Abstract
A highly efficient DNA sequencing strategy was developed on the basis of the bacteriophage Mu in vitro DNA transposition reaction. In the reaction, an artificial transposon with a chloramphenicol acetyltransferase (cat) gene as a selectable marker integrated into the target plasmid DNA containing a 10.3-kb mouse genomic insert to be sequenced. Bacterial clones carrying plasmids with the transposon insertions in different positions were produced by transforming transposition reaction products into Escherichia coli cells that were then selected on appropriate selection plates. Plasmids from individual clones were isolated and used as templates for DNA sequencing, each with two primers specific for the transposon sequence but reading the sequence into opposite directions, thus creating a minicontig. By combining the information from overlapping minicontigs, the sequence of the entire 10,288-bp region of mouse genome including six exons of mouse Kcc2 gene was obtained. The results indicated that the described methodology is extremely well suited for DNA sequencing projects in which considerable sequence information is on demand. In addition, massive DNA sequencing projects, including those of full genomes, are expected to benefit substantially from the Mu strategy.
[The sequence data reported in this paper have been submitted to the GenBank data library under accession no. AJ011033.]
During the last decade, the sequenced DNA information has accumulated with increasing pace. The yeast genome, as well as a variety of microbial genomes, has already been sequenced entirely, and projects destined to sequence several eukaryotic genomes including those of man, mouse, fly, worm, and plant are being completed in the near future (Ash 1997; Koonin 1997; Beck and Sterk 1998). Nevertheless, the need for DNA sequencing is not expected to be diminished. For example, comparative genome analyses, between species or within species, that will yield a wealth of information concerning principles of life, will necessarily involve massive DNA sequencing.
Efficient DNA sequencing strategy that will yield sequence information reliably and rapidly is a necessity for a sequencing project. In principle, the strategies used so far can be divided into three main categories: (1) random, (2) directed, and (3) transposon-based strategies. The most widely used strategy, especially for large-scale DNA sequencing projects, is a random shotgun approach (for review, see Hunkapiller et al. 1992; Fraser and Fleischmann 1997). Directed strategies, typified by primer-walking and nested deletions, have been used mainly to sequence DNA regions of moderate size (for review, see Ansorge et al. 1997). Transposon-based strategies combine features of random and directed approaches and can be used efficiently in various types of sequencing projects.
Several DNA sequencing strategies based on in vivo DNA transposition reactions have been characterized (Adachi et al. 1987; Phadnis et al. 1989; Strathman et al. 1991; Kasai et al. 1992; Berg et al. 1993). However, in vivo approaches require special host strains and multiple steps of manipulation that limit the usefulness of these strategies. In addition, in some of the cases, a stringency of the target-site selectivity was not optimal for sequencing purposes.
In vitro DNA transposition-based sequencing strategy has also been described and shown to be highly useful, especially for sequencing repetitive DNA (Devine and Boeke 1994; Devine et al. 1997). The system utilized a customized artificial transposon in combination with integrase activity present in purified yeast Ty1 retrotransposon virus-like particles. However, no DNA sequencing strategy that would utilize a single transposase protein in in vitro conditions has yet been described. Such a system would be advantageous: Relative simplicity of protein purification compared with that of protein complexes, as well as at least potentially more preferable stability characteristics of proteins, make a transposase-catalyzed reaction an attractive choice for sequencing purposes.
Mu DNA transposition reaction is one of the best-characterized transposition reactions (Mizuuchi 1992 a,b). In vivo, and in certain reaction conditions with plasmid substrates in vitro, the reaction is relatively complex and involves several protein and DNA cofactors (for reviews, see Chaconas et al. 1996; Lavoie and Chaconas 1996) of which the phage-encoded MuB protein affects target selection (Adzuma and Mizuuchi 1988, 1989; Mizuuchi and Mizuuchi 1993). However, with certain DNA substrates and reaction conditions in vitro the requirements for efficient transposition reaction are substantially relaxed (Craigie and Mizuuchi 1986, 1987; Mizuuchi and Mizuuchi 1989; Baker and Mizuuchi 1992; Savilahti et al. 1995). The minimal macromolecular components for in vitro transposition into intermolecular targets are the MuA transposase, the donor DNA, and the target DNA (Savilahti et al. 1995). The chemical steps of Mu transposition proceed within a protein–DNA complex called Mu transpososome that in its core contains the critical components, a tetramer of MuA transposase and Mu DNA ends (Craigie and Mizuuchi 1987; Surette et al. 1987; Lavoie et al. 1991; Baker and Mizuuchi 1992; Baker et al. 1993; Savilahti et al. 1995). Initially, the Mu transpososome core is assembled from four molecules of MuA and two Mu DNA ends, and this underlining architecture is then maintained throughout the two chemical steps of transposition. First, the transposon DNA is cleaved at its 3′ ends (called the donor cleavage). Second, the target DNA is cut at staggered positions and the target 5′ ends are joined into the 3′ ends of the transposon DNA in a concerted reaction (called strand transfer). In vitro, the first of the chemical steps can also be bypassed artificially by use of a modified donor DNA substrate. It was shown that Mu donor DNA that was cut precisely at Mu 3′ ends by a restriction endonuclease was efficiently utilized in transferring these precut 3′ ends into the target DNA (Craigie and Mizuuchi 1987). In turn, this result brought about an opportunity to use the Mu transposition reaction in a simple manner for genetic manipulation.
