.4 mm; and FOV = 25 cm. The image voxel size was 0.98 × 0.98 × 1.4 mm.
Image preprocessing
An automated Brain Surface Algorithm (BSE) from Brainsuite [35] was applied, along with manual editing, to remove skull and other non-brain tissues. We then corrected intensity inhomogeneity caused by nonuniformities in the radio frequency (RF) receiver coils using the N3 bias field algorithm proposed by Sled et al. [36]. To adjust for global differences in brain positioning, orientation, and scaling across individuals, all scans were affinely registered to the stereotactic space defined by the International Consortium for Brain Mapping (ICBM-53) [37] using a 9-parameter (9P) transformation (3 translations, 3 rotations, and 3 scales). All follow-up scans were then aligned to the same subject’s baseline scan, and both were aligned to the ICBM space. Globally aligned images were resampled in an isotropic space of 230 voxels along each axis (x-, y-, and z-dimensions) with an interpolated voxel size of 1 mm3.
To inspect the quality of the 9-parameter registrations, we used a 3D visualization tool called REGISTER, which automatically overlays the arbitrary slice geometry of each scan pair in ICBM space. All scans were confirmed to have satisfactory alignment without noticeable distortion or mismatch.
Jacobian maps quantifying structural changes over time
To quantify 3D patterns of volumetric brain atrophy over time for each subject, an individual change map, or Jacobian map, was computed by non-linearly registering the follow-up scan to the baseline scan with mutual information based unbiased registration algorithm [38]. Mutual information is a measure of how much information one random variable has about another and it can be used to align images of different modalities, without requiring knowledge of the relationship (joint intensity distribution) of the two registered images [39, 40]. The unbiased image registration technique computes deformation fields by penalizing statistical bias in the resulting Jacobian maps, thus eliminating skew from the distribution of the corresponding Jacobian determinants, and has been shown to perform favorably in recovering true physiological changes in serial MRI data [41].
For each subject, a local tissue growth/atrophy map was obtained by calculating the local Jacobian determinant (i.e., “expansion factor”) of the deformation field, which measures progressive volume contraction (Jacobian < 1) or volume expansion (Jacobian > 1). To adjust for variable time differences between scans, individual Jacobian maps were normalized by dividing each map (percent tissue change) by its corresponding inter-scan interval (in years) to create the annualized Jacobian maps, which represent the average change over 1 year. All results and statistical analyses are based on the annualized Jacobian maps.
Regions of interest
The regions of interest (ROI), comprised of frontal, temporal, parietal, and occipital lobes, and the hippocampus, were manually hand-traced by a trained anatomist on the ICBM template using the Brainsuite software program [35]. Brain slices depicting the ROIs for the temporal lobe regions and the hippocampus for both hemispheres are shown in Fig. 1.
Fig. 1.
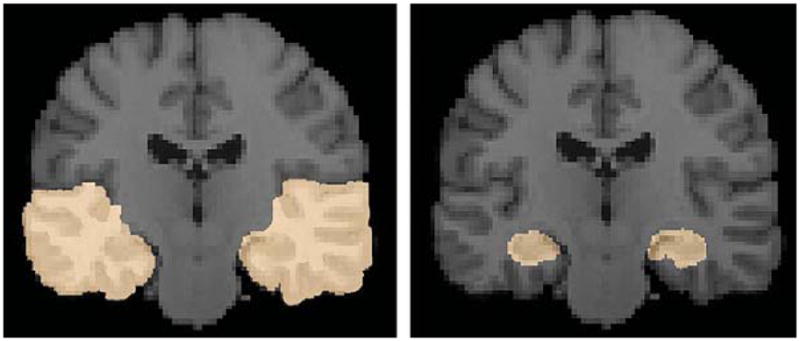
Regions of interest of the temporal lobe regions (left panel) and the hippocampus (right panel) of both hemispheres are shown. The ROIs were manually delineated on the ICBM template by a trained anatomist using the Brainsuite software program [32].
Statistical analyses
To illustrate systematic difference in atrophic rates between ε2 and ε4 carriers, we constructed voxel-wise statistical maps based on the Student’s t-statistic. The annualized Jacobian maps of ε2 and ε4 groups were compared using permutation-based two sample t tests to assess overall significance of group differences inside each ROI, corrected for multiple comparisons [42–44]. In brief, a null distribution for the group differences in atrophic rates (Jacobian values) at each voxel was constructed using 10,000 random permutations of the data. For each test, the subjects’ genetic status (ε2 vs. ε4 carriers) was randomly permutated and voxel-wise t-tests were conducted to identify voxels more significant than p = 0.05. The volume of voxels inside a mask (i.e., temporal lobes) more significant than p = 0.05 was computed for the real experiment and for the random assignments. A ratio, describing the fraction of the time the t-statistic was more extreme in the randomized tests than the original test, was calculated to yield an overall p-value for the significance of the map (corrected for multiple comparison by permutation). A numeric summary, the mean atrophy rate for all voxels within each ROI, was computed for each person, to summarize the annual change within the ROI. This approach has been used extensively in prior work [28, 29].
