Abstract
Voltage-dependent sodium channels are believed to have evolved from calcium channels at the origin of the nervous system. A search of the genome of a single-celled choanoflagellate (the sister group of animals) identified a gene that is homologous to animal sodium channels and has a putative ion selectivity filter intermediate between calcium and sodium channels. Searches of a wide variety of animal genomes, including representatives of each basal lineage, revealed that similar homologs were retained in most lineages. One of these, the Placozoa, does not possess a nervous system. We cloned and sequenced the full choanoflagellate channel and parts of two placozoan channels from mRNA, showing that they are expressed. Phylogenetic analysis clusters the genes for these channels with other known sodium channels. From this phylogeny we infer ancestral states of the ion selectivity filter and show that this state has been retained in the choanoflagellate and placozoan channels. We also identify key gene duplications and losses and show convergent amino acid replacements at important points along the animal lineage.
Keywords: eumetazoan, inactivation gate, pore motif
Early animals radiated explosively in the Precambrian (1). This radiation was facilitated by the previous evolution of genes for cell adhesion that presaged the evolution of multicellularity (2). Another key animal innovation was the nervous system, which is present in all but a few animals (i.e., sponges and placozoans). Rapid, specific, long-distance communication among excitable cells is achieved in bilaterian animals and a few jellyfish (cnidarians) through the use of action potentials (APs) in neurons generated by voltage-dependent sodium (Nav) channels. Voltage-dependent calcium (Cav) channels evolved in single-celled eukaryotes and were used for intracellular signaling. It has been hypothesized that Nav channels were derived from Cav channels at the origin of the nervous system (3), thereby conferring the ability to conduct action potentials without interfering with intracellular calcium. This view was reinforced by the apparent lack of sodium currents in sponges (4).
To test this hypothesis, we searched newly available genome databases from two animals with simple nerve nets (the sea anemone Nematostella vectensis and the ctenophore Mnemiopsis leidyi), a placozoan with no nervous system (Trichoplax adhaerens), a sponge (Amphimedon queenslandica), a single-celled eukaryote (the choanoflagellate, Monosiga brevicollis), as well as fungi and additional single-celled eukaryotes for homologs of Cav and Nav channels. We then verified the expression of these genes in M. brevicollis and T. adhaerens and examined amino acid changes in these genes throughout the history of animal evolution.
Choanoflagellates are widely distributed unicellular protists (5, 6) that form the sister group to the multicellular animals (7). Placozoans are an early-diverging animal lineage that has been proposed to be sister to the eumetazoans, that is, to all animals with nervous systems (8). However, phylogenetic placement of the basal animal lineages is not yet fully resolved (9–12), and many aspects of placozoan life cycles remain unknown (13–15). Choanoflagellates and placozoans have received considerable attention due to their possession of numerous genes once thought to be exclusive to eumetazoans (2, 16–18).
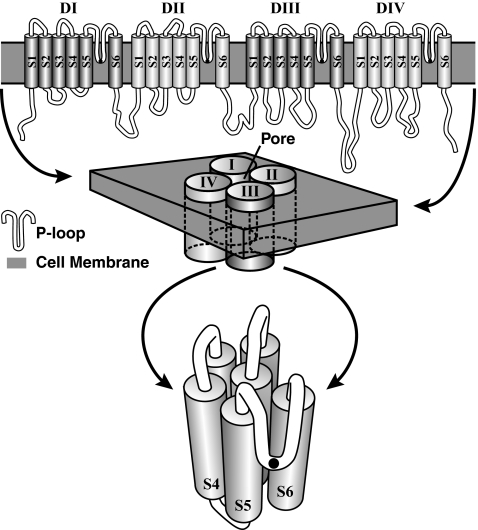
Cav and Nav channels have four domains, each of which has a pore loop (Fig. 1). A single amino acid at the deepest part of each pore loop is responsible for ion selectivity in the pore. Cav channels have acidic residues (E and D) in the pore of domains I–IV (usually E/E/E/E or E/E/D/D). Selectivity for sodium, on the other hand, is based on the residues D/E/K/A in the pore. Sodium channels also have a cytoplasmic loop between their third and fourth domains that swings up and occludes the channel pore just milliseconds after activation (Fig. 1). This fast inactivation makes sodium signaling reliable on the millisecond time scale, and mutations at this region in human Nav channel genes cause many well-known pathologies (19). Calcium channels do not have a similar motif at the homologous region. Because of the differences in the amino acids responsible for ion selectivity, and because proteins are likely to be under strong evolutionary constraints along every point of their evolution (20), it has been suggested that channels with intermediate pore sequences may exist in extant taxa (3, 21), and some invertebrate channels have been proposed as representatives of these intermediate states (21, 22). The phylogenetic relationships of these channels are not clear however (22–24), and no suggestion of an ancestral metazoan pore state has been put forth.
Fig. 1.
Hypothetical secondary structure of a sodium-channel protein. (Top) Transmembrane domains (DI–DIV), their component segments (S1–S6), and their connecting loops (in white). The pore loops (P loop), which dip down into the membrane, form the ion-selectivity filter. The inactivation gate resides on the long loop between DIII/S6 and DIV/S1. (Middle) How the domains cluster to form the protein and its pore. (Bottom) Fine structure of one of the domains with the pore loop in the foreground. The black dots on the pore loops in the Top and Bottom represent the location of the amino acids, which makes up the pore motif.
Our objective was to find voltage-gated ion-channel genes in basal animals and their close unicellular relatives, determine whether the genes are expressed in a few key species, and analyze the evolutionary history of the genes for Nav and Cav channels. We examined pore motifs, inactivation gate sequence, and inactivation gate secondary structure and then mapped these states onto our phylogeny. This work provides a unique view of Nav and Cav channel evolution and the evolution of excitable tissues in animals.
Results
Sodium Channel Homologs in Early-Diverging Animals and Choanoflagellates.
We found that the genomes of M. brevicollis, T. adhaerens, N. vectensis, and M. leidyi contain genes for ion channels that group with the Nav family (Fig. 2), and we used these genomic sequences as references for further analyses. We found pairs of Nav paralogs in Trichoplax, Nematostella, and Mnemiopsis, which we name α and β. The genome of the sponge A. queenslandica did not contain Nav homologs but did have one gene for a Cav channel. No Nav homologs were found in the genomes of Aspergillus niger, Saccharomyces cerevisiae, or any other fungi in the Joint Genome Institute database. We sequenced the entire ORF of an mRNA transcript from Monosiga and partial transcripts from the two genes in Trichoplax, thereby demonstrating that these genes are expressed. The genes have a pore motif D/E/E/A that is intermediate between Cav and Nav channels and is the same as some previously described invertebrate channels (21, 23).
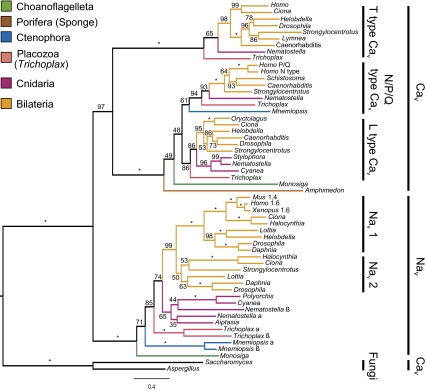
Fig. 2.
Maximum likelihood phylogeny of Nav and Cav channels. Bootstrap scores are indicated on branches, with stars indicating scores of 100%. Clades corresponding to major ion channel groups are detailed on the Right.
For one of the paralogs, Trichoplax β, only three of the four domains typical to Cav and Nav channels were found in the genome, likely due to a problem with the genome assembly. It is unlikely that a three-domain protein could function as an ion channel alone, but it is not yet known whether the genome sequencing effort simply missed part of the genome, whether our BLAST analysis misidentified the exons for the last domain, or whether it is actually a splice variant or some other regulatory transcript. The ctenophore Nav homologs and the sponge Cav channel are missing amino acids in the putative pore regions, perhaps also due to incomplete assembly.
Four overlapping segments from choanoflagellate mRNA were compiled to yield 4,589 nucleotides, which we believe includes the whole ORF. This sequence was 93.4% identical to the reference sequence obtained with BLAST (SI Materials and Methods and Fig. S1).
Sequencing of the Trichoplax genes yielded 868 bp from the Trichoplax α gene, which aligned to the reference with 92.7% identity, and 1,062 bp from the Trichoplax β gene, which aligned with 91.0% identity. Many of the mismatches in the Trichoplax β segment are from indeterminate nucleotides and may be due to the fact that this segment was sequenced directly from the PCR products rather than from cloned genes. Although further confirmation of the exact sequences is needed, the presence of these sequences in the mRNA demonstrates that both Trichoplax genes are indeed transcribed.
Phylogenetic Analyses.
We performed maximum likelihood (ML) analyses on a data set consisting of our sequenced choanoflagellate gene, a putative Cav gene from Monosiga, and Nav and Cav genes from all major animal lineages and two fungal species, Aspergillus and Saccharomyces (Fig. 2). The phylogenetic placement of the ion channel genes agrees with the well supported parts of the phylogeny for animals, choanoflagellates, and fungi (8, 9, 12), and the topology was robust to analyses on other platforms and removal of taxa (SI Materials and Methods). The placement of the Amphimedon Cav channel as basal to Monosiga Cav (Fig. 2) is probably an artifact due to long-branch attraction (LBA). This seems likely because the Amphimedon branch is long, and Monosiga Cav is a partial sequence. The placement of these two sequences within Cav channels is not consistent with an LBA artifact, however, and is strongly supported by bootstrap analysis, indicating that these are true Cav channels. The fungal Cav channels were resolved as the sister group to all animal and choanoflagellate channels. These results support the hypothesis that Nav genes evolved from Cav genes, because the Nav family emerges from within animal and fungal Cav channels.
Our phylogeny supports the view that placozoans, which have the simplest animal body plan, branched off the animal stem after ctenophores and are therefore likely to be secondarily simplified. This scenario was found in both Nav and N/P/Q type Cav genes (SI Materials and Methods).
Bootstrapping scores indicate strong support for critical nodes of the Cav/Nav gene phylogeny. The position of the choanoflagellate Nav channel gene at the base of animal Nav channel genes was supported in 100% of the bootstrap replicates. The bootstrapping analysis also provides strong support for the monophyly of known groups of Cav and Nav channel genes, including the bilaterian Nav 1 clade and the three major groups of Cav channels. The clades containing channels with pore motifs D/E/E/A in both Cnidaria and Bilateria were less well supported in the bootstrap analysis (50–65% of replicates).
Secondary Structure.
Analysis of secondary structure of the inactivation gate revealed conservation of two important α-helices across the Nav channel family (Fig. S2). This structure was absent in Cav channels, including the yeast Cav channel.
Discussion
Rooting the Nav and Cav Gene Families.
The choanoflagellate M. brevicollis and the placozoan T. adhaerens express ion-channel genes that group phylogenetically with previously described sodium channels (Fig. 2) and have key molecular signatures of sodium channels (Fig. 3). Others have proposed that Nav channels evolved from an ancient Cav channel resembling the T-type channels (3) and that there may therefore be extant channels that have properties midway between Cav and Nav channels (21). Candidates for such channels have been proposed (21, 22), but the origin and genetic history of Nav channels have remained obscure. Our phylogenies show that the Nav ion channel family originated not only before the advent of the nervous system, but probably even before the advent of multicellularity. These results support the idea that Nav channels arose from Cav channels, but push back this divergence date to at least the common ancestor of animals and choanoflagellates. This demonstrates that complex systems like excitable tissues can evolve by coopting existing genes for new functions, rather than by de novo evolution of new genes.
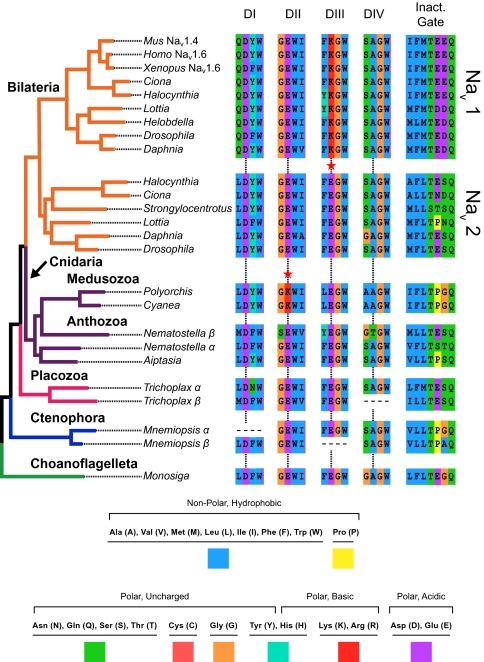
Fig. 3.
Phylogeny of Nav channels with key amino acid sequences mapped to their corresponding taxa. Taxa are color coded the same as in Fig. 2. The amino acids are alignments of the pore loops of all four domains (DI–DIV) and the critical inactivation particle on the inactivation gate. The critical amino acids in the pore are indicated by the vertical lines, and there are red stars next to convergent lysines (red K). Note the functional conservation of the hydrophobic triplet called the “inactivation particle” (first three amino acids on the inactivation gate).
Voltage-Gated Ion Channels and the Animal Phylogeny.
The phylogenetic placement of basal animal lineages (sponges, ctenophores, placozoans, and cnidarians) is not yet fully clear, although some placements are less controversial than others. The placement of sponges as sister to all other animals, and of cnidarians as sister to bilaterians, are fairly consistent results (12). The placements of ctenophores and placozoans, however, are less certain (SI Materials and Methods). Our results are consistent with the traditional phylogenetic placement of sponges and cnidarians, but place ctenophores, which have a nervous system, outside of the placozoans, cnidarians, and bilaterians (Figs. 2 and 4). This would suggest that placozoans have lost their nervous system or, much less likely, that the nervous system evolved twice in ctenophores and the cnidarian–bilaterian ancestor. Although our analysis has relatively strong bootstrap support, it has sparse taxon sampling, which has been shown to meaningfully affect phylogenetic inference (11, 25) and cannot therefore be considered a decisive species phylogeny.
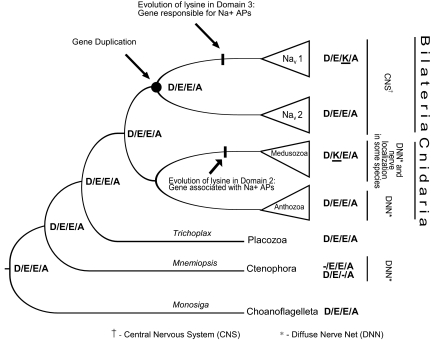
Fig. 4.
Simplified gene tree of the Nav family showing inferred ancestral states of the pore motifs. The gene duplication leading to the bilaterian Nav 1 and Nav 2 clades is noted, as are the points where we reconstruct fixation of lysines (K) in pore loops. Taxonomic information and information about the nervous system is also given. The Nematostella β and Trichoplax β genes have been left out for simplicity, but their addition would not change the proposed ancestral states. Pore states for both Mnemiopsis genes are shown because neither has a complete pore motif.
Also interesting is the apparent loss of Nav homologs in the sponge Amphimedon, an event that may reflect the sedentary lifestyle of these animals. Electrical impulse conduction has not been shown in demosponges, the group that includes Amphimedon, but it has been shown in a hexactinellid sponge (4). Hexactinellids differ drastically from demosponges in terms of morphology; further analysis of hexactinellids will be needed to determine whether Nav homologs have been retained in this group.
Genetic History—Bilateria.
Our results help clarify the diversity of pore states observed in animal Nav channels. The topology of our tree suggests that D/E/E/A is the ancestral pore sequence of the Nav gene family and that genes with this motif have been retained in every metazoan lineage that we examined, except for sponges, vertebrates, and the cnidarian subgroup Medusozoa (Figs. 3 and 4). The topology of the Nav 1 and Nav 2 clades supports the hypothesis that a gene duplication occurred around the time of the bilaterian radiation and before the split of protostomes and deuterostomes (24). The Nav 1 duplicate evolved a pore motif D/E/K/A and underwent further duplications in early tetrapods, creating the genes for Nav 1.1–1.9 in mammals (26). The other duplicate retained the ancestral pore motif and was lost in vertebrates.
Genetic History—Cnidaria.
Cnidarians diverged before the bilaterian gene duplication and do not have D/E/K/A channels, but the medusozoans have an amino acid substitution in the second domain pore loop, resulting in a clade of channels with the pore motif D/K/E/A. Although the topology of cnidarian channels with glutamic acid (E) in the second domain was not well supported, the clade of D/K/E/A channels was repeatedly found to represent a derived state and was monophyletic with 100% support. In species-tree analyses, the medusozoans share a common ancestor that is not shared with the anthozoans (8–12). The medusozoan subgroups represented here are Hydrozoa (Polyorchis) and Scyphozoa (Cyanea), both of which have D/K/E/A in the pore, whereas the anthozoan representatives (Aiptasia and Nematostella) both have D/E/E/A channels (Fig. 3). Our Nav tree is therefore consistent with proposed species trees and suggests a lysine (K) substitution in the common ancestor of medusozoans (Fig. 4). There is also a Nematostella channel whose pore sequence D/E/E/T is unique among sampled ion channels.
Sodium-based APs have been reported in both Cyanea (27) and Polyorchis (28), whereas APs in anthozoans and ctenophores seem to be carried mostly by calcium (29, 30). The pore motif D/K/E/A has been shown to be less selective for sodium than the D/E/K/A pore but more so than the D/E/E/A pore (21, 31–33). Channels with D/E/E/A have a higher affinity for calcium than sodium. The convergence to lysine in different domains of medusozoan and bilaterian ion channels may therefore have resulted from similar evolutionary pressure for sodium selectivity, as this would allow for less disruption of calcium homeostasis because Ca2+ is used for intracellular signaling in eukaryotes (3). Some medusozoans have concentrated nerve clusters and complex sense organs, which likely emerged convergently with the bilaterian central nervous system, as such nerve concentration is absent in anthozoans (34). It is not known whether the Nav genes function in these organs.
Evolution of Sodium Selectivity and Fast Inactivation.
The pore sequence D/E/E/A is intermediate between Cav channel and Nav channel pore motifs. It may also have an intermediate selectivity between calcium and sodium. The function of D/E/E/A channels in such a wide range of organisms and the reason for their apparent loss in medusozoans and vertebrates remains unknown. Mutation studies of the DSC1 channel (called Drosophila Nav 2 here) showed an effect in olfactory behavior in flies (35), but no function for these channels has been suggested in other organisms. The widespread retention of these channels suggests that they probably have important, yet possibly divergent, functions (e.g., not all lineages with D/E/E/A channels have olfaction). The sea urchin Strongylocentrotus purpuratus is only known to have an Nav 2 ortholog (24).
Hydrophobic sites on the domain III/IV linker that are critical for inactivation (19) are functionally conserved in all of the sodium channels that we investigated here, albeit with a wide range of different amino acid combinations at homologous sites (Fig. 3). Secondary structure of the inactivation gate is also relatively conserved. Two helices on either side of the hydrophobic triad that forms the “inactivation particle” have been predicted before and may act to stabilize and direct the inactivation particle as it swings up and binds to the channel (36, 37). These two helices are present across the Nav family, but not in the Cav families (Fig. S2). These findings suggest that all of the Nav homologs presented here may include an inactivation gate, even in the single-celled choanoflagellate.
Nav Channels in the Animal Genetic Repertoire.
This study adds to the growing evidence that much of the genetic repertoire for animal development, cell signaling, and even the nervous system was already present in the common ancestor of choanoflagellates and animals. Choanoflagellates have genes for cell-adhesion proteins (2, 38), tyrosine kinases and related proteins (2), proteins related to the postsynaptic density of neurons (18), and a remarkable complement of calcium signaling proteins (17). Some choanoflagellate species have a colonial life stage (7), and these genes may function in colony maintenance.
The function of sodium channel homologs in choanoflagellates or placozoans is unknown. They may create calcium-based APs, as suggested by the presence of such APs in ctenophores (30), but there are other possibilities. Both organisms can inhabit coastal marine areas with abundant fresh water runoff (5, 13). Trichoplax is restricted to warm coastal waters and is known to be sensitive to lowered salinity (13). It is possible that the channels act as osmosensors or osmoregulators in these organisms. Choanoflagellates have a long flagellum that they use to swim and to capture prey, and Trichoplax has a ciliated ventral layer that it uses for gliding across surfaces. It is possible that the channels control flagellar or ciliary beating through the influx of calcium, which triggers actin, or sodium, which is known to mediate flagellar motors in bacteria (39). Trichoplax has a layer of contractile fiber cells that form a syncytium and seem to function as muscle and a nervous system simultaneously (40). It is possible that the channels function in this dual purpose tissue.
Functional assays of Nav-channel homologs will shed light on their biological function and on the evolution of Nav channels as a whole. Determining the ion selectivity of these channels is critical to understanding how sodium selectivity can evolve from calcium selectivity by sequential mutations. Gaining insight into the function of these channels will not only enlighten the history of this protein's “adaptive walk” (20), it will also help elucidate the evolution of the nervous system.
Materials and Methods
Sources of RNA.
M. brevicollis and T. adhaerens were cultured in the laboratory using previously described and publicly available protocols (41). Placozoans were provided by Andreas Heyland. Choanoflagellate cells were fed on the bacteria present in the inoculum, and the placozoans were fed Cryptomonas sp. (LB 2423) from the University of Texas at Austin collection of algae. To extract RNA from M. brevicollis, we mixed and centrifuged 2 mL of the culture medium at 4 °C. Whole RNA was extracted using a RNA STAT-60 kit (Tel-Test) and then stored at –20 °C. The same protocol was used to isolate and store RNA from 15 T. adhaerens individuals that had been kept in algae-free seawater for 2 d to reduce the chance of contamination with algal RNA.
Gene Amplification and Sequencing.
Specific primers were designed from the BLAST sequences for RT and PCR reactions. RT reactions were conducted with a SuperScript II kit (Invitrogen) using both specific and poly-T primers to prevent bacterial RNA contamination. Primers are reported in SI Materials and Methods. PCR reactions were carried out with the following cycle for 39 repetitions: Denaturation at 94° (30 s), annealing at a primer-specific temperature (30 s), and elongation at 72° (1 min/kb). This cycle was preceded by an initial denaturation at 94° for 3 min 10 s and followed by a final elongation at 72° for 7 min. PCR products were visualized and purified with gel electrophoresis and then cloned using a TOPO cloning kit (Invitrogen) and One Shot Top 10 (Invitrogen) chemically competent Escherichia coli. We sequenced the M. brevicollis gene in four overlapping segments using vector-specific primers after cloning.
Sequence Analysis.
We performed a maximum likelihood phylogenetic analysis using the translated mRNA sequence from M. brevicollis and amino acid sequences from online databases for the other organisms. The latter were obtained either from catalogued, known channels or from BLAST searches of available genomes (for accession numbers, see Table S1). Amino acid sequences were aligned using the E-INS-I strategy in MAFFT (42). We used the Guidance algorithm available on the Guidance server to remove columns that had a score below 0.377 from the alignment (43). Maximum likelihood phylogenetic analysis and bootstrapping were performed in Garli (44), using a model of amino acid replacement selected using the Akaike Information Criterion in ProtTest (45). The model of protein evolution selected in the ProtTest analysis was WAG + I + G + F (Whelan and Goldman model, with invariant sites, parameter for gamma-distributed rate heterogeneity, and amino acid frequencies matched to the observed data). The maximum likelihood tree was obtained using Garli set to use the WAG + I + G + F model. The full amino acid alignment was analyzed for four search repetitions operating across 5 million generations each. A total of 100 bootstrap samples were collected using a halved topological termination condition, as recommended in the Garli manual, and a stop time of 1 million generations. All bootstrap outputs were analyzed in PAUP (46).
Secondary structure of the inactivation gate region was examined using the online server PsiPred (47), the results of which are reported in Fig. S2.
Supplementary Material
Acknowledgments
We thank Andreas Heyland (University of Guelph, Guelph, ON, Canada) for providing stocks of Trichoplax; Nicole King and Vicki Pearse for helpful discussion and advice; Marianna Grenadier for figure editing; and Emily McTavish, Thomas Keller, and Shannon Hedtke for advice on computational analysis. We thank Andy Baxevanis and Joseph Ryan for providing sequences from the M. leidyi draft genome. We also thank Bertil Hille and Edmund Brodie III for helpful suggestions on the manuscript. Laboratory work was funded by National Institutes of Health Grant R01GM084879.
Footnotes
The authors declare no conflict of interest.
Data deposition: Accession numbers and relevant databases are provided in Table S1. The sequences reported in this paper have been deposited in the GenBank database (accession nos. JF827087, JF905561, JF905562 and JF905563).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106363108/-/DCSupplemental.
References
- 1.Rokas A, Krüger D, Carroll SB. Animal evolution and the molecular signature of radiations compressed in time. Science. 2005;310:1933–1938. doi: 10.1126/science.1116759. [DOI] [PubMed] [Google Scholar]
- 2.King N, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hille B. Ionic Channels of Excitable Membranes. Sunderland, MA: Sinauer; 1984. pp. 720–722. [Google Scholar]
- 4.Leys SP, Mackie GO, Meech RW. Impulse conduction in a sponge. J Exp Biol. 1999;202:1139–1150. doi: 10.1242/jeb.202.9.1139. [DOI] [PubMed] [Google Scholar]
- 5.King N. Choanoflagellates. Curr Biol. 2005;5:113–114. doi: 10.1016/j.cub.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Caron DA, Worden AZ, Countway PD, Demir E, Heidelberg KB. Protists are microbes too: A perspective. ISME J. 2009;3:4–12. doi: 10.1038/ismej.2008.101. [DOI] [PubMed] [Google Scholar]
- 7.Carr M, Leadbeater BSC, Hassan R, Nelson M, Baldauf SL. Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc Natl Acad Sci USA. 2008;105:16641–16646. doi: 10.1073/pnas.0801667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Philippe H, et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- 9.Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 10.Hejnol A, et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Biol Sci. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pick KS, et al. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol Biol Evol. 2010;27:1983–1987. doi: 10.1093/molbev/msq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philippe H, et al. Resolving difficult phylogenetic questions: Why more sequences are not enough. PLoS Biol. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearse VB, Voight O. Field biology of placozoans (Trichoplax): Distribution, diversity, biotic interactions. Integr Comp Biol. 2007;47:677–692. doi: 10.1093/icb/icm015. [DOI] [PubMed] [Google Scholar]
- 14.Signorovitch AY, Dellaporta SL, Buss LW. Molecular signatures for sex in the Placozoa. Proc Natl Acad Sci USA. 2005;102:15518–15522. doi: 10.1073/pnas.0504031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eitel M, Schierwater B. The phylogeography of the Placozoa suggests a taxon-rich phylum in tropical and subtropical waters. Mol Ecol. 2010;19:2315–2327. doi: 10.1111/j.1365-294X.2010.04617.x. [DOI] [PubMed] [Google Scholar]
- 16.Srivastava M, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 17.Cai X. Unicellular Ca2+ signaling ‘toolkit’ at the origin of metazoa. Mol Biol Evol. 2008;25:1357–1361. doi: 10.1093/molbev/msn077. [DOI] [PubMed] [Google Scholar]
- 18.Alié A, Manuel M. The backbone of the post-synaptic density originated in a unicellular ancestor of choanoflagellates and metazoans. BMC Evol Biol. 2010;10:34. doi: 10.1186/1471-2148-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldin AL. Mechanisms of sodium channel inactivation. Curr Opin Neurobiol. 2003;13:284–290. doi: 10.1016/s0959-4388(03)00065-5. [DOI] [PubMed] [Google Scholar]
- 20.Maynard-Smith J. Natural selection and the concept of a protein space. Science. 1970;225:563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- 21.Zhou W, Chung I, Liu Z, Goldin AL, Dong K. A voltage-gated calcium-selective channel encoded by a sodium channel-like gene. Neuron. 2004;42:101–112. doi: 10.1016/s0896-6273(04)00148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spafford JD, Spencer AN, Gallin WJ. A putative voltage-gated sodium channel alpha subunit (PpSCN1) from the hydrozoan jellyfish, Polyorchis penicillatus: Structural comparisons and evolutionary considerations. Biochem Biophys Res Commun. 1998;244:772–780. doi: 10.1006/bbrc.1998.8332. [DOI] [PubMed] [Google Scholar]
- 23.Nagahora H, et al. Diversity of voltage-gated sodium channels in the ascidian larval nervous system. Biochem Biophys Res Commun. 2000;275:558–564. doi: 10.1006/bbrc.2000.3290. [DOI] [PubMed] [Google Scholar]