In this paper we describe a highly efficient and easy methodology for DNA sequencing that is based on the bacteriophage Mu in vitro transposition reaction. As a test case, we used the system to determine a 10,288-bp sequence from a mouse genomic locus containing a Kcc2 gene. The respective protein, KCC2, is a K+/Cl− cotransporter that is abundantly expressed in brain and suggested to be involved in extruding chloride in mature neurons (Payne et al. 1996). We are currently producing mice with targeted inactivation of Kcc2 and, for this purpose, the sequence of the mouse genomic fragment was needed. Bacteriophage Mu in vitro DNA transposition system proved to be very useful for DNA sequencing purposes: (1) The reaction in vitro is simple; only one protein component, the MuA transposase, is needed in addition to donor DNA, target DNA, and buffer. (2) The transposition reaction is highly efficient and reproducible in in vitro conditions. (3) The artificial cat–Mu transposon contains an easily selectable marker, chloramphenicol acetyltransferase gene (cat), that allows convenient selection of integrants. (4) Mu in vitro transposition produces relatively even distribution of integrations into the target DNA.
RESULTS
Experimental Outline
We developed a DNA sequencing strategy that is based on bacteriophage Mu in vitro DNA transposition reaction (Fig. 1). In the reaction, the artificial cat–Mu transposon integrated in vitro into the target plasmid DNA to be sequenced. The reaction products were then transformed into Escherichia coli cells and the bacteria were selected for antibiotic resistance to identify clones that carried target plasmids with cat–Mu transposon insertions. From the selected clones, a sequence on both sides of the inserted transposon was retrieved by use of two primers specific for the cat–Mu sequence. Each clone thus created a minicontig. The final sequence was obtained by assembling the overlapping minicontigs.
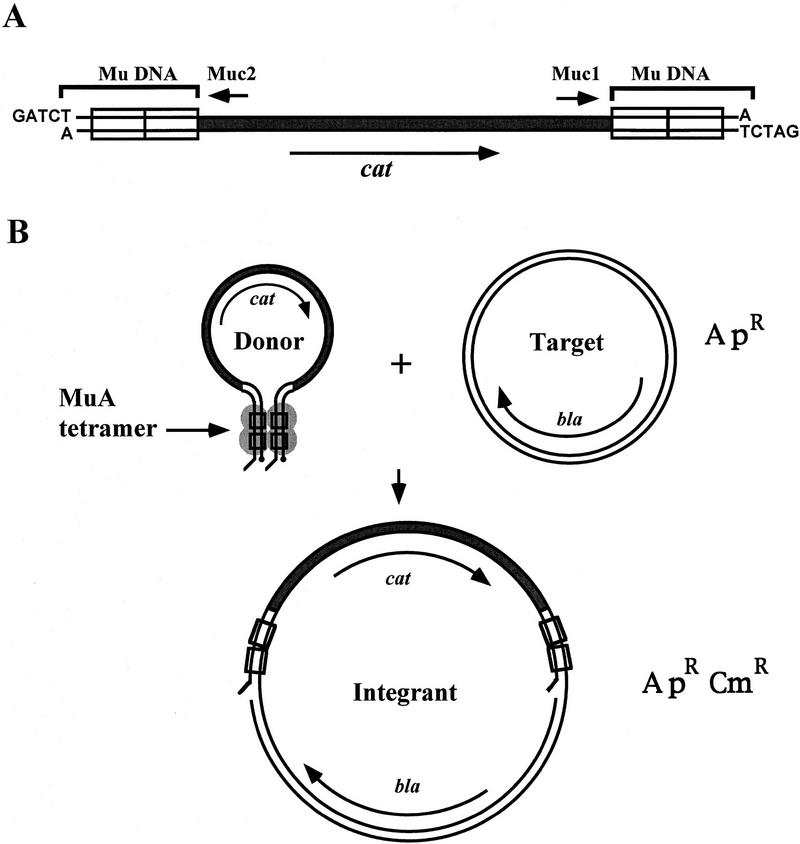
Figure 1.
