Abstract
Two major bottlenecks in elucidating the structure and function of membrane proteins are the difficulty of producing large quantities of functional receptors, and stabilizing them for a sufficient period of time. Selecting the right surfactant is thus crucial. Here we report using peptide surfactants in commercial Escherichia coli cell-free systems to rapidly produce milligram quantities of soluble G protein-coupled receptors (GPCRs). These include the human formyl peptide receptor, human trace amine-associated receptor, and two olfactory receptors. The GPCRs expressed in the presence of the peptide surfactants were soluble and had α-helical secondary structures, suggesting that they were properly folded. Microscale thermophoresis measurements showed that one olfactory receptor expressed using peptide surfactants bound its known ligand heptanal (molecular weight 114.18). These short and simple peptide surfactants may be able to facilitate the rapid production of GPCRs, or even other membrane proteins, for structure and function studies.
Keywords: in vitro translation, label-free
Membrane proteins play vital roles in all living systems. Approximately 30% of genes in almost all sequenced genomes code for membrane proteins (1–3). However, our detailed understanding of membrane protein structure and function lags far behind that of soluble proteins. Indeed, as of April 2011, there are over 72,000 structures in the Protein Data Bank. Of these, only 280 are unique membrane proteins, and only six are unique G protein-coupled receptors (GPCRs). This surprising disparity is due to bottlenecks at nearly every stage of experimentation, from large-scale membrane protein production to X-ray crystal diffraction.
Recent advances have overcome several bottlenecks in studying membrane proteins. Commercial development of nanoliter drop-setting robots and a wide variety of kits have made crystal screening less laborious. The development of worldwide accessible synchrotron facilities, and microfocus beamlines capable of collecting data from crystals < 10–60 μm, have overcome the bottlenecks in data collection. Likewise, rapid advancements in computing power and an increase in open access software development have made the determination of structures a much less daunting task. However, inexpensive large-scale production of soluble and nonaggregated membrane proteins still remains a formidable challenge. Likewise, systematic surfactant screens still remain one of the most time-consuming and expensive experimental tasks. To overcome these bottlenecks, the discovery or invention of simple and broadly useful surfactants is crucial.
Several cell-based membrane protein production systems have been developed, but they are costly and require months to generate sufficient quantities of protein. Commercial cell-free systems are alternative methods of producing membrane proteins. However, to produce nonaggregated membrane proteins, optimal selection of surfactants is critical: The newly produced membrane proteins must not only fold correctly, they must also remain soluble and biologically functional. Until now, finding an appropriate surfactant for specific membrane proteins has been laborious, because even highly related proteins may react differently to the same detergent. It would be advantageous if a class of simple surfactants could be used for stabilizing diverse membrane proteins.
We previously reported the invention of a class of short peptide surfactants (4–8). We also reported that these peptides effectively stabilized the transmembrane protein glycerol-3-phosphate dehydrogenase (9), the photosystem-I protein complex (10, 11), and the G protein-coupled receptor bovine rhodopsin (12). We recently asked if these peptides could also be used in cell-free production of membrane proteins, particularly bioengineered olfactory receptors (13), trace amine-associated receptors (14), and formyl peptide receptors (15–17).
Here we report using commercially available Escherichia coli cell-free kits with our peptide surfactants (Fig. 1) to rapidly produce four GPCRs in milligram quantities. The surfactant property of the peptides was necessary to yield soluble GPCRs. They not only solubilized and stabilized the GPCRs, they also facilitated the proper folding of these receptors. Moreover, the cell-free produced olfactory receptor mOR103-15 was able to bind its odorant ligand heptanal [molecular weight (MW) 114.18], suggesting that the receptor is biologically functional. Our results indicate that these short and simple peptide surfactants may be a general class of surfactants that can facilitate the production of a wider range of GPCRs, and perhaps other membrane proteins.
Fig. 1.
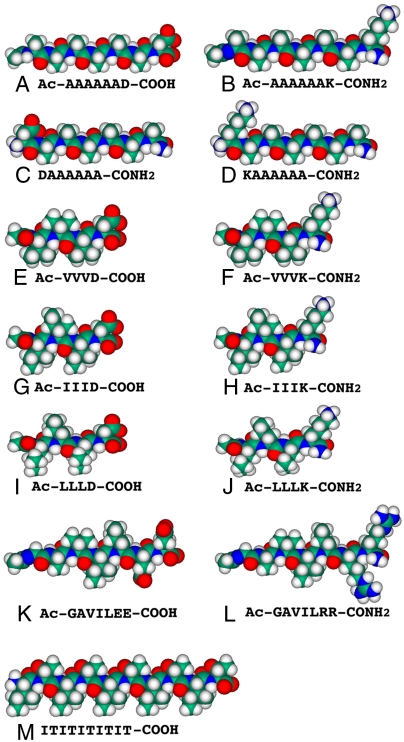
Molecular models of the peptide surfactants and nonsurfactant peptide at pH 7.5. The peptide sequences are under each molecular model. Aspartic acid (D) and glutamic acid (E) are negatively charged whereas arginine (R) and lysine (K) are positively charged. The hydrophobic tails of the peptide surfactants consist of glycine (G), alanine (A), valine (V), isoleucine (I), and leucine (L). Each peptide surfactant is approximately 2–2.5 nm long, similar to biological phospholipids. Color code: teal, carbon; red, oxygen; blue, nitrogen; and white, hydrogen.
Results
We previously achieved expression of olfactory receptors hOR17-4, mOR23, and mS51, using the Roche wheat germ cell-free protein production technology (18). However, the Roche system is expensive and requires a special apparatus to use, making it unaffordable for most laboratories. To attract more people to study membrane proteins, a rapid, affordable, and commercially available production method is needed. In this study, we were able to produce milligram quantities of four GPCRs using commercial E. coli cell-free systems. The receptors include the olfactory receptors (ORs) hOR17-210 and mOR103-15, the human formyl peptide receptor 3 (hFPR3) (15–17), and the human trace amine-associate receptor 5 (hTAAR5) (14).
The Effect of Peptide Surfactants for Cell-Free GPCR Expression and Solubility.
We systematically screened 12 peptide surfactants with hOR17-210, mOR103-15, hFPR3, and hTAAR5 in a commercial E. coli cell-free expression system. Western blotting was used to quantify protein expression. Fig. 2 shows the results for hFPR3 and hOR17-210. Very little soluble protein was produced without a peptide surfactant. Although high yields were consistently achieved with several peptides, the total yield depended on both the specific protein and the specific peptide. For example, the peptide Ac-GAVILRR-NH2 consistently yielded low levels of receptor, whereas high levels were consistently achieved with the peptide Ac-A6D-OH. In contrast, receptor yields with Ac-I3K-NH2 and Ac-A6K-NH2 were variable. Low expression levels in the presence of the nonsurfactant peptide (IT)5 demonstrate the necessity of using a surfactant with the GPCRs. However, the low expression levels in the presence of several surfactant peptides indicate that a surfactant character for the peptides is necessary, but not sufficient.
Fig. 2.
Western blotting detection of cell-free produced GPCRs. After the reactions, the samples were centrifuged. The supernatant containing the solubilized protein was removed, and the pellet was resuspended in an equivalent volume of buffer. The solubilized protein and resuspended pellets were detected by Western blotting. As controls, reactions with no peptide or the nonsurfactant peptide (IT)5 were performed. (A) Western blot of solubilized hFPR3 in the presence of different peptides. (B) Western blot of solubilized hOR17-210 produced in the presence of different peptides. Similar results were obtained with hTAAR5 and mOR103-15.
Fig. 3 shows that the peptide surfactants solubilized the tested GPCRs, but with varying effectiveness. This efficacy depended on both the peptide and the specific GPCR. The receptors hFPR3 and hTAAR5 consistently had 60–90% of the total synthesized monomer solubilized, whereas hOR17-210 and mOR103-15 were highly soluble in the presence of only three peptides. The peptides KA6-NH2, Ac-A6K-NH2, and Ac-V3K-NH2 were able to solubilize more than 70% of at least three GPCRs, whereas Ac-A6K-NH2 and Ac-A6D-OH were able to solubilize 80–90% of two GPCRs. The peptides Ac-GAVILRR-NH2, DA6-NH2, and Ac-I3D-OH were least efficient at solubilizing the GPCRs, particularly the olfactory receptors. The controls demonstrated that the presence of a surfactant was necessary for GPCR solubilization. Interestingly, the peptide (IT)5, which has alternating hydrophobic isoleucine and hydrophilic serine residues, was also able to solubilize some proteins. However, unlike the peptide surfactants, it seemed to inhibit protein synthesis.
Fig. 3.
Comparison of the solubility of four GPCRs in 12 different peptide surfactants. Each GPCR was expressed in the presence of surfactant peptides using an E. Coli cell-free protein expression system. Upon completion, reactions were centrifuged to separate solubilized protein from aggregate. The soluble fraction was removed, and the pellet was resuspended in an equivalent volume of buffer. Solubilized and nonsolubilized protein fractions were assayed using a Western blot; relative band intensities were used to calculate the percentage of solubilized protein. As controls, reactions with no peptide or with the nonsurfactant peptide (IT)5 were performed. The presence of any peptide increased the fraction of solubilized protein. However, significant quantities of solubilized protein were only obtained in the presence of surfactant peptides.
The Maximum Yields of Cell-Free Produced GPCRs.
Western blotting was used to estimate protein yields from the cell-free reactions using a known protein concentration as a standard reference. Fig. 4 shows that cell-free synthesis in the presence of peptide surfactants can yield a few milligrams of receptor per 10 mL reaction, and may thus be a good alternative for large-scale membrane protein production. However, as with the solubilization results, the protein yield depends on both the specific GPCR and the specific peptide surfactant used (Fig. 4 and Table S1). At least approximately 2 mg of three purified GPCRs could be produced in a 10 mL reaction with three of the peptide surfactants. The peptide Ac-A6D-OH yielded the largest amounts of protein, with a maximum yield of approximately 4.8 mg for mOR103-15. Five other peptides were able to exceed 2.5 mg of purified receptors per 10 mL reaction for at least one GPCR: Ac-I3D-OH, Ac-L3K-NH2, Ac-V3D-OH, Ac-V3K-NH2, and Ac-GAVILEE-OH. The peptide Ac-GAVILRR-NH2 consistently gave low protein yields for all receptors except hTAAR5. Overall, at least 2 mg of each purified receptor could be synthesized in a 10 mL reaction in the presence of at least one of the peptide surfactants. The control experiments demonstrate that the presence of a peptide is necessary for protein synthesis. The (IT)5 control further indicates that surfactant properties are necessary for sufficient GPCR yields.
Fig. 4.
The maximum expected yields of soluble receptors produced in the presence of peptide surfactants. (A) The maximum expected yields of solubilized monomer for each GPCR in the presence of each peptide or control condition. To determine the expected yields, solubilized protein and protein with a known concentration were assayed on a Western blot. The relative intensities of the known protein sample and the test samples were used to calculate the maximum protein yields. Surfactant peptides increased the yield of the solubilized monomeric form of the tested GPCRs. (B) The maximum yield of the monomeric form of each tested GPCR in a 10 mL reaction. Results from the most effective surfactant peptide are shown; it is compared to the maximum expected yield without peptide, or with a nonsurfactant peptide. The yield for each receptor varies with peptide surfactants used.
Our results show that the presence of peptide surfactants is necessary to achieve both high solubility and high protein yields (Figs. 3 and 4). They further suggest that the peptide surfactants may have a preference for different receptors, perhaps due to the nature of their composition and structure. For example, the different yields in Ac-A6D-OH vs. DA6-NH2, or Ac-A6K-NH2 vs. KA6-NH2 suggest that the ionic character of the peptide is important. Our results suggest that ionic peptides yield more protein than zwitterionic peptides when the head group is negatively charged. However, when the head group is positively charged, a trend with the tested GPCRs is not visible. It is also possible that the peptide batch or structural phase can affect their ability to act as surfactants. Several investigators have observed that the peptides form dynamic structures in aqueous phase, changing between micelles, vesicles, nanotubes, and other structures (4–8, 19, 20). It is possible that different mesoscale peptide phases affect protein production and solubility, and more research is required to elucidate these effects. Because a large number of diverse peptide surfactants can be designed, it may be possible to engineer a peptide that works well for a specific receptor.
Secondary Structural Analysis Using CD.
CD was used to verify that the cell-free produced receptors were properly folded, and that the peptide surfactants did not significantly affect the receptors’ structures. In these experiments, the GPCRs were purified with fos-choline-14 (FC-14) because the peptide CD signal interferes with and overwhelms the receptor signals, and also because FC-14 has successfully been used with diverse GPCRs (21–24). Fig. 5 shows the CD spectra of each receptor. These purified receptors have characteristic α-helical structures, with signature minima at 220 and 208 nm. Because GPCRs are expected to have seven transmembrane helical domains, these CD spectra indicate that the receptors are properly folded.
Fig. 5.
CD spectra of four GPCRs produced in the presence of peptide surfactants using a commercial E. coli cell-free system. (A) hFPR3 produced in Ac-VVVK-CONH2. (B) hTAAR5 produced in Ac-IIIK-CONH2. (C) mOR103-15 produced in Ac-VVVD-OH. (D) hOR17-210 produced in Ac-AAAAAAD-OH. These receptors all have characteristic α-helical spectra with valleys at 208 and 222 nm. Because GPCRs have 7-transmembrane α-helical domains, the CD spectra indicate that the receptors are properly folded.
Ligand Binding Measurements of Purified GPCRs Using Microscale Thermophoresis.
Next, we tested whether the purified GPCRs were able to bind to their ligands. Microscale thermophoresis, a technique, which uses temperature gradients to measure ligand binding, was used because it can detect interactions between small ligands and large receptors in free solution (25–27). Because odorants are less than 300 Da whereas their receptors are greater than 300 kDa, mass-based measurements like quartz crystal microbalance cannot be used. As negative controls, boiled receptors were used to rule out the possibility of nonspecific binding or surfactant interference.
Fig. 6 shows that peptide-produced mOR103-15 bound its ligand heptanal (MW 114.18), whereas the boiled control did not. The peptide surfactant-produced sample displayed a typical sigmoidal binding curve, with a half maximal effective concentration (EC50) of approximately 0.9 µM. In contrast, the boiled control did not show any significant differences between low and high ligand concentrations. These results suggest that peptide surfactants can be used to produce soluble and functional GPCRs.
Fig. 6.
Microscale thermophoresis ligand binding measurements of mOR103-15 produced in Ac-VVVD-OH. (A) Peptide-produced mOR103-15 has a sigmoidal shape, suggesting that the receptor binds to its ligand heptanal (MW 114.18). The measured EC50 is approximately 0.9 µM. (B) The boiled control showed fluctuations around a centerline, suggesting that binding observed with the normal receptor is not a result of nonspecific binding to the surfactant or denatured receptor. All curves were normalized to the fraction of bound receptor. Open circles show the mean measurements from three experiments; the lines through the points are the best-fit curves using the Hill equation.
Discussion
Peptide Surfactants for Membrane Protein Studies.
Surfactants play an indispensible role in the study of membrane proteins, as their presence is necessary to solubilize and stabilize these proteins outside of their native membrane environment. The choice of surfactant is crucial. Previous studies showed that the most successfully used surfactants in crystallization experiments have short hydrophobic tails about 6–12 carbons in length (28). However, the surfactants that are often best for crystallization can destabilize some membrane proteins. Careful surfactant screening of individual membrane proteins is thus necessary.
Surfactant peptides are a class of molecules with properties similar to those of surfactants that have successfully been used for crystallization. They have hydrophilic heads comprised of 1–2 residues, and hydrophobic tails 3–6 residues long. They are about 2–3 nm in length, and their ionic character and strength can be controlled by selecting appropriate amino acids or by capping the termini. Additionally, they have shown potential for use with membrane proteins (9–12). They have been used to solubilize glycerol-3-phosphate dehydrogenase, and to stabilize bovine rhodopsin and subunits of the photosystem I protein complex (9–12).
To evaluate the potential of peptide surfactants to be used with membrane proteins, we screened four GPCRs with 12 different peptides. Our current study suggests that these peptides may represent a class of surfactants that can aid in the solubilization and stabilization of GPCRs. The milligram protein yields we obtained further suggest that cell-free protein synthesis performed in the presence of peptide surfactants is a rapid, efficient, and relatively cost-effective means of producing the milligram quantities of membrane proteins necessary for structure and function studies. CD spectroscopy measurements of the GPCRs showed that the peptides facilitate proper protein folding, whereas microscale thermophoresis indicates that the peptide-produced proteins are functional.
Perspective.
The studies reported here suggest that peptide surfactants are versatile, and can be used with a wider range of GPCRs. Their ease of use with commercially available protein production kits further underscores their value: They can now be used to produce milligram quantities of properly folded and functional GPCRs. Future experiments need to be performed to determine whether a single peptide or a cocktail of several peptides are optimal surfactants for a given membrane protein. Future experiments are also needed to determine whether these surfactants can be useful for membrane protein crystallization. Our studies and previous research (9–12) suggest that peptide surfactants are a promising material not only for membrane protein studies, but also in the development of GPCR-based nanodevices.
Materials and Methods
Peptide Design and Synthesis.
Peptide surfactants are comprised of a hydrophilic head and a hydrophobic tail. Three kinds of surfactant peptides were designed and used in this study: cationic, anionic, and zwitterionic. The ionic nature of the peptides was controlled by acetylation at the N terminus, amidation at the C terminus, or both. It was further controlled by choosing positively charged lysine or negatively charged asparatic acid for the head group. The length of hydrophobic tail ranged from three to six amino acids. As a control, the nonsurfactant peptide (IT)5 was also designed and used. Molecular models of the surfactant peptides and control are shown in Fig. 1.
All peptides were commercially synthesized and purified by CPC Scientific, Inc. The peptides were received as a powder and dissolved in Milli-Q water before use. Each peptide suspension was sonicated, and the pH value was adjusted above 7.0 with NaOH or HCl solutions to increase peptide solubility. The suspension was then filtered through a 0.22 μm filter to remove insoluble particles. The filtered peptide solution was kept at room temperature (RT) as a stock solution.
Determination of CACs of Peptide Surfactants.
To determine the critical aggregation concentration (CAC) of the peptides, two methods were used: electrical conductivity and surface tension. For each peptide surfactant, 30 serial dilutions were prepared in Milli-Q water (resistivity > 18 MΩ·cm). The CACs of the cationic peptide surfactants (Ac-AAAAAAK-CONH2, KAAAAAA-CONH2, Ac-VVVK-CONH2, Ac-IIIK-CONH2, Ac-LLLK-CONH2, and Ac-GAVILRR-CONH2) were determined by electrical conductivity measurements using a conductivity meter (model DDS-307) at 25.0 + 0.1 °C. Because of their relatively low solubility in Milli-Q water, the anionic and zwitterionic peptide solutions (Ac-AAAAAAD, Ac-VVVD, Ac-IIID, Ac-LLLD, Ac-GAVILEE, and DAAAAAA-CONH2) were adjusted to pH 9.0 before surface tension measurements were employed to determine their CACs. The surface tension measurements were carried out with the EasyDyne tensiometer (Kruss) at 25.0 ± 0.1 °C by using the Wilhelmy plate method. The values of surface tension γ were determined after a period of 10 min to ensure equilibration conditions. The CACs are shown in Table S2.
GPCR DNA Template Design and Construction.
Protein sequences of the following four GPCRs were obtained from the National Center for Biotechnology Information online database: hOR17-210 (UniProt release no. Q8WZA6), mOR103-15 (NP release no. 035113.1), hTAAR5 (NP release no. 003958.2), and hFPR3 (NP release no. 002021.3). To adapt the GPCRs for detection and purification, the rho1D4 epitope tag (TETSQVAPA) was added to their C termini. Codons of the engineered GPCRs were optimized for expression in E. coli. The genes were then commercially synthesized and subcloned into the pIVEX2.3d vector (Roche Diagnostics Corp.) by GENEART. The final constructs were verified by DNA sequencing and were used for all subsequent studies.
Cell-Free GPCR Production.
E. coli-based cell-free expression kits were purchased from Invitrogen (catalog no. K9900-97, Qiagen 32506). To compensate for the lack of a natural membrane, surfactant peptides were added directly to the reactions. The experimental concentrations of each peptide surfactant for each GPCR are shown in Table S2. A final reaction volume of 25 μL was used for all screens.
Systematic Screen of Peptide Surfactants.
Six pairs of surfactant peptides were used in the cell-free production of GPCRs. As a control, the nonsurfactant peptide (IT)5 was also used. The surfactant peptides were all tested at concentrations above their determined or estimated CACs. Cell-free reactions were performed according to the manufacturers instructions. Briefly, plasmid DNA, peptide, and the reaction reagents were incubated at 33 °C and 260 rpm for 30 min. A feed buffer was then added, and the reaction was incubated for an additional 90 min. The samples were then centrifuged at 10,000 rpm for 5 min. The supernatant containing the solubilized protein was removed, and the pellet was resuspended in an equivalent volume of PBS. The quantity of each protein produced and solubilized was determined with Western blotting.
Western Blotting Detection of rho1D4 Tagged GPCRs.
Samples were prepared and loaded in Novex 10% Bis-Tris SDS-PAGE gels (Invitrogen) according to the manufacturer’s protocol, with the exception that the samples were incubated at RT prior to loading as boiling causes membrane protein aggregation. The Full-Range Rainbow Molecular Weight Marker (GE Healthcare) was loaded as the protein size standard. After the samples were resolved on the SDS-PAGE gels, they were transferred to a 0.45 μm nitrocellulose membrane, blocked in milk [5% nonfat dried milk in Tris-buffered saline plus Tween-20 (TBST), 1 h, RT], and incubated with a rho1D4 primary antibody (1∶3,000 in TBST, 1 h, RT). The GPCRs were then detected with a goat anti-mouse HRP-conjugated secondary antibody (Pierce) (1∶5,000 in TBST, 1 h, RT) and visualized using the ECL Plus kit (GE Healthcare). Western blotting images were captured using a FluorChem gel documentation system (Alpha Innotech).
Immunoaffinity Purification.
We used the rho1D4 monoclonal antibody (Cell Essentials) chemically linked to CNBr-activated Sepharose 4B beads (GE Healthcare). The rho1D4 elution peptide Ac-TETSQVAPA-CONH2 was synthesized by CPC Scientific, Inc. Solubilized protein from the cell-free reactions was mixed with a rho1D4-coupled sepharose bead slurry (binding capacity 0.7 mg/mL) and rotated overnight at 4 °C to capture the GPCRs. The beads were pelleted by centrifugation at 1,400 g for 1 min. The beads were then washed with wash buffer (PBS + 0.2% FC-14) until spectrophotometer readings indicated that all excess protein had been removed (mg/mL < 0.01). The captured GPCRs were eluted with elution buffer (PBS + 0.2% FC-14+800 μM elution peptide). Elutions were performed until spectrophotometer readings indicated no more protein was present (mg/mL < 0.01). The protein was then concentrated using a 30 kDa molecular weight cut-off filter column (Millipore) and its concentration was measured using the NanoDrop 1000 spectrophotometer (Thermo Scientific).
Secondary Structure Analysis by CD Spectroscopy.
Spectra were recorded on a CD spectrometer (Aviv Associates model 410) at 15 °C over the wavelength range of 195 to 250 nm with a step size of 1 nm and an averaging time of 4 s. Spectra for purified GPCRs were blanked to wash buffer. Spectra were collected with a 111-QS quartz sample cell (Hellma) with a path length of 1 mm. The spectra were smoothed using an averaging filter with a span of five.
Microscale Thermophoresis Ligand Binding Measurements.
Binding interactions between the purified mOR103-15 and the ligand heptanal (MW 114.18) were measured using microscale thermophoresis as described (24–27). Briefly, for each tested receptor, a titration series with constant receptor concentration and varying ligand heptanal (MW 114.18) concentrations was prepared in a final solution of 10% DMSO and 0.2% FC-14 in PBS. Approximately 1.5 μL of each sample was loaded in a fused silica capillary (Polymicro Technologies). An infrared laser diode was used to create a 0.12 K/μm temperature gradient inside the capillaries [Furukawa FOL1405-RTV-617-1480, wavelength λ = 1,480 nm, 320 mW maximum power (AMS Technologies AG)]. Tryptophan fluorescence was excited with an ultraviolet light emitting diode (285 nm), and was measured with a 40x SUPRASIL synthetic quartz substrate microscope objective, numerical aperture 0.8 (Partec). The local receptor concentration in response to the temperature gradient was detected with a photon counter photomultiplier tube P10PC (Electron Tubes, Inc.). All measurements were performed at RT. The Hill equation (n = 2) was fit to the data to determine the EC50 value for each sample; the EC50 value is the concentration at which half of the GPCR sample is bound to its ligand.
Supplementary Material
Acknowledgments.
We thank members of Zhang lab for stimulating discussions. This work is supported in part by Defense Advanced Research Projects Agency HR0011-09-C-0012, the Center for NanoScience and the NanoSystems Initiative Munich. X.W. is also supported by a fellowship from China University of Petroleum (East China).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018185108/-/DCSupplemental.
References
- 1.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loll PJ. Membrane protein structural biology: The high throughput challenge. J Struct Biol. 2003;142:144–153. doi: 10.1016/s1047-8477(03)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Nilsson J, Persson B, von Heijne G. Comparative analysis of amino acid distributions in integral membrane proteins from 107 genomes. Proteins. 2005;60:606–616. doi: 10.1002/prot.20583. [DOI] [PubMed] [Google Scholar]
- 4.Vauthey S, Santoso S, Gong H, Watson N, Zhang S. Molecular self-assembly of surfactant-like peptides to form nanotubes and nanovesicles. Proc Natl Acad Sci USA. 2002;99:5355–5360. doi: 10.1073/pnas.072089599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santoso S, Hwang W, Hartman H, Zhang S. Self-assembly of surfactant-like peptides with variable glycine tails to form nanotubes and nanovesicles. Nano Lett. 2002;2:687–691. [Google Scholar]
- 6.von Maltzahn G, Vauthey S, Santos S, Zhang S. Positively charged surfactant-like peptides self-assemble into nanostructures. Langmuir. 2003;19:4332–4337. [Google Scholar]
- 7.Nagai A, Nagai Y, Qu H, Zhang S. Self-assembling behaviors of lipid-like peptides A6D and A6K. J Nanosci Nanotechnol. 2007;7:2246–2252. doi: 10.1166/jnn.2007.647. [DOI] [PubMed] [Google Scholar]
- 8.Khoe U, Yang Y, Zhang S. Self-assembly of nano-donut structure from coneshaped designer lipid-like peptide surfactant. Langmuir. 2009;25:4111–4114. doi: 10.1021/la8025232. [DOI] [PubMed] [Google Scholar]
- 9.Yeh JI, Du S, Tordajada A, Paulo J, Zhang S. Peptergent: Peptide detergents that improve stability and functionality of a membrane protein glycerol-3-phosphate dehydrogenase. Biochemistry. 2005;44:16912–16919. doi: 10.1021/bi051357o. [DOI] [PubMed] [Google Scholar]
- 10.Kiley P, Zhao X, Bruce BD, Baldo M, Zhang S. Self-assembling peptide detergents stabilize isolated photosystem I on a dry surface for an extended time. PLoS Biol. 2005;3:1181–1186. doi: 10.1371/journal.pbio.0030230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto K, Koutsopoulos S, Vaughn M, Bruce BD, Zhang S. Designer lipid-like peptide surfactants stabilize functional photosystem I membrane complex in solution. J Phys Chem. 2009;115:75–83. doi: 10.1021/jp8021425. [DOI] [PubMed] [Google Scholar]
- 12.Zhao X, et al. Designer lipid-like peptides significantly stabilize G protein-coupled receptor bovine rhodopsin. Proc Natl Acad Sci USA. 2006;103:17707–17712. doi: 10.1073/pnas.0607167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaillard I, Rouquier S, Giorgi D. Olfactory receptors. Cell Mol Life Sci. 2004;61:456–469. doi: 10.1007/s00018-003-3273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindemann L, et al. Trace amine-associated receptors form structurally and functionally distinct subfamilies of novel G protein-coupled receptors. Genomics. 2005;85:372–385. doi: 10.1016/j.ygeno.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Migeotte I, Communi D, Parmentier M. Formyl peptide receptors: A promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine Growth Factor Rev. 2006;17:501–519. doi: 10.1016/j.cytogfr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Bao L, Gerard NP, Eddy RL, Jr, Shows TB, Gerard C. Mapping of genes for the human C5a receptor (C5AR), human FMLP receptor (FPR), and two FMLP receptor homologue orphan receptors (FPRH1, FPRH2) to chromosome 19. Genomics. 1992;13:437–440. doi: 10.1016/0888-7543(92)90265-t. [DOI] [PubMed] [Google Scholar]
- 17.Rivière S, Challet L, Fluegge D, Spehr M, Rodriguez I. Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature. 2009;459:574–577. doi: 10.1038/nature08029. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser L, et al. Efficient cell-free production of olfactory receptors: Detergent optimization, structure, and odorant binding analyses. Proc Natl Acad Sci USA. 2008;105:15726–15731. doi: 10.1073/pnas.0804766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, et al. Molecular self-assembly and applications of designer peptide amphiphiles. Chem Soc Rev. 2010;39:3480–3498. doi: 10.1039/b915923c. [DOI] [PubMed] [Google Scholar]
- 20.Yaghmur A, Laggner P, Zhang S, Rappolt M. Tuning curvature and stability of monoolein bilayers by designer lipid-like peptide surfactants. PLoS One. 2007;2:e479. doi: 10.1371/journal.pone.0000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cook BL, Ernberg KE, Chung H, Zhang S. Study of a synthetic human olfactory receptor 17-4: Expression and purification from an inducible mammalian cell line. PLoS One. 2008;3:e2920. doi: 10.1371/journal.pone.0002920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cook BL, et al. Large-scale production and study of a synthetic G protein-coupled receptor: Human olfactory receptor 17-4. Proc Natl Acad Sci USA. 2009;106:11925–11930. doi: 10.1073/pnas.0811089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren H, et al. High-level production, solubilization, and purification of synthetic human GPCR chemokine receptors CCR5, CCR3, CXCR4, and CX3CR1. PLoS One. 2009;4:e4509. doi: 10.1371/journal.pone.0004509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leck K-J, Zhang S, Hauser CAE. Study of bioengineered zebra fish olfactory receptor 131-2: Receptor purification and secondary structure analysis. PLoS One. 2010:e15027. doi: 10.1371/journal.pone.0015027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duhr S, Braun D. Why molecules move along a temperature gradient. Proc Natl Acad Sci USA. 2009;103:19678–19682. doi: 10.1073/pnas.0603873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baaske P, Wienken CJ, Reineck P, Duhr S, Braun D. Optical thermophoresis for quantifying the buffer dependence of aptamer binding. Angew Chem Int Ed Engl. 2010;49:2238–2241. doi: 10.1002/anie.200903998. [DOI] [PubMed] [Google Scholar]
- 27.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1 doi: 10.1038/ncomms1093. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 28.le Maire M, Champeil P, Moller JV. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim Biophys Acta. 2000;1508:86–111. doi: 10.1016/s0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.