RESULTS
Individual Jacobian maps were averaged for the ε2 and ε4 groups to demonstrate the average amount of annual brain tissue loss (in blue colors) and ventricular enlargement (in red colors) for each group (Fig. 2). As expected, the maps displaying the pattern of progressive brain changes were not clearly distinguishable between the ε2 and ε4 carriers since both groups were comprised of healthy, cognitively intact elderly subjects. However, closer inspection reveals subtle but visible changes showing greater ventricular expansion and temporal lobe atrophy in the ε4 group relative to the ε2 carriers.
Fig. 2.
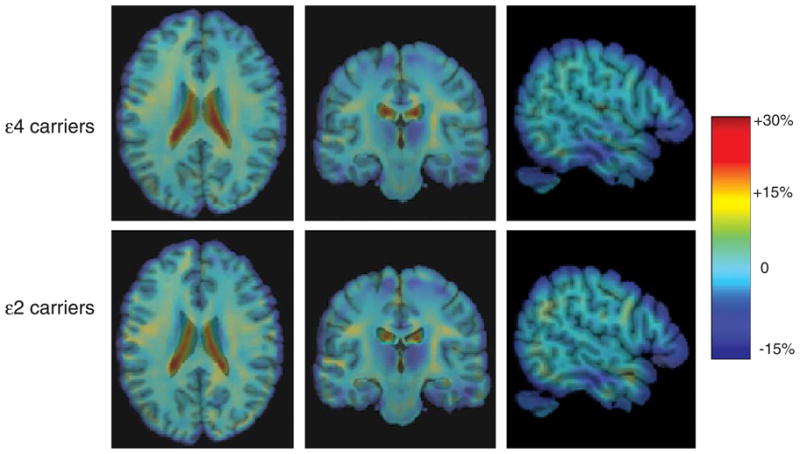
Jacobian maps showing the mean annual rate of atrophy of brain tissue (in blue color) and ventricular enlargement (in red color) for the ε4 carriers (top row) and the ε2 carriers (bottom row). These tissue changes are shown as percentages, relative to the baseline scan, and are computed within each individual before averaging across subjects in the group.
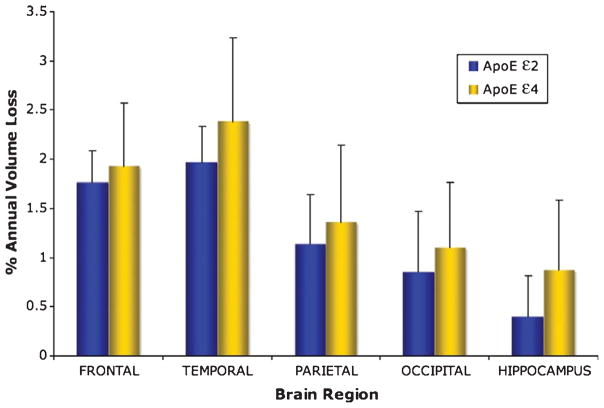
Mean annualized Jacobian values within each ROI, normalized to indicate annualized rates of atrophy (in percent volume loss per year), were computed for each subject, and group averages for each genotype are graphically depicted in Fig. 3. In general, a consistent trend was observed with the ε4 group demonstrating greater rates of atrophy across all regions of interest; however, independent t-tests of these numerical values did not reveal any statistically significant group differences (p > 0.06).
Fig. 3.
Bar plots showing percent volume loss per year in the brain regions of interest for ApoE ε2 and ε4 groups. The group differences were not statistically significant for any region (p > 0.06).
Permutation tests, using suprathreshold percentages, provide a detailed 3D visualization of the local atrophy profile. Significantly greater atrophy rates were observed for ε4 carriers compared to ε2 group for temporal lobes (p = 0.048) and more specifically, the hippocampus (p = 0.016). The p-value maps representing the regions of significant genotypic group difference are presented in Fig. 4. Interestingly, when the right and left hippocampus were examined separately, the difference between ε4 and ε2 groups was greater in the right hippocampus (p = 0.008) than the left hippocampus (p = 0.096). This is consistent with our previous findings that permutation testing, using the suprathreshold volume of statistics in a map, may be more powerful for detecting group differences than performing univariate tests on regional averages (numeric summaries) derived from the maps.
Fig. 4.
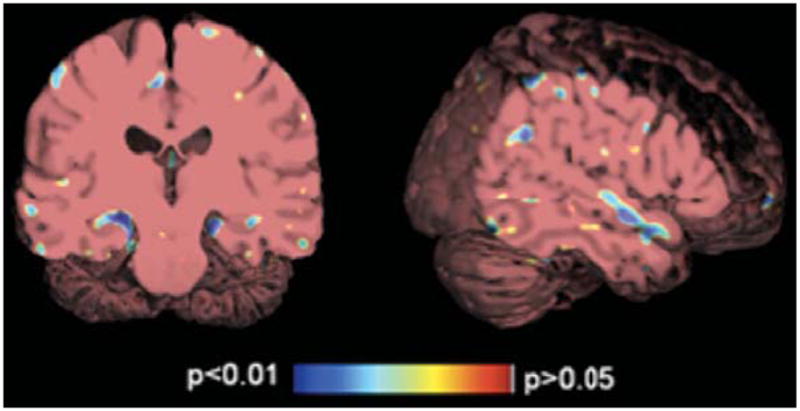
Significance maps showing that the ε4 carriers demonstrate significantly greater annual atrophy rate (as indicated in blue) in the hippocampus (left panel) and the superior temporal gyrus (right panel) than the ε2 group.
DISCUSSION
Using TBM, the results from the present longitudinal study demonstrate that cognitively intact adults with the ApoE ε4 allele showed significantly increased rate of temporal lobe and hippocampal atrophy than those with the ε2 genotype. Furthermore, the rate of volume loss in the right hippocampus was significantly greater than the left hippocampus. While not statistically significant, greater percent volume loss was observed in the frontal, parietal, and occipital lobes of the ε4 group, indicating that ongoing atrophy is widely distributed but significantly greater in areas that are among the first to be involved with AD pathology.
Longitudinal neuroimaging studies of the effects of ApoE ε4 genotype on brain structural changes typically combine individuals with the ε3ε3 and ε2ε3 genotypes into a single “non-carrier” group with inter-scan intervals ranging from 17 months to 3.5 years [15–18]. The present study focused on the comparison between ApoE ε2 and ε4 carriers due to the contrasting effects of these two genotypes on the age of onset for AD. The extended time interval of approximately 5 years between MRI examinations also allows for brain structural volume differences to emerge in cognitively intact, high functioning “younger-elderly” individuals. Present findings confirm and extend existing literature reporting accelerated hippocampal atrophy in ε4 carriers [15, 17] as well as contribute to the mounting evidence of decreased brain atrophy in ε2 carriers. While these results may be suggestive of a protective role of the ε2 genotype against the development of AD [2, 5, 9, 10], this cannot be conclusively demonstrated without the inclusion of an ε3ε3 group, since it is possible that individuals with the ε3ε3 genotype may show the same attenuated atrophy rate as the ε2 group or they may atrophy at a rate that is intermediate to that of ε2 and ε4 carriers. Future studies that include ε3ε3 subjects can better elucidate the effects of each specific genotype on hippocampal volume and allow more definitive conclusions regarding the potential protective effect of the ε2 allele.
The asymmetric finding of more severe right hippocampal atrophy in association with the ε4 allele has been repeatedly demonstrated in non-demented subjects [11–13, 45–47] and patients with AD [48–51]. However, some studies have reported the absence of hemispheric differences in hippocampal volume in relation to ApoE genotype [20, 21]. The observation of a “normal” asymmetry favoring the right hippocampus has been reported in several MRI studies, and a recent meta-analysis confirmed that the right hippocampal volume is reliably larger than the left in normal adults [52]. Therefore, the absence or reversal of such a discrepancy was postulated to be a possible indicator of existing or impending pathology [45, 46, 48] although the functional implication for the differential hemispheric vulnerability in the hippocampus remains incompletely understood.
The finding of significantly greater volume loss associated with ApoE ε4 genotype in this healthy “younger-elderly” sample cannot be easily explained given that neuronal loss is not associated with aging [53, 54]. On the other hand, the process of age-related myelin breakdown and loss has been thoroughly demonstrated [55–61]. When compared with other structural indices of brain health, age-related degenerative changes are most pronounced in late-myelinating regions [62], such as the frontal lobe white matter (FLwm) that contain higher proportions of smaller thinly-myelinated axons [58, 63, 64].
Histopathological studies have demonstrated white matter degeneration in AD, independent of gray matter lesions or vascular disease [65, 66]. Specifically, myelin staining is reduced in the perforant pathway, the main input fibers projecting neocortical information from the entorhinal cortex to the granule cells of the dentate gyrus in the hippocampal formation [66]. Absence of myelin due to poor development can reduce overall hippocampal volume despite normal numbers of neurons [67, 68], and the reverse is also possible as hippocampal volume can increase despite neuronal loss [69]. ApoE is the principal brain cholesterol transporter and is essential to the process of “recycling” cholesterol by helping reclaim it from damaged myelin and supplying it for rapid membrane biogenesis during re-myelination [70, 71]. The number of ApoE molecules necessary for this recycling process is lowest in ε4 carriers and highest in individuals with the ε2 allele [72, 73]. Therefore, the mechanism underlying the connection between the ApoE gene and the pathophysiology of AD, namely hippocampal atrophy, may be better understood by examining the role of ApoE in myelin maintenance and repair.
The strengths of the present study include the longitudinal design in which each subject acts as his/her own control and measurement of intra-individual rates of change yields greater sensitivity for detection of subtle brain changes over time. Furthermore, TBM may have an advantage over standard ROI-based, manual tracing methods for detecting subtle or highly localized differences. Several study limitations should also be acknowledged. The absence of a ε3ε3 group limits the interpretation on the protective effects of the ε2 allele. Another important limitation is the small sample size, thus replication with larger sample sizes is warranted; however, the number of participants in the present study is comparable to most longitudinal MRI studies of the ApoE effect on brain changes with sample sizes ranging from 25 to 42 participants [15–18]. The heterozygous makeup of the ε4 group may limit the generalizability of the findings to homozygous ε4 carriers, but several cross-sectional studies have demonstrated a dose effect such that homozygous ε4 groups exert a greater influence on brain changes than heterozygous ε4 carriers and non-carriers [13, 22]. Another limitation is the difference in MR instrument for the baseline and follow-up scans due to machine change, but the registration method for fluid alignment in our version of TBM optimizes the mutual information between images, which is a cost function that is deliberately designed to be robust under changes in image contrast. It is also important to emphasize that the study design is balanced such that the scanner change affects all subjects and is not specific to one genetic group. The maps of annual change thus reflect both aging and the scanner effect; this would be a concern for interpreting age-related change but should not be a problem for interpreting modulators or correlates of change including ApoE genotype. In addition, while the relatively strict inclusion criteria (no AD family history, no cardiovascular disease, no obesity) reduce the number of factors that may contribute to the brain changes, it also makes the results less generalizable to the overall US population.
Analysis of serial MRI scans using TBM appears to be sensitive in tracking age-related progression of brain changes and the modifying effects of the ApoE gene on this process. The ApoE ε4 gene appears to exert its impact via accelerated atrophy that is not reliably detected upon cross-sectional study. Additionally, greater right hippocampal reduction may suggest that ApoE genotypic risk is manifested in a reduction in normal hippocampal asymmetry. Therefore, longitudinal decline in hippocampal volume as well as hippocampal asymmetry may hold more promise as possible biomarkers of an approaching cognitive decline than absolute differences within either left or right hippocampus examined at only one time point.
Acknowledgments
This work was supported by grant K23-AG028727 from the National Institute of Aging (NIA), a grant from the Alzheimer’s Association (NIRG-07-60424), the Alzheimer’s Disease Research Center grant P50 AG-16570, and Alzheimer’s Disease Research Center of California grant. Further support for this study came from NIH grants (MH51928; MH6357-01A1; MH066029-01A2; AG027342-01A2; U54 RR021813 to GB). Algorithm development for this study was also funded by the NIA, NIBIB, the National Library of Medicine, and the National Center for Research Resources (AG016570, EB01651, LM05639, RR019771 to PT).
Footnotes
This data was presented in part at the 11th annual meeting of the International Conference for Alzheimer’s Disease, Chicago, IL.
Authors’ disclosures available online (http://www.jalz.com/disclosures/view.php?id=639).
References
- 1.Seshadri S, Drachman DA, Lippa CF. Apolipoprotein E epsilon4 allele and the lifetime risk of Alzheimer’s disease. What physicians know, and what they should know. Arch Neurol. 1995;52:1074–1079. doi: 10.1001/archneur.1995.00540350068018. [DOI] [PubMed] [Google Scholar]
- 2.Ashford JW. Apoe genotype effects on Alzheimer’s disease onset and epidemiology. J Mol Neurosci. 2004;23:157–166. doi: 10.1385/JMN:23:3:157. [DOI] [PubMed] [Google Scholar]
- 3.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Dujin CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. Apoe and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 4.Poirier J, Davignon J, Bouthillier D, Kogan S, Bertrand P, Gauthier S. Apolipoprotein E polymorphism and Alzheimer’s disease. Lancet. 1993;342:697–699. doi: 10.1016/0140-6736(93)91705-q. [DOI] [PubMed] [Google Scholar]
- 5.Raber J, Huang Y, Ashford JW. ApoE genotype accounts for the vast majority of AD risk and AD pathology. Neurobiol Aging. 2004;25:641–650. doi: 10.1016/j.neurobiolaging.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 6.Deary IJ, Whiteman MC, Pattie A, Starr JM, Hayward C, Wright AF, Visscher PM, Tynan MC, Whalley LJ. Apolipoprotein E gene variability and cognitive functions at age 79: a follow-up of the Scottish mental survey of 1932. Psychol Aging. 2004;19:367–371. doi: 10.1037/0882-7974.19.2.367. [DOI] [PubMed] [Google Scholar]
- 7.Staehelin HB, Perrig-Chiello P, Mitrache C, Miserez AR, Perrig WJ. Apolipoprotein E genotypes and cognitive functions in healthy elderly persons. Acta Neurol Scand. 1999;100:53–60. doi: 10.1111/j.1600-0404.1999.tb00723.x. [DOI] [PubMed] [Google Scholar]
- 8.Wilson RS, Bienias JL, Berry-Kravis E, Evans DA, Bennett DA. The apolipoprotein E epsilon2 allele and decline in episodic memory. J Neurol Neurosurg Psychiatry. 2002;73:672–677. doi: 10.1136/jnnp.73.6.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, Jr, Rimmler JB, Locke PA, Conneally PM, Schmader KE, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7:180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- 10.Khachaturian AS, Corcoran CD, Mayer LS, Zandi PP, Breitner JC. Apolipoprotein E epsilon4 count affects age at onset of Alzheimer disease, but not lifetime susceptibility: The Cache County Study. Arch Gen Psychiatry. 2004;61:518–524. doi: 10.1001/archpsyc.61.5.518. [DOI] [PubMed] [Google Scholar]
- 11.den Heijer T, Oudkerk M, Launer LJ, van Duijn CM, Hofman A, Breteler MM. Hippocampal, amygdalar, and global brain atrophy in different apolipoprotein E genotypes. Neurology. 2002;59:746–748. doi: 10.1212/wnl.59.5.746. [DOI] [PubMed] [Google Scholar]
- 12.Tohgi H, Takahashi S, Kato E, Homma A, Niina R, Sasaki K, Yonezawa H, Sasaki M. Reduced size of right hippocampus in 39- to 80-year-old normal subjects carrying the apolipoprotein E epsilon4 allele. Neurosci Lett. 1997;236:21–24. doi: 10.1016/s0304-3940(97)00743-x. [DOI] [PubMed] [Google Scholar]
- 13.Lemaitre H, Crivello F, Dufouil C, Grassiot B, Tzourio C, Alperovitch A, Mazoyer B. No epsilon4 gene dose effect on hippocampal atrophy in a large MRI database of healthy elderly subjects. Neuroimage. 2005;24:1205–1213. doi: 10.1016/j.neuroimage.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 14.Tupler LA, Krishnan KR, Greenberg DL, Marcovina SM, Payne ME, MacFall JR, Charles HC, Doraiswamy PM. Predicting memory decline in normal elderly: genetics, MRI, and cognitive reserve. Neurobiol Aging. 2007;28:1644–1656. doi: 10.1016/j.neurobiolaging.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Cohen RM, Small C, Lalonde F, Friz J, Sunderland T. Effect of apolipoprotein E genotype on hippocampal volume loss in aging healthy women. Neurology. 2001;57:2223–2228. doi: 10.1212/wnl.57.12.2223. [DOI] [PubMed] [Google Scholar]
- 16.Jak AJ, Houston WS, Nagel BJ, Corey-Bloom J, Bondi MW. Differential cross-sectional and longitudinal impact of Apoe genotype on hippocampal volumes in nondemented older adults. Dement Geriatr Cogn Disord. 2007;23:382–389. doi: 10.1159/000101340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moffat SD, Szekely CA, Zonderman AB, Kabani NJ, Resnick SM. Longitudinal change in hippocampal volume as a function of apolipoprotein E genotype. Neurology. 2000;55:134–136. doi: 10.1212/wnl.55.1.134. [DOI] [PubMed] [Google Scholar]
- 18.Du AT, Schuff N, Chao LL, Kornak J, Jagust WJ, Kramer JH, Reed BR, Miller BL, Norman D, Chui HC, Weiner MW. Age effects on atrophy rates of entorhinal cortex and hippocampus. Neurobiol Aging. 2006;27:733–740. doi: 10.1016/j.neurobiolaging.2005.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt H, Schmidt R, Fazekas F, Semmler J, Kapeller P, Reinhart B, Kostner GM. Apolipoprotein E ε4 allele in the normal elderly: neuropsychologic and brain MRI correlates. Clin Genet. 1996;50:293–299. doi: 10.1111/j.1399-0004.1996.tb02377.x. [DOI] [PubMed] [Google Scholar]
- 20.Reiman EM, Uecker A, Caselli RJ, Lewis S, Bandy D, de Leon MJ, De Santi S, Convit A, Osborne D, Weaver A, Thibodeau SN. Hippocampal volumes in cognitively normal persons at genetic risk for Alzheimer’s disease. Ann Neurol. 1998;44:288–291. doi: 10.1002/ana.410440226. [DOI] [PubMed] [Google Scholar]
- 21.Bigler ED, Tate DF, Miller MJ, Rice SA, Hessel CD, Earl HD, Tschanz JT, Plassman B, Welsh-Bohmer KA. Dementia, asymmetry of temporal lobe structures, and apolipoprotein E genotype: relationships to cerebral atrophy and neuropsychological impairment. J Int Neuropsychol Soc. 2002;8:925–933. doi: 10.1017/s1355617702870072. [DOI] [PubMed] [Google Scholar]
- 22.Chen K, Reiman EM, Alexander GE, Caselli RJ, Gerkin R, Bandy D, Domb A, Osborne D, Fox N, Crum WR, Saunders AM, Hardy J. Correlations between apolipoprotein E epsilon4 gene dose and whole brain atrophy rates. Am J Psychiatry. 2007;164:916–921. doi: 10.1176/ajp.2007.164.6.916. [DOI] [PubMed] [Google Scholar]
- 23.Persson J, Lind J, Larsson A, Ingvar M, Cruts M, van Broeckhoven C, Adolfsson R, Nilsson LG, Nyberg L. Altered brain white matter integrity in healthy carriers of the Apoe epsilon4 allele: a risk for AD? Neurology. 2006;66:1029–1033. doi: 10.1212/01.wnl.0000204180.25361.48. [DOI] [PubMed] [Google Scholar]
- 24.Honea RA, Vidoni E, Harsha A, Burns JM. Impact of Apoe on the healthy aging brain: a voxel-based MRI and DTI study. J Alzheimers Dis. 2009;18:533–564. doi: 10.3233/JAD-2009-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nierenberg J, Pomara N, Hoptman MJ, Sidtis JJ, Ardekani BA, Lim KO. Abnormal white matter integrity in healthy apolipoprotein E epsilon4 carriers. Neuroreport. 2005;16:1369–1372. doi: 10.1097/01.wnr.0000174058.49521.16. [DOI] [PubMed] [Google Scholar]
- 26.Bartzokis G, Lu PH, Geschwind DH, Edwards N, Mintz J, Cummings JL. Apolipoprotein E genotype and age-related myelin breakdown in healthy individuals: implications for cognitive decline and dementia. Arch Gen Psychiatry. 2006;63:63–72. doi: 10.1001/archpsyc.63.1.63. [DOI] [PubMed] [Google Scholar]
- 27.Ashburner J, Friston KJ. Human Brain Function. Academic Press; 2003. Morphometry. [Google Scholar]
- 28.Hua X, Leow AD, Levitt JG, Caplan R, Thompson PM, Toga AW. Detecting brain growth patterns in normal children using tensor-based morphometry. Hum Brain Mapp. 2007;30:209–219. doi: 10.1002/hbm.20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua X, Leow AD, Lee S, Klunder AD, Toga AW, Lepore N, Chou YY, Brun C, Chiang MC, Barysheva M, Jack CR, Jr, Bernstein MA, Britson PJ, Ward CP, Whitwell JL, Borowski B, Fleisher AS, Fox NC, Boyes RG, Barnes J, Harvey D, Kornak J, Schuff N, Boreta L, Alexander GE, Weiner MW, Thompson PM Alzheimer’s Disease Neuroimaging, Initiative. 3D characterization of brain atrophy in Alzheimer’s disease and mild cognitive impairment using tensor-based morphometry. Neuroimage. 2008;41:19–34. doi: 10.1016/j.neuroimage.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gogtay N, Lu A, Leow AD, Klunder AD, Lee AD, Chavez A, Greenstein D, Giedd JN, Toga AW, Rapoport JL, Thompson PM. Three-dimensional brain growth abnormalities in childhood-onset schizophrenia visualized by using tensor-based morphometry. Proc Natl Acad Sci U S A. 2008;105:15979–15984. doi: 10.1073/pnas.0806485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4. Washington: APA; 1994. [Google Scholar]
- 32.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 33.Delis DC, Kramer JH, Kaplan E, Ober B. The California Verbal Learning Test. The Psychological Corporation; San Antonio: 1987. [Google Scholar]
- 34.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state.” A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 35.Shattuck DW, Leahy RM. BrainSuite: an automated cortical surface identification tool. Med Image Anal. 2002;6:129–142. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- 36.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 37.Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leow AD, Yanovsky I, Chiang MC, Lee AD, Klunder AD, Lu A, Becker JT, Davis SW, Toga AW, Thompson PM. Statistical properties of Jacobian maps and the realization of unbiased large-deformation nonlinear image registration. IEEE Trans Med Imaging. 2007;26:822–832. doi: 10.1109/TMI.2007.892646. [DOI] [PubMed] [Google Scholar]
- 39.D’Agostino E, Maes F, Vandermeulen D, Suetens P. A viscous fluid model for multimodal non-rigid image registration using mutual information. Med Image Anal. 2003;7:565–575. doi: 10.1016/s1361-8415(03)00039-2. [DOI] [PubMed] [Google Scholar]
- 40.Wells WM, 3rd, Viola P, Atsumi H, Nakajima S, Kikinis R. Multi-modal volume registration by maximization of mutual information. Med Image Anal. 1996;1:35–51. doi: 10.1016/s1361-8415(01)80004-9. [DOI] [PubMed] [Google Scholar]
- 41.Yanovsky I, Leow AD, Lee S, Osher SJ, Thompson PM. Comparing registration methods for mapping brain change using tensor-based morphometry. Med Image Anal. 2007;13:679–700. doi: 10.1016/j.media.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bullmore ET, Suckling J, Overmeyer S, Rabe-Hesketh S, Taylor E, Brammer MJ. Global, voxel, and cluster tests, by theory and permutation, for a difference between two groups of structural MR images of the brain. IEEE Trans Med Imaging. 1999;18:32–42. doi: 10.1109/42.750253. [DOI] [PubMed] [Google Scholar]
- 43.Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW. Dynamics of gray matter loss in Alzheimer’s disease. J Neurosci. 2003;23:994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soininen H, Partanen K, Pitkanen A, Hallikainen M, Hanninen T, Helisalmi S, Mannermaa A, Ryynanen M, Koivisto K, Riekkinen P., Sr Decreased hippocampal volume asymmetry on MRIs in nondemented elderly subjects carrying the apolipoprotein E epsilon4 allele. Neurology. 1995;45:391–392. doi: 10.1212/wnl.45.2.391. [DOI] [PubMed] [Google Scholar]
- 46.Geroldi C, Laakso MP, DeCarli C, Beltramello A, Bianchetti A, Soininen H, Trabucchi M, Frisoni GB. Apolipoprotein E genotype and hippocampal asymmetry in Alzheimer’s disease: a volumetric MRI study. J Neurol Neurosurg Psychiatry. 2000;68:93–96. doi: 10.1136/jnnp.68.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plassman BL, Welsh-Bohmer KA, Bigler ED, Johnson SC, Anderson CV, Helms MJ, Saunders AM, Breitner JC. Apolipoprotein E epsilon4 allele and hippocampal volume in twins with normal cognition. Neurology. 1997;48:985–989. doi: 10.1212/wnl.48.4.985. [DOI] [PubMed] [Google Scholar]
- 48.Barnes LL, Wilson RS, Bienias JL, Schneider JA, Evans DA, Bennett DA. Sex differences in the clinical manifestations of Alzheimer disease pathology. Arch Gen Psychiatry. 2005;62:685–691. doi: 10.1001/archpsyc.62.6.685. [DOI] [PubMed] [Google Scholar]
- 49.Bigler ED, Lowry NJ, Anderson CV, Johnson SC, Terry J, Steed M. Dementia, quantitative neuroimaging, and apolipoprotein E genotype. Am J Neuroradiol. 2000;21:1857–1868. [PMC free article] [PubMed] [Google Scholar]
- 50.Hashimoto M, Yasuda M, Tanimukai S, Matsui M, Hirono N, Kazui H, Mori E. Apolipoprotein E epsilon4 and the pattern of regional brain atrophy in Alzheimer’s disease. Neurology. 2001;57:1461–1466. doi: 10.1212/wnl.57.8.1461. [DOI] [PubMed] [Google Scholar]
- 51.Lehtovirta M, Laakso MP, Soininen H, Helisalmi S, Mannermaa A, Helkala EL, Partanen K, Ryynanen M, Vainio P, Hartikainen P, Riekkinen PJ., Sr Volumes of hippocampus, amygdala and frontal lobe in Alzheimer patients with different apolipoprotein E genotypes. Neuroscience. 1995;67:65–72. doi: 10.1016/0306-4522(95)00014-a. [DOI] [PubMed] [Google Scholar]
- 52.Pedraza O, Bowers D, Gilmore R. Asymmetry of the hippocampus and amygdala in MRI volumetric measurements of normal adults. J Int Neuropsychol Soc. 2004;10:664–678. doi: 10.1017/S1355617704105080. [DOI] [PubMed] [Google Scholar]
- 53.Gomez-Isla T, Hollister R, West H, Mui S, Growdon JH, Petersen RC, Parisi JE, Hyman BT. Neuronal loss correlates with but exceeds neurofibrillary tangles in Alzheimer’s disease. Ann Neurol. 1997;41:17–24. doi: 10.1002/ana.410410106. [DOI] [PubMed] [Google Scholar]
- 54.Peters A, Morrison JH, Rosene DL, Hyman BT. Are neurons lost from the primate cerebral cortex during normal aging? Cereb Cortex. 1998;8:295–300. doi: 10.1093/cercor/8.4.295. [DOI] [PubMed] [Google Scholar]
- 55.Peters A. Structural changes in the normally aging cerebral cortex of primates. Prog Brain Res. 2002;136:455–465. doi: 10.1016/s0079-6123(02)36038-2. [DOI] [PubMed] [Google Scholar]
- 56.Kemper T. Neuroanatomical and neuropathological changes during aging and dementia. In: Albert M, Knoefel J, editors. Clinical Neurology of Aging. 2. Oxford University Press; New York: 1994. [Google Scholar]
- 57.Tang Y, Nyengaard JR, Pakkenberg B, Gundersen HJ. Age-induced white matter changes in the human brain: a stereological investigation. Neurobiol Aging. 1997;18:609–615. doi: 10.1016/s0197-4580(97)00155-3. [DOI] [PubMed] [Google Scholar]
- 58.Marner L, Nyengaard JR, Tang Y, Pakkenberg B. Marked loss of myelinated nerve fibers in the human brain with age. J Comp Neurol. 2003;462:144–152. doi: 10.1002/cne.10714. [DOI] [PubMed] [Google Scholar]
- 59.Peters A, Rosene DL, Moss MB, Kemper TL, Abraham CR, Tigges J, Albert MS. Neurobiological bases of age-related cognitive decline in the rhesus monkey. J Neuropathol Exp Neurol. 1996;55:861–874. doi: 10.1097/00005072-199608000-00001. [DOI] [PubMed] [Google Scholar]
- 60.Peters A, Sethares C, Killiany RJ. Effects of age on the thickness of myelin sheaths in monkey primary visual cortex. J Comp Neurol. 2001;435:241–248. doi: 10.1002/cne.1205. [DOI] [PubMed] [Google Scholar]
- 61.Bartzokis G, Sultzer D, Lu PH, Nuechterlein KH, Mintz J, Cummings J. Heterogeneous age-related breakdown of white matter structural integrity: implications for cortical “disconnection” in aging and Alzheimer’s disease. Neurobiol Aging. 2004;25:843–851. doi: 10.1016/j.neurobiolaging.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 62.Kochunov P, Thompson PM, Lancaster JL, Bartzokis G, Smith S, Coyle T, Royall DR, Laird A, Fox PT. Relationship between white matter fractional anisotropy and other indices of cerebral health in normal aging: tract-based spatial statistics study of aging. Neuroimage. 2007;35:478–487. doi: 10.1016/j.neuroimage.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 63.Bartzokis G. Age-related myelin breakdown: a developmental model of cognitive decline and Alzheimer’s disease. Neurobiol Aging. 2004;25:5–18. doi: 10.1016/j.neurobiolaging.2003.03.001. [DOI] [PubMed] [Google Scholar]
- 64.Salat DH, Tuch DS, Greve DN, van der Kouwe AJ, Hevelone ND, Zaleta AK, Rosen BR, Fischl B, Corkin S, Rosas HD, Dale AM. Age-related alterations in white matter microstructure measured by diffusion tensor imaging. Neurobiol Aging. 2005;26:1215–1227. doi: 10.1016/j.neurobiolaging.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 65.Brun A, Englund E. A white matter disorder in dementia of the Alzheimer type: a pathoanatomical study. Ann Neurol. 1986;19:253–262. doi: 10.1002/ana.410190306. [DOI] [PubMed] [Google Scholar]
- 66.Hyman BT, van Hoesen GW, Kromer LJ, Damasio AR. Perforant pathway changes and the memory impairment of Alzheimer’s disease. Ann Neurol. 1986;20:472–481. doi: 10.1002/ana.410200406. [DOI] [PubMed] [Google Scholar]
- 67.Beck KD, Powell-Braxton L, Widmer HR, Valverde J, Hefti F. Igf1 gene disruption results in reduced brain size, CNS hypomyelination, and loss of hippocampal granule and striatal parvalbumin-containing neurons. Neuron. 1995;14:717–730. doi: 10.1016/0896-6273(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 68.Abraham J, Vincze A, Jewgenow I, Veszpremi B, Kravjak A, Gomori E, Seress L. Myelination in the human hippocampal formation from midgestation to adulthood. Int J Dev Neurosci. 2010;28:401–410. doi: 10.1016/j.ijdevneu.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 69.Cotel MC, Bayer TA, Wirths O. Age-dependent loss of dentate gyrus granule cells in APP/PS1KI mice. Brain Res. 2008;1222:207–213. doi: 10.1016/j.brainres.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 70.Boyles JK, Zoellner CD, Anderson LJ, Kosik LM, Pitas RE, Weisgraber KH, Hui DY, Mahley RW, Gebicke-Haerter PJ, Ignatius MJ. A role for apolipoprotein E, apolipoprotein A-I, and low density lipoprotein receptors in cholesterol transport during regeneration and remyelination of the rat sciatic nerve. J Clin Invest. 1989;83:1015–1031. doi: 10.1172/JCI113943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goodrum JF. Cholesterol from degenerating nerve myelin becomes associated with lipoproteins containing apolipoprotein E. J Neurochem. 56:2082–2086. doi: 10.1111/j.1471-4159.1991.tb03469.x. [DOI] [PubMed] [Google Scholar]
- 72.Poirier J. Apolipoprotein E, cholesterol transport and synthesis in sporadic Alzheimer’s disease. Neurobiol Aging. 2005;26:355–361. doi: 10.1016/j.neurobiolaging.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 73.Riddell DR, Zhou H, Atchison K, Warwick HK, Atkinson PJ, Jefferson J, Xu L, Aschmies S, Kirksey Y, Hu Y, Wagner E, Parratt A, Xu J, Li Z, Zaleska MM, Jacobsen JS, Pangalos MN, Reinhart PH. Impact of apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J Neurosci. 2008;28:11445–11453. doi: 10.1523/JNEUROSCI.1972-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]