A schematic representation of the utilized in vitro transposition system. (A) cat–Mu minitransposon. The linear transposon fragment is shown to be released from its vector plasmid by BglII digestion; it is therefore in precut configuration. (Rectangles) MuA binding sites; (small arrows) primers used in sequencing. (B) The reaction. MuA forms with the transposon ends a tetrameric transposition complex, a transpososome, that integrates the minitransposon into the target DNA. (ApR, CmR) Ampicillin and chloramphenicol resistance, respectively. (Long arrows) Genes coding for β-lactamase (bla) and CAT (cat); (•) reactive donor DNA 3′ ends.
CAT–Mu Artificial Transposon
The CAT–Mu artificial transposon that was used as a donor DNA in an in vitro transposition reaction contains the cat gene as a selectable marker and flanking sequences with inverted 50-bp segments of Mu right end sequence (Fig. 1A). This 1.2-kb transposon fragment was released from its vector plasmid by BglII digestion that leaves four nucleotide 5′-overhangs flanking the transposon sequence and thus generates the precut end configuration. The transposon end configuration has been shown to have an influence on Mu transpososome core assembly and stability; precut molecules with overhangs were among the most efficient in these respects (Savilahti et al. 1995; Savilahti and Mizuuchi 1996). In addition, precut donor molecules are readily available for the second chemical step of transposition, the strand-transfer reaction (Craigie and Mizuuchi 1987; Savilahti et al. 1995).
Transposition Reaction
We performed in vitro transposition reaction using a cat–Mu transposon fragment as a donor DNA and supercoiled plasmid pSR11 that contains the mouse genomic insert to be sequenced as a target DNA (Fig. 1B). In the reaction conditions used, most of the primary reaction products are open circular target molecules with an integrated transposon in different positions (data not shown). The molar ratio of donor to target was adjusted to avoid multiple transposon integrations into a single target molecule; such products would have been useless for sequencing purposes.
A portion of the reaction products was transformed into electrocompetent E. coli DH5α cells and plated onto selection plates containing chloramphenicol and ampicillin. The yield of transformants was 540 colonies per 50 ng of target DNA with electrocompetent DH5α cells that showed transformation efficiency of ∼107 cfu/μg pUC19 DNA. DNA from a control reaction without MuA did not produce colonies.
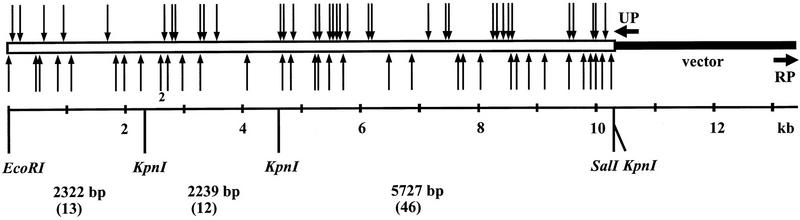
Plasmids were prepared from 95 randomly chosen colonies by a quick boiling method. A restriction analysis was then performed with EcoRI–KpnI double digestion to identify the clones that had the transposon integrated into the cloned insert and not into the vector backbone of the plasmid (Fig. 2). Of the clones examined, 93 exhibited a clear restriction pattern consistent with a single transposon insertion in the plasmid. Plasmids from 79 clones contained the transposon in the cloned mouse DNA portion and 14 in the vector portion of the molecule. For the 71 selected clones, sequencing quality plasmid preparations were then generated and sequencing was performed with two cat–Mu specific primers that read the sequence from within the transposon but toward opposite directions (Tables 1 and 2). A total of 10,288 bp of sequence was retrieved (deposited in GenBank; accession no. AJ011033). On the average, ∼500 bp could be read with one primer. Thus, an average length of a minicontig, connected from the data of two sequencing runs on the same template, was ∼1000 bp. The location and orientation of each minicontig representing the 71 sites of cat–Mu insertions in plasmid pSR11 mouse DNA insert region are shown in Figures 2 and 3. The cat gene of the transposon was in the same relative orientation as the KCC2 gene in 36 clones; in 35 clones the orientation was opposite.
Figure 2.
Distribution of cat–Mu integration sites in the plasmid pSR11. (Open bar) Mouse genomic insert; (arrows) transposon integration sites: Above the bar are indicated the transposons that contained the cat gene in the same orientation as the Kcc2 gene; below, the transposons are in the opposite orientation. (RP, UP) Primers used to sequence the insert ends. The restriction sites used in cloning and screening of integrants are indicated. Also shown are lengths of the restriction fragments (in bp) produced by EcoRI–KpnI double digestion as well as the number of sequenced clones for each restriction fragment (in parenthesis).