- 24.Hill AS, et al. Ion channel clustering at the axon initial segment and node of Ranvier evolved sequentially in early chordates. PLoS Genet. 2008;4:e1000317. doi: 10.1371/journal.pgen.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hedtke SM, Townsend TM, Hillis DM. Resolution of phylogenetic conflict in large data sets by increased taxon sampling. Syst Biol. 2006;55:522–529. doi: 10.1080/10635150600697358. [DOI] [PubMed] [Google Scholar]
- 26.Zakon HH, Jost MC, Lu Y. Expantion of voltage-dependent Na+ channel gene family in early tetrapods coincided with the emergence of terrestriality and increased brain complexity. Mol Biol Evol. 2011;28:1415–1424. doi: 10.1093/molbev/msq325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson PA, Schwab WE. Action potential in neurons of motor nerve net of Cyanea (Coelenterata) J Neurophysiol. 1983;50:671–683. doi: 10.1152/jn.1983.50.3.671. [DOI] [PubMed] [Google Scholar]
- 28.Spencer AN, Satterlie RA. The action potential and contraction in subumbrellar swimming muscle of Polyorchis penicillatus (Hydromedusae) J Comp Physiol. 1981;144:401–407. [Google Scholar]
- 29.White GB, et al. Structure of a putative sodium channel from the sea anemone Aiptasia pallida. Invert Neurosci. 1998;3:317–326. doi: 10.1007/BF02577691. [DOI] [PubMed] [Google Scholar]
- 30.Bilbaut A, Nicaise MH, Meech RW. In: Evolution of the First Nervous Systems. Anderson PAV, editor. New York: Plenum; 1989. pp. 299–314. [Google Scholar]
- 31.Heinemann SH, Terlau H, Stühmer W, Imoto K, Numa S. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 1992;356:441–443. doi: 10.1038/356441a0. [DOI] [PubMed] [Google Scholar]
- 32.Lipkind GM, Fozzard HA. Voltage-gated Na channel selectivity: The role of the conserved domain III lysine residue. J Gen Physiol. 2008;131:523–529. doi: 10.1085/jgp.200809991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlief T, Schönherr R, Imoto K, Heinemann SH. Pore properties of rat brain II sodium channels mutated in the selectivity filter domain. Eur Biophys J. 1996;25:75–91. doi: 10.1007/s002490050020. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe H, Fujisawa T, Holstein TW. Cnidarians and the evolutionary origin of the nervous system. Dev Growth Differ. 2009;51:167–183. doi: 10.1111/j.1440-169X.2009.01103.x. [DOI] [PubMed] [Google Scholar]
- 35.Kulkarni NH, Yamamoto AH, Robinson KO, Mackay TFC, Anholt RRH. The DSC1 channel, encoded by the smi60E locus, contributes to odor-guided behavior in Drosophila melanogaster. Genetics. 2002;161:1507–1516. doi: 10.1093/genetics/161.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sirota FL, Pascutti PG, Anteneodo C. Molecular modeling and dynamics of the sodium channel inactivation gate. Biophys J. 2002;82:1207–1215. doi: 10.1016/S0006-3495(02)75477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Catterall WA. From ionic currents to molecular mechanisms: The structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 38.King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- 39.Fukuoka H, Wada T, Kojima S, Ishijima A, Homma M. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol Microbiol. 2009;71:825–835. doi: 10.1111/j.1365-2958.2008.06569.x. [DOI] [PubMed] [Google Scholar]
- 40.Rassat R, Ruthmann A. Trichoplax adhaerens F.E. Schulze (Placozoa) in the scanning electron microscope. Zoomorphologie. 1979;93:59–72. [Google Scholar]
- 41.The King Lab Resources. Available at http://kinglab.berkeley.edu/resources/; accessed on February 19, 2010.
- 42.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Penn O, et al. Guidance: A web server for assessing alignment confidence scores. Nucleic Acids Res. 2010;38:W23–W28. doi: 10.1093/nar/gkq443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zwickl DJ. Austin, TX: University of Texas; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD dissertation. [Google Scholar]
- 45.Abascal F, Zardoya R, Posada D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 46.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2003. Version 4. [Google Scholar]
- 47.Bryson K, et al. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.