Abstract
Rust fungi are some of the most devastating pathogens of crop plants. They are obligate biotrophs, which extract nutrients only from living plant tissues and cannot grow apart from their hosts. Their lifestyle has slowed the dissection of molecular mechanisms underlying host invasion and avoidance or suppression of plant innate immunity. We sequenced the 101-Mb genome of Melampsora larici-populina, the causal agent of poplar leaf rust, and the 89-Mb genome of Puccinia graminis f. sp. tritici, the causal agent of wheat and barley stem rust. We then compared the 16,399 predicted proteins of M. larici-populina with the 17,773 predicted proteins of P. graminis f. sp tritici. Genomic features related to their obligate biotrophic lifestyle include expanded lineage-specific gene families, a large repertoire of effector-like small secreted proteins, impaired nitrogen and sulfur assimilation pathways, and expanded families of amino acid and oligopeptide membrane transporters. The dramatic up-regulation of transcripts coding for small secreted proteins, secreted hydrolytic enzymes, and transporters in planta suggests that they play a role in host infection and nutrient acquisition. Some of these genomic hallmarks are mirrored in the genomes of other microbial eukaryotes that have independently evolved to infect plants, indicating convergent adaptation to a biotrophic existence inside plant cells.
Keywords: comparative genomics, plant pathogen, basidiomycete, evolution, rust disease
Rust fungi (Pucciniales, Basidiomycota) are a diverse group of plant pathogens composed of more than 120 genera and 6,000 species, and are one of the most economically important groups of pathogens of native and cultivated plants (1, 2). Puccinia graminis, the causal agent of stem rust, has caused devastating epidemics wherever wheat is grown (3), and a new highly virulent strain (Ug99) threatens wheat production worldwide (4). Similarly, epidemics of poplar leaf rust, caused by Melampsora spp., is a major constraint on the development of bioenergy programs based on domesticated poplars (5) as a result of the lack of durable host resistance (6, 7). Rust fungi are obligate biotrophic parasites with a complex life cycle that often includes two phylogeneticaly unrelated hosts (2). They have evolved specialized structures, haustoria, formed within host tissue to efficiently acquire nutrients and suppress host defense responses (8). Molecular features driving adaptations to an obligate biotrophic association with plant hosts are unknown. Whether the convergent biotrophic adaptation observed in bacterial parasites (9) and other lineages of microbial eukaryotes (e.g., microsporidia) (10) has led to functional specializations at the genome level (i.e., gene gain or loss, regulation of gene expression) remains to be determined. The recent report of the genome sequence of Blumeria graminis, an ascomycete biotroph pathogen responsible for barley powdery mildew, revealed a genome size expansion caused by transposon proliferation concomitant with dramatic reduction in gene content, i.e., genes encoding sugar-cleaving enzymes, transporters and assimilatory enzymes for inorganic nitrate and sulfur (11). Similarly, gene losses were observed in the genome of the oomycete Hyaloperonospora arabidopsidis, a biotroph parasite infecting Arabidopsis thaliana, as well as the diversification of genes encoding RXLR effector-like secreted proteins (12). Despite their phylogenetic distance, these two pathogens forming haustoria seem to share striking adaptation convergences to biotrophy. To determine the genetic features underlying pathogenesis and biotrophic ability of rust pathogens, we report here the genome sequences of the rust fungi Melampsora larici-populina and P. graminis f. sp. tritici.
Results and Discussion
Genome Sequencing, Gene Family Annotation, and Expression Analysis.
We have sequenced the dikaryotic genomes of the poplar leaf rust fungus, M. larici-populina, and of the wheat stem rust fungus, P. graminis f. sp. tritici, by a Sanger whole-genome shotgun strategy (SI Text, Genome Sequencing and Assembly). The overall assembly sizes of the haploid genomes of M. larici-populina and P. graminis f. sp. tritici are 101.1 Mb and 88.6 Mb, respectively (Table 1). These genomes are much larger than the other sequenced basidiomycete genomes (13, 14), but no evidence for whole-genome duplication or large-scale dispersed segmental duplications was observed. The expanded size results from a massive proliferation of transposable elements (TEs), which account for nearly 45% of both assembled genomes (Figs. S1 and S2 and Dataset S1, Tables S3 and S4). Class I LTR retroelements are more abundant in P. graminis f. sp. tritici, whereas class II terminal inverted repeat (TIR) DNA transposons are prominent in M. larici-populina. Timing of TE activity by using sequence divergence of extant copies suggests that a major wave of retrotransposition in the M. larici-populina and P. graminis f. sp. tritici lineages occurred more than 1 Mya (SI Text, Repeat Analysis).
Table 1.
Assembly statistics for the dikaryotic genome of M. larici-populina (Mlp) 98AG31 and P. graminis f. sp. tritici (Pgt) CDL75-36–700-3, race SCCL
| Parameter | Mlp* | Pgt |
| Sequence coverage | 6.9 | 12 |
| Scaffold total, Mb | 101.1 | 88.6 |
| Scaffolds | 462 | 392 |
| Scaffold N50 length, Mb† | 1.1 | 0.97 |
| Scaffold N50† | 27 | 30 |
| Assembly in scaffolds > 50 kb, % | 96.5 | 97.1 |
| Contig sequence total, Mb | 97.7 | 81.5 |
| Contigs | 3,254 | 4,557 |
| Contig N50 length, kb† | 112.3 | 39.5 |
| Contig N50† | 265 | 546 |
| Base quality ≥ Q40, % | 98.3 | 96.3 |
| Gap content, % | 3.4 | 8 |
| GC content, % | 41 | 43.3 |
| Protein coding genes | 16,399 | 17,773 |
| Mean coding sequence length, nt | 1,565 | 1,075 |
| Mean exon number per gene | 4.92 | 4.7 |
| Mean exon length, nt | 247 | 175 |
| Mean intron length, nt | 118 | 133 |
| Mean intergenic length, nt | 4,356 | 3,328 |
| tRNAs | 253 | 428 |
*Statistics for Mlp are based on the ”main genome scaffolds” of the assembly; the ”repetitive,” “excluded,” and ”altHaplotype” scaffolds for Mlp (Dataset S1, Table S2) were not included.
†The N50 metric corresponds to the N largest scaffolds required to capture half of the total sequence. The N50 length is that of the smallest scaffold in the N50 set.
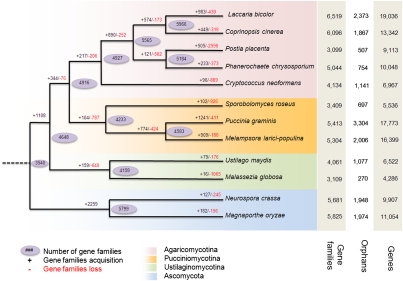
We predicted 16,399 and 17,773 protein-coding genes in M. larici-populina and P. graminis f. sp. tritici, respectively. The size of these proteomes is similar to the symbiotic basidiomycete Laccaria bicolor (14), but strikingly larger than the corn smut fungi Ustilago maydis and Sporisorium reilianum, two nonobligate pathogenic biotrophs that encode only approximately 6,500 proteins (15, 16). Among the predicted proteins, only 41% and 35% in M. larici-populina and P. graminis f. sp. tritici, respectively, showed significant sequence similarity to documented proteins (BLASTP E-value ≤ 10−5; SI Text, Gene Prediction and Annotation and Fig. S3). To investigate protein evolution in M. larici-populina and P. graminis f. sp. tritici, we constructed families containing both orthologues and paralogues from a diverse set of ascomycetous and basidiomycetous fungi (SI Text, Multigene Families and Evolutionary Analysis of Multigene Families). The two genomes shared 3,984 orthologous Tribe-MCL families, which comprised 7,959 P. graminis f. sp. tritici genes and 7,875 M. larici-populina genes; 26% of the predicted protein families were lineage-specific, whereas 774 gene families were unique to these two rust fungi. Expansion of protein family sizes was prominent in both M. larici-populina and P. graminis f. sp. tritici (Fig. 1, Fig. S4, and Dataset S1, Tables S6–S8); several expanded protein families are lineage-specific, suggesting that important protein-coding innovation occurred in these lineages. Of the 5,045 M. larici-populina genes that have an orthologue in P. graminis f. sp. tritici (best reciprocal BLASTP hit, E-value ≤ 10−5), very few showed conservation of neighboring orthologues, suggesting there is little synteny between the genomes (SI Text, Lack of Genome Duplication and Synteny Between M. larici-populina and P. graminis f. sp. tritici, and Fig. S5). This is likely because of the expansion of the TE and massive reshuffling of the genomes as a result of recombination between TEs. In addition, within the rust fungi, M. larici-populina and P. graminis f. sp. tritici represent very divergent phylogenetic lineages (1). Gene family expansions also occurred in those genes coding for oligopeptide membrane transporters (OPTs; Dataset S1, Table S18), copper/zinc superoxide dismutase (SOD; Dataset S1, Table S23), different types of glycosyl hydrolases, lipases, and peptidases, and several groups of predicted signaling genes, including kinases and transcription factors (Figs. S4 and S6–S8). Several gene families encoding leucine-rich repeat domain-containing proteins were expanded (Fig. S4A), and are potentially involved in protein–protein interactions in rust fungi. Different types of helicases are also represented in expanded gene families of rust fungi and could allow for an increased capability for DNA maintenance and repair. Strikingly, both rust fungi have expanded lineage-specific gene families encoding zinc-finger proteins (Fig. S4B), with significantly overrepresented nucleic acid binding and zinc ion binding gene ontology terms in both genes sets, which represent potential transcription factors (SI Text, Multigene Families and Evolutionary Analysis of Multigene Families, and Dataset S1, Tables S7 and S8). These results suggest that rust fungi possess a diverse potential to regulate and repair nucleic acid; targeted work will be required to decipher the roles of these proteins during the interaction with plant hosts. Although proliferation of TEs might have contributed to the expansion of gene families in rust fungi, no specific localization of particular gene families was identified in TE-rich regions of rust genomes, such as reported for effectors in other plant pathogens (17,18, 19) (SI Text, Multigene Families and Evolutionary Analysis of Multigene Families).
Fig. 1.
Predicted pattern of gene families gain and loss in representative fungal genomes. The figure represents the total number of protein families in each species or node estimated by Dollo parsimony principle. The numbers on the branches of the phylogenetic tree correspond to expanded (Left, black), contracted (Right, red), or inferred ancestral (oval) protein families along each lineage by comparison with the putative pan-proteome. For each species, the number of gene families, orphan genes, and the total gene number are indicated on the right.
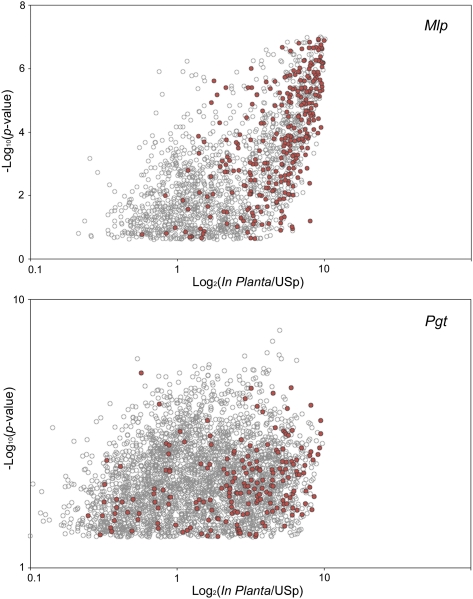
Seventy and 54% of the predicted genes of M. larici-populina and P. graminis f. sp. tritici, respectively, were detected by custom microarray transcript profiling of resting and germinating urediniospores, as well as infected leaves (SI Text, Whole-Genome Exon Oligoarrays). A significant proportion of the detected transcripts (18%) is differentially expressed (fold ratio ≥ 10.0; P < 0.05) in infected leaves, whereas only approximately 8.0% are specifically expressed in planta (SI Text, Whole-Genome Exon Oligoarrays). Transcripts coding for secreted peptidases and lipases, transporters of hexoses, amino acids, and oligopeptides, and carbohydrate-cleaving enzymes, such as chitin deacetylases and cutinases (Tables 2 and 3 and Dataset S1, Tables S12 and S16), are strikingly enriched (≥10-fold) in planta. However, the most highly up-regulated transcripts in planta (≥100-fold) are mainly comprised of lineage-specific transcripts, including those coding for small secreted proteins (SSPs; Fig. 2 and Dataset S1, Tables S10 and S14). These in planta-induced, lineage-specific genes are likely involved in the specific relationship established between these rust fungi and their respective hosts.
Table 2.
Selection of M. larici-populina (Mlp) genes strongly up-regulated during poplar leaf infection
| Best BLAST hit |
Expression level |
96 hpi/urediniospores |
|||||
| Mlp ID | Function | Pgt ID | GenBank accession no. | 96 hpi | Urediniospores | Fold-change | P value |
| 89465 | Aspartic peptidase A1, secreted | PGTG_10570 | XP_001881739 | 44,063 | 38* | 1159.6 | 3.42 × 10−5 |
| 94889 | Lipase, secreted | PGTG_15782 | XP_749106 | 27,318 | 36* | 758.9 | 1.72 × 10−4 |
| 123524 | SSP, RTP homologue | PGTG_18022 | ABS86408 | 49,354 | 68* | 725.8 | 8.53 × 10−4 |
| 106755 | Glycosyl hydrolase, GH16, secreted | No hit | No hit | 25,530 | 57* | 447.9 | 7.42 × 10−5 |
| 88574 | Oligopeptide transporter, OPT | PGTG_17016 | XP_001394363 | 38,726 | 88* | 440.1 | 1.40 × 10−4 |
| 86448 | Transporter, AEC (Auxin Efflux Carrier) family | PGTG_06747 | XP_759229 | 17,984 | 42* | 428.2 | 1.33 × 10−4 |
| 112330 | α-Glycosidase, secreted, GH47 | PGTG_09507 | XP_001881296 | 14,561 | 41* | 355.2 | 3.92 × 10−5 |
| 36184 | Amino acid permease, PIG2 homologue | PGTG_15547 | XP_001873273 | 10,319 | 34* | 303.5 | 2.10 × 10−4 |
| 95696 | Alanine aminotransferase | PGTG_07510 | XP_001837651 | 11,018 | 37* | 297.8 | 3.84 × 10−4 |
| 53832 | Thiamin biosynthesis enzyme, THI4 homologue | PGTG_01304 | Q9UVF8 | 52,910 | 194 | 272.8 | 1.14 × 10−4 |
| 39287 | SSP, Cro r I homologue | No hit | AAF87492 | 7,916 | 30* | 263.9 | 0.026 |
| 64764 | SSP, HESP-376 homologue | No hit | No hit | 7,596 | 35* | 217.1 | 1.26 × 10−3 |
| 89463 | Subtilisin protease S8A, secreted | PGTG_18581 | XP_001877576 | 18,072 | 87* | 207.8 | 1.15 × 10−4 |
| 40379 | Sugar transporter HXT1, MFS | PGTG_15147 | XP_001874568 | 12,387 | 61* | 203.1 | 2.64 × 10−4 |
| 91040 | β-Glycosidase, endoglucanase, GH5 | PGTG_17056 | XP_001875020 | 7,212 | 36* | 200.4 | 5.13 × 10−4 |
| 124202 | Secreted protein, AvrM-B homologue | No hit | ABB96259 | 3,764 | 27* | 139.5 | 4.12 × 10−4 |
| 67013 | Thiamin biosynthesis enzyme THI1 homologue | PGTG_10151 | ABK96768 | 35,825 | 274 | 130.8 | 1.51 × 10−4 |
| 48366 | Carotenoid ester lipase, secreted | PGTG_13346 | XP_001875752 | 14,890 | 121 | 123.1 | 1.26 × 10−3 |
| 40488 | Chitin deacetylase, CE4 | PGTG_09635 | XP_774611 | 3,704 | 39* | 95 | 1.10 × 10−3 |
| 109896 | Secreted protein related to plant expansins | PGTG_19856 | XP_771894 | 4,998 | 52* | 96.2 | 4.34 × 10−3 |
| 60884 | Glycosyltransferase GT18 | PGTG_01151 | XP_001884748 | 3,889 | 41* | 94.9 | 8.36 × 10−4 |
| 87910 | Oligopeptide transporter, OPT | PGTG_15138 | XP_001834544 | 12,366 | 160 | 77.3 | 6.03 × 10−5 |
| 39227 | Zinc transporter, CDF | PGTG_14264 | CAE00445 | 3,210 | 43* | 74.7 | 5.67 × 10−3 |
| 25498 | Chitin deacetylase, CE4 | PGTG_09635 | XP_774611 | 4,541 | 61* | 74.5 | 3.41 × 10−3 |
| 55212 | SSP, HESP-735 homologue | No hit | ABB96276 | 2,221 | 33* | 67.4 | 5.20 × 10−4 |
Up-regulation in poplar infected leaves is assessed by comparing transcripts profiles to those from resting urediniospores. Poplar leaves were infected by M. larici-populina urediniospores and left for 96 hours postinoculation (hpi) under controlled conditions. At this stage, poplar rust pathogen has formed many haustoria in planta and sporulation has not yet occurred. Expression values are the means of three biological replicates for 96 hpi and urediniospores. Based on statistical analysis of normalized fluorescence levels, a gene was considered significantly regulated if it met two criteria (1): t test P value, 0.05 (ArrayStar; DNAStar); infected poplar leaves at 96 hpi versus urediniospores fold change > 10. Genes were selected on the basis of homology to a function, and hypothetical proteins or genes without homology of unknown function (exception of SSPs homologues of candidate rust pathogen effectors) were discarded. The complete list of significantly regulated genes is detailed in Dataset S1, Table S12.
*Below background expression level.
Table 3.
Selection of P. graminis f. sp. tritici (Pgt) genes strongly up-regulated during wheat infection
| Best BLAST hit |
Expression levels |
Wheat/urediniospores |
|||||
| Pgt ID | Function | Mlp ID | GenBank accession no. | Wheat | Urediniospores | Fold-change | P value |
| PGTG_12502 | Amino acid permease | 113062 | No hit | 31,670 | 68* | 467.2 | 0.004 |
| PGTG_15174 | Differentiation-related protein Infp | No hit | AAD38996 | 23,002 | 50* | 466.3 | 0.002 |
| PGTG_07532 | Amino acid permease | 113062 | No hit | 13,666 | 47* | 293.8 | 0.005 |
| PGTG_07938 | Invertase | 44167 | CAG26671 | 18,901 | 70* | 271 | 3.63 × 10−4 |
| PGTG_17720 | Zinc finger, C2H2 type | No hit | No hit | 31,604 | 175 | 180.9 | 0.004 |
| PGTG_16569 | Multicopper oxidase, secreted | 112024 | BAG50320 | 18,825 | 114* | 166.6 | 0.012 |
| PGTG_15026 | Lipase | 96073 | XP_001273241 | 21,088 | 229 | 92.4 | 1.22 × 10−6 |
| PGTG_10570 | Aspartic protease, secreted | 89871 | No hit | 3,493 | 46* | 76.1 | 0.04 |
| PGTG_05667 | Cu/Zn SOD, secreted | 73483 | XP_002418001 | 10,257 | 138* | 74.7 | 0.004 |
| PGTG_11683 | Major intrinsic protein | 106246 | No hit | 8,738 | 118* | 74.6 | 4.73 × 10−4 |
| PGTG_19191 | Serine carboxypeptidase, secreted | 49959 | EEY14780 | 6,156 | 86* | 71.8 | 0.017 |
| PGTG_11725 | Endo-1,4-β-glucanase, secreted, GH5 | 47207 | AAR29981 | 6,503 | 100* | 65.3 | 0.038 |
| PGTG_08842 | Thiamine monophosphate synthase/TENI | 63716 | No hit | 7,343 | 117* | 63.1 | 7.47 × 10−4 |
| PGTG_10915 | Major intrinsic protein | 89561 | No hit | 41,747 | 686 | 61 | 0.006 |
| PGTG_05491 | MFS sugar transporter, putative | 86594 | XP_002480590 | 28,494 | 478 | 59.8 | 0.006 |
| PGTG_15162 | Endo-β-mannanase, GH5 | 86044 | ABR27262 | 6,992 | 123* | 57.3 | 0.009 |
| PGTG_02527 | Chitin synthase N-terminal, GT2 | 73345 | ABB70409 | 33,954 | 766 | 44.4 | 8.40 × 10−4 |
| PGTG_06309 | Plasma membrane (H+) ATPase | 44104 | CAA05841 | 10,443 | 272 | 38.5 | 0.003 |
| PGTG_01889 | Lipase, secreted | 91294 | No hit | 13,249 | 348 | 38.2 | 9.55 × 10−4 |
| PGTG_15889 | Aspartic peptidase A1 | 34644 | XP_001880663 | 4,880 | 128* | 38.2 | 0.019 |
| PGTG_15122 | Chitinase, GH18 | 75188 | CAQ51152 | 15,175 | 415 | 36.6 | 2.63 × 10−4 |
| PGTG_12200 | MFS monocarboxylate transporter | 86626 | XP_001267950 | 1,636 | 49* | 34 | 0.012 |
| PGTG_15888 | Aspartic peptidase A1 | 34644 | XP_001880663 | 2,159 | 77* | 28.2 | 0.021 |
| PGTG_18584 | Hexose transporter HXT1 | 38418 | CAC41332 | 8,629 | 378 | 22.9 | 0.006 |
Up-regulation in infected wheat is assessed by comparing transcripts profiles to those from resting urediniospores. Wheat leaves were infected by P. graminis f. sp. tritici urediniospores and left for 8 d after inoculation under controled conditions. At this stage, wheat rust pathogen has started to sporulate and macroscopic flecking are visible. Expression values are the means of three biological replicates for 8 dpi and urediniospores. Based on statistical analysis of normalized fluorescence levels, a gene was considered significantly regulated if it met two criteria (1): t test P value, 0.05 (using mattes in Matlab); infected wheat at 8 dpi versus urediniospores fold-change > 10. Genes were selected on the basis of homology to a function, and hypothetical proteins or genes without homology of unknown function were discarded. The complete list of significantly regulated genes is detailed in Dataset S1, Table S16.
*Below background expression level.
Fig. 2.
Expression profiling of candidate rust effector genes. Global gene expression in infected poplar or wheat leaves (in planta) versus resting urediniospores (USp) in M. larici-populina (Mlp, Top) and P. graminis f. sp. tritici (Pgt, Bottom). Log2-fold change ratio of significantly induced genes in planta compared with resting urediniospores plotted versus the P value. Gray circles, significantly in planta-induced genes; red circles, SSP encoding genes.
Rust Fungi Secretomes Contain Species-Specific Candidate Effectors.
Microbial pathogens have evolved highly advanced mechanisms to engage their hosts in intimate contact and sabotage host immune responses by secreting effector proteins into host cells to target regulators of defense (20–22). Most SSPs that are specifically produced during plant infection are likely to be effectors that manipulate host cells to facilitate parasitic colonization, such as by suppressing plant innate immunity or enhancing nutrient availability (21). In silico gene prediction and manual annotation of SSP genes in M. larici-populina genome identified a set of 1,184 SSPs (SI Text, Effector/Secretome, and Dataset S1, Table S17), of which 74% are lineage-specific. P. graminis f. sp. tritici contains a similar number of 1,106 SSP genes, of which 84% are lineage-specific. In M. larici-populina, a total of 812 SSPs are organized in 169 families of two to 111 members (Dataset S1, Table S17); the largest family contains a highly conserved 10-cysteine pattern (Fig. S6A). In P. graminis f. sp. tritici, a total of 593 SSPs are organized in 164 families of two to 44 members and the largest family contains a highly conserved eight-cysteine pattern (Fig. S6B). Four of these proteins show evidence of haustorial expression in wheat rust, providing additional evidence that they are potentially effectors. This expansion of SSP genes in rust fungi is striking as SSP families account for approximately 10% of the expanded families in both rust genomes. Between 50% and 56% of the lineage-specific SSP genes are supported by ESTs or expression detected on the custom oligoarrays, which provide evidence to support these predicted genes of unknown function; additional genes could be specifically expressed during the sexual phase of the lifecycle (23), which was not explored here. Both M. larici-populina and P. graminis f. sp. tritici require an alternate host to complete their lifecycle and achieve sexual reproduction, and successful infection of the alternate host may involve a different set of SSP genes. Homologues of known effectors from Melampsora lini, such as haustorially expressed secreted proteins (HESPs) and the avirulence factors AvrM, AvrL567, AvrP123, and AvrP4 from the flax rust fungus M. lini (8, 21), and the rust-transferred protein RTP1 from the bean rust pathogen (22), are present among highly up-regulated M. larici-populina transcripts (Table 2 and Dataset S1, Tables S10–S12). Interestingly, whereas 19 of the 21 M. lini HESPs (24) showed significant similarity to M. larici-populina SSP genes (BLASTP E-value ≤ 10−5), only nine showed similarity to P. graminis f. sp. tritici SSP genes, suggesting the presence of effector genes conserved in the Melampsoraceae and not shared within the Pucciniales order. By contrast, homologues of Uromyces fabae RTP1 were detected in the poplar and the wheat rust genomes—three and seven, respectively—indicating the presence of conserved effectors families in the Pucciniales. Recently, [Y/F/W]xC motifs were reported in the N-terminal region of secreted proteins in the ascomycete B. graminis, an obligate biotroph of wheat, as well as in Puccinia spp. (25). Systematic search for these motifs in the poplar leaf and the wheat stem rust fungi showed that they were indeed abundant in SSPs but not restricted to the N-terminal region as in B. graminis (Fig. S6A; SI Text, Effector/Secretome). These motifs were also present in nonsecreted proteins related to zinc binding and nucleic acid binding (SI Text, Effector/Secretome), suggesting these motifs are also conserved in other cysteine-rich proteins. At least 43% of M. larici-populina and 40% of P. graminis f. sp. tritici SSPs are expressed in infected leaves. In P. graminis f. sp. tritici, PGTG_17547 matches the highest number of haustorial ESTs, and is similar in sequence to a predicted secreted protein (ADA54575) from the wheat stripe rust fungus, Puccinia striiformis (25); this protein is lineage-specific, sharing no significant similarity with proteins outside the Pucciniales. In both rust species, one highly in planta-expressed SSP [PGTG_13212, Joint Genome Institute (JGI) ID no. 85525] is similar in sequence to HESP-735 from the flax rust pathogen (24) (Dataset S1, Tables S12 and S14). SSPs are highly overrepresented in the mostly highly induced genes; 50 and 29 SSPs belong to the top 100 most highly transcriptionally up-regulated in infected poplar and wheat leaves compared with M. larici-populina and P. graminis f. sp. tritici urediniospores, respectively (Fig. 2 and Dataset S1, Tables S10 and S14). Most up-regulated SSP transcripts in planta were lineage-specific, as only 16% have an orthologue in both rust species, suggesting that these sequences are evolving at a very high rate. The specific location remains to be determined, whether up-regulated SSPs are expressed in infection hyphae and/or haustoria, and whether they remain in the cell wall or extrahaustorial matrix or are adressed to specific compartments of the host cell where they interact with their target proteins as shown for avirulence proteins in M. lini (8, 21). Similarly, some of the predicted SSP genes not expressed in urediniospores or in planta could act as specialized effectors during infection of the alternate host.
Rust Fungi Carbohydrate-Active Enzymes Set.
Gene families encoding host-targeted, hydrolytic enzymes acting on plant biopolymers, such as proteinases, lipases, and several sugar-cleaving enzymes (carbohydrate-active enzymes; CAZymes) (26), are highly up-regulated in both rust pathogen transcriptomes in planta (Tables 2 and 3 and Dataset S1, Tables S12 and S16), suggesting that the invading hypha is penetrating the host cells by using these degrading enzymes. The comparison of the glycoside hydrolase (GH), glycosyltransferases (GTs), polysaccharide lyase (PL), and carbohydrate esterase (CE) of 21 sequenced fungi (Fig. S8) revealed that M. larici-populina and P. graminis f. sp. tritici have a relatively smaller set of GH-encoding genes (173 and 158 members, respectively; SI Text, Annotation of Putative CAZymes, and Dataset S1, Table S19); this content is similar to that in the basidiomycete symbiont L. bicolor (14), but much fewer than in hemibiotrophic or necrotrophic phytopathogens (e.g., Magnaporthe oryzae) and saprotrophs (including Neurospora crassa; Coprinopsis cinerea; Schizophyllum commune) (27). This set of CAZymes is strikingly larger than the repertoire of the biotroph U. maydis (100 members) (15). In evolving a biotrophic lifestyle, the rust fungi have lost several secreted hydrolytic GH and PL enzymes acting on plant cell wall (PCW) polysaccharides (Fig. S8) and they are lacking the cellulose-binding carbohydrate-binding module 1 (CBM1). However, they show a moderate expansion of a few GHs cleaving plant celluloses and hemicelluloses (e.g., GH7, GH10, GH12, GH26, and GH27) compared with the biotroph U. maydis or the hemibiotroph M. oryzae. These enzymes, together with in planta up-regulated and expanded α-mannosidase (GH47) and β-1,3-glucanase (GH5) transcripts (Dataset S1, Tables S12 and S16), may play a key role in the initial stages of host colonization, i.e., penetration of the parenchyma cells. A different set of enzymes, induced chitin deacetylases (CE4) present in P. graminis f. sp. tritici, M. larici-populina, and the symbiont L. bicolor (14), are likely involved in fungal cell wall remodeling and may play a role in the alteration of the fungal cell wall surface during infection to conceal invading hypha from the host (28).
Expanded Rust Transporters Gene Families Are Expressed During Host Infection.
Acquisition of nutrients, including carbohydrates and amino acids, is crucial to the success of rust pathogen biotrophic interactions established by invading hyphae forming haustoria within the host plant (21, 29, 30). The repertoire of membrane transporters (SI Text, Transporters, and Dataset S1, Table S18) in M. larici-populina and P. graminis f. sp. tritici contains homologues of the hexose transporter HXT1, amino acid transporters AAT1, AAT2, and AAT3, and H+-ATPases from the bean rust pathogen (U. fabae), known to be highly up-regulated during the interaction with its host plant. In addition, M. larici-populina and P. graminis f. sp. tritici genomes display an increased genetic potential for peptide uptake with 22 and 21 OPT genes, respectively, whereas other basidiomycete genomes contain only five to 16 OPT genes (Dataset S1, Table S18). OPT genes that are transcriptionally up-regulated in planta (Dataset S1, Tables S12 and S16), are likely involved in the transport of peptides released by the action of the induced proteinases (aspartic peptidase, subtilisin) expressed in infected leaf tissues. The Major Facilitator Superfamily (MFS) gene family is reduced in the M. larici-populina and P. graminis f. sp. tritici genomes compared with other basidiomycetes (Dataset S1, Table S18), but many MFS transcripts are, however, highly expressed in planta including two HXT1 homologues. Consistent with in planta expression of M. larici-populina and P. graminis f. sp. tritici invertase genes (Dataset S1, Tables S12 and S16), no homologue of the sucrose transporter Srt1 recently described in U. maydis (29) was identified, supporting the preferential uptake of host hexoses by invading rust pathogen hyphae (30). The increased activity of membrane transporters provides the needed fuel for the high primary metabolism activity observed in the invading rust fungi (Dataset S1, Tables S12 and S16). Strikingly, both rust fungi showed expansion of genes encoding auxin efflux carriers compared with other basidiomycetes (SI Text, Transporters), several of which are strongly expressed during plant infection. In addition, homologues of U. maydis auxin synthesis genes are also expressed during host infection. The potential for synthesis of auxin-like compounds that could regulate plant growth or development, as well as the expansion of strongly expressed auxin efflux carriers in rust fungi, suggests that fungal auxins could affect host hormone signaling and defense response or PCW integrity during rust infection.
Nitrate and Sulfate Assimilation Pathway Deficiencies in Rust Fungi.
Based on the inability of rust fungi to grow in vitro, we hypothesized that the M. larici-populina and P. graminis f. sp. tritici genomes may lack genes typically present in saprotrophic basidiomycetes. Major anabolic pathways of primary metabolism were manually inspected for potential deficiencies. Although the enzymes of the NH4+ assimilation pathway were identified, several genes involved in nitrate assimilation were lacking in both rust pathogen genomes; the nitrate/nitrite porter and the nitrite reductase are missing from the nitrate assimilation gene cluster found in other fungi (31). Loss of another pathway was specific to one of the genomes; genes required to perform the primary sulfate assimilation were identified in M. larici-populina whereas they were not found in P. graminis f. sp. tritici. The latter fungus lacks both α- and β-subunits of sulfite reductase (SiR), whereas the M. larici-populina β-subunit of SiR is missing the transketolase domain present in other fungal SiRs. The apparent incapacitation of nitrate and sulfate assimilation pathways in both rust fungi is consistent with their obligate biotrophic lifestyle, as they depend on reduced nitrogen (NH4+ or amino acids) and sulfur from plant cells. These metabolic deficiencies have also been found in other plant pathogens that represent two independent evolutionary lineages of obligate biotrophy in the oomycete (H. arabidopsidis) and ascomycete (B. graminis) lineages (11, 12).
Conclusions
Little is known about how obligate biotrophic rust fungi invade their hosts and avoid or suppress defense responses. The genome sequences of the poplar leaf and wheat stem rust fungi provide an unparalleled opportunity to address questions related to the obligate biotrophic lifestyle. The genetic changes that brought about the evolution of obligate biotrophy from nonbiotrophic progenitors remain obscure. Our comparisons of M. larici-populina and P. graminis f. sp. tritici to other saprotrophic, pathogenic, and symbiotic basidiomycetes indicate that developmental innovations in the rust fungi lineages did not involve major changes in the ancestral repertoire of conserved proteins with known function. However, gene family expansions observed for OPT, auxin efflux carriers, SOD, and signaling elements could reflect specific adaptations to this extreme parasitic lifestyle of these fungi. Similarly, lineage-specific gene families encoding zinc finger proteins, which may act as transcription factors during plant–rust interactions, suggest that these allow for different transcriptional programs in the two fungi. Analysis of these genomes revealed that the largest innovation of gene content encompasses the large set of lineage-specific, expanding gene families, which may enable developmental innovation and adaptation. Further, our analysis shows that the colonization of the host leaf, differentiation of pathogenic structures, and control of the plant immune system can be associated with a large-scale invention of lineage-specific proteins. For example, the rich repertoire of candidate effector-like SSPs could underlie the coevolution and adaptation of these obligate pathogens to the plant immune system. In contrast to obligate bacterial biotrophs and microsporidial fungal parasites, which frequently undergo gene loss and genome compaction (9, 10), the rust pathogen genomes are among the largest fungal genomes sequenced so far, as a result of expanded gene families and massive proliferation of TEs. No large-scale gene loss was observed in M. larici-populina and P. graminis f. sp. tritici, but some losses of clear impact, including the deletion of genes apparently not essential for the obligate biotrophic lifestyle (nitrate and sulfur assimilation), and a reduced set of PCW polysaccharide degrading enzymes, are genomic hallmarks of rust fungi, and, more broadly, of biotrophic pathogens as a group (11, 12). A deeper understanding of the complex array of the factors, including effector-like SSPs, affecting host–pathogen interactions and coevolution could enable efficient targeting of parasite-control methods in agricultural and forest ecosystems.
Materials and Methods
Genome Sequencing, Assembly, and Annotation.
The dikaryotic M. larici-populina 98AG31 and P. graminis f. sp. tritici CDL 75–36-700–3 (race SCCL) strains (SI Text, Background Information) were sequenced by whole-genome sequencing and were assembled into predicted 101.1-Mb and 88.6-Mb genomes, respectively (SI Text, Genome Sequencing and Assembly). The protein-coding genes were predicted with a combination of automated gene callers, ESTs produced from each rust fungus, and filtering dubious genes with similarity to TEs (SI Text, Gene Prediction and Annotation). In total, the gene sets included 16,399 and 17,773 predicted genes for M. larici-populina and P. graminis f. sp. tritici, respectively; these were the basis for multigene family analyses. The M. larici-populina genome sequence can be accessed at http://jgi.doe.gov/Melampsora and the P. graminis f. sp. tritici genome sequence can be accessed at http://www.broadinstitute.org/annotation/genome/puccinia_group/MultiHome.html.
Microarray Analysis of Gene Expression in Urediniospores and Rust-Infected Plants.
For both M. larici-populina and P. graminis f. sp. tritici, gene expression was assessed in resting and in vitro germinating urediniospores of the sequenced rust strains, as well as in respective host plant tissues at late stages of infection, by using specific custom 70-mer oligoarrays (SI Text, Whole-Genome Exon Oligoarrays). Methods for RNA isolation, probe synthesis and hybridization, and data capture and analyses are described in SI Text, Whole-Genome Exon Oligoarrays, and the data can be accessed in the Gene Expression Omnibus (GEO) database (GSE23097 for M. larici-populina and GSE25020 for P. graminis f. sp. tritici).
Supplementary Material
Acknowledgments
We thank M.-P. Oudot-LeSeq for the initial M. larici-populina TE annotation; B. Hilselberger for database construction, C. Commun and H. Niculita-Hirzel for the annotation of the M. larici-populina secretome and mating-type genes, respectively; and Jerry Johnson for technical assistance. The work conducted on M. larici-populina by the Joint Genome Institute of the US Department of Energy is supported by the Office of Science of the US Department of Energy under Contract DE-AC02-05CH11231. This project was also funded by grants from the Institut National de la Recherche Agronomique and the Région Lorraine Council (to F.M. and S.D.) and a grant from Natural Resources Canada (to R.C.H.), The sequencing of P. graminis f. sp. tritici was funded by the US National Science Foundation and conducted by the Broad Institute Sequencing Platform. The work of Y.-C.L., P.R., and Y.V.d.P. was supported by Interuniversity Attraction Pole P6/25 (BioMaGNet).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 8921.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AECX00000000 (M. larici-populina 98AG31) and AAWC01000000 (Puccinia graminis f. sp. tritici)]; the data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo [accession nos. GSE23097 (M. larici-populina 98AG31) and GSE25020 (Puccinia graminis f. sp. tritici)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019315108/-/DCSupplemental.
References
- 1.Aime MC, et al. An overview of the higher level classification of Pucciniomycotina based on combined analyses of nuclear large and small subunit rDNA sequences. Mycologia. 2006;98:896–905. doi: 10.3852/mycologia.98.6.896. [DOI] [PubMed] [Google Scholar]
- 2.Cummins GB, Hiratsuka Y. Illustrated Genera of Rust Fungi. 3rd Ed. St. Paul: APS Press; 2004. [Google Scholar]
- 3.Leonard KJ, Szabo LJ. Stem rust of small grains and grasses caused by Puccinia graminis. Mol Plant Pathol. 2005;6:99–111. doi: 10.1111/j.1364-3703.2005.00273.x. [DOI] [PubMed] [Google Scholar]
- 4.Stokstad E. Plant pathology. Deadly wheat fungus threatens world's breadbaskets. Science. 2007;315:1786–1787. doi: 10.1126/science.315.5820.1786. [DOI] [PubMed] [Google Scholar]
- 5.Rubin EM. Genomics of cellulosic biofuels. Nature. 2008;454:841–845. doi: 10.1038/nature07190. [DOI] [PubMed] [Google Scholar]
- 6.Duplessis S, Major I, Martin F, Séguin A. Poplar and pathogen interactions: insights from Populus genome-wide analyses of resistance and defense gene families and gene expression profiling. Crit Rev Plant Sci. 2009;28:309–334. [Google Scholar]
- 7.Gérard PR, Husson C, Pinon J, Frey P. Comparison of genetic and virulence diversity of Melampsora larici-populina populations on wild and cultivated poplar and influence of the alternate host. Phytopathology. 2006;96:1027–1036. doi: 10.1094/PHYTO-96-1027. [DOI] [PubMed] [Google Scholar]
- 8.Dodds PN, et al. Effectors of biotrophic fungi and oomycetes: Pathogenicity factors and triggers of host resistance. New Phytol. 2009;183:993–1000. doi: 10.1111/j.1469-8137.2009.02922.x. [DOI] [PubMed] [Google Scholar]
- 9.Ochman H, Moran NA. Genes lost and genes found: Evolution of bacterial pathogenesis and symbiosis. Science. 2001;292:1096–1099. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- 10.Corradi N, Pombert JF, Farinelli L, Didier ES, Keeling PJ. The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nat Commun. 2010;1:77. doi: 10.1038/ncomms1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spanu PD, et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330:1543–1546. doi: 10.1126/science.1194573. [DOI] [PubMed] [Google Scholar]
- 12.Baxter L, et al. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science. 2010;330:1549–1551. doi: 10.1126/science.1195203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuomo CA, Birren BW. The fungal genome initiative and lessons learned from genome sequencing. Methods Enzymol. 2010;470:833–855. doi: 10.1016/S0076-6879(10)70034-3. [DOI] [PubMed] [Google Scholar]
- 14.Martin F, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- 15.Kämper J, et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 2006;444:97–101. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- 16.Schirawski J, et al. Pathogenicity determinants in smut fungi revealed by genome comparison. Science. 2010;330:1546–1548. doi: 10.1126/science.1195330. [DOI] [PubMed] [Google Scholar]
- 17.Haas B, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 18.Raffaele S, et al. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science. 2010;330:1540–1543. doi: 10.1126/science.1193070. [DOI] [PubMed] [Google Scholar]
- 19.Rouxel T, et al. Effector diversification within compartments of the Leptosphaeria maculans genome affected by repeat-induced point mutations. Nature Commun. 2011;2:202. doi: 10.1038/ncomms1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panstruga R, Dodds PN. Terrific protein traffic: the mystery of effector protein delivery by filamentous plant pathogens. Science. 2009;324:748–750. doi: 10.1126/science.1171652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis JG, Rafiqi M, Gan P, Chakrabarti A, Dodds PN. Recent progress in discovery and functional analysis of effector proteins of fungal and oomycete plant pathogens. Curr Opin Plant Biol. 2009;12:399–405. doi: 10.1016/j.pbi.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Voegele RT, Hahn M, Mendgen K. The uredinales: cytology, biochemistry, and molecular biology. In: Deising HB, editor. The Mycota V: Plant Relationships. Berlin: Spinger; 2009. pp. 69–98. [Google Scholar]
- 23.Xu J, et al. Gene discovery in EST sequences from the wheat leaf rust fungus Puccinia triticina sexual spores, asexual spores and haustoria, compared to other rust and corn smut fungi. BMC Genomics. 2011;12:161. doi: 10.1186/1471-2164-12-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Catanzariti A-M, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell. 2006;18:243–256. doi: 10.1105/tpc.105.035980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godfrey D, et al. Powdery mildew fungal effector candidates share N-terminal Y/F/WxC-motif. BMC Genomics. 2010;11:317. doi: 10.1186/1471-2164-11-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohm RA, et al. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol. 2010;28:957–963. doi: 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
- 28.El Gueddari NE, Rauchhaus U, Moerschbacher BM, Deising HB. Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 2002;156:103–112. [Google Scholar]
- 29.Wahl R, Wippel K, Goos S, Kämper J, Sauer N. A novel high-affinity sucrose transporter is required for virulence of the plant pathogen Ustilago maydis. PLoS Biol. 2010;8:e1000303. doi: 10.1371/journal.pbio.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voegele RT, Struck C, Hahn M, Mendgen K. The role of haustoria in sugar supply during infection of broad bean by the rust fungus Uromyces fabae. Proc Natl Acad Sci USA. 2001;98:8133–8138. doi: 10.1073/pnas.131186798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slot JC, Hibbett DS. Horizontal transfer of a nitrate assimilation gene cluster and ecological transitions in fungi: a phylogenetic study. PLoS ONE. 2007;2:e1097. doi: 10.1371/journal.pone.0001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.