Table 1.
Sequencing the Mouse KCC2 Locus with the Mu System
| Plasmid | pSR11 |
| Insert size (kb) | 10.3 |
| Vector size (kb) | 2.9 |
| Minitransposon used | cat–Mu |
| E. coli cells used | |
| Strain | DH5α |
| Competence (cfu/μg pUC19 DNA) | ∼107 |
| Plasmid introduced into cells by | electroporation |
| Fraction of standard reaction used in electroporation | 1:10 |
| Number of ApR/CmR colonies obtained | 540 |
| Number of clones screened by restriction analysis | 95 (100%) |
| two plasmids apparent | 2a (2%) |
| one plasmid apparent | 93 (98%) |
| Clones with one transposon | 93 (98%) |
| in vector | 14 (15%) |
| in insert | 79 (83%) |
| Number of clones sequenced | 71 (75%) |
| Total sequenced (bp) | 68,143 |
| Final sequence (bp) | 10,288 |
| Average read per sequence reaction | |
| (nucleotide) | 494 |
| Redundancy (fold) | 6.6 |
Sequences retrieved using RP and UP primers are included.
These clones exhibited a restriction pattern consistent with cotransformation of two plasmids.
Table 2.
Statistics of Insertions
| Insert region (kb) | No of insertions |
|---|---|
| 0–1 | 8 |
| 1–2 | 3 |
| 2–3 | 9 |
| 3–4 | 4 |
| 4–5 | 6 |
| 5–6 | 11 |
| 6–7 | 4 |
| 7–8 | 5 |
| 8–9 | 9 |
| 9–10 | 8 |
| 10–10.288a | 4 |
The last region is 288 bp in length. Mean interval distance = 146.5; s.d. of mean interval distance = 144.0; minimum interval distance = 0; maximum interval distance = 570; mean no. of insertions per kb target DNA = 6.9; Orientation: forward 36 (50.7%); reverse 35 (49.3%). n = 71.
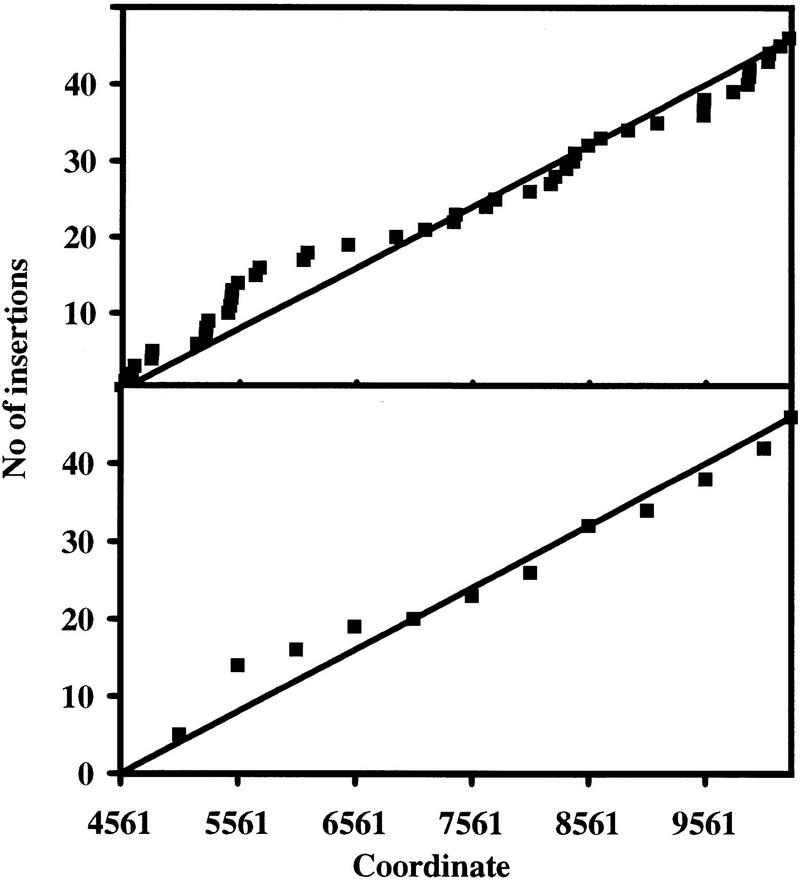
Figure 3.
Distribution of 46 Mu transposon insertions in the 5727-bp KpnI fragment. Insertions into this fragment were screened by restriction analysis and appropriate clones were randomly chosen for sequence analysis. The cumulative number of insertions is plotted against the coordinates in the final sequence. (Top) 1-bp resolution; (bottom) 500-bp resolution. On the basis of this analysis, the distribution appears to be relatively even; totally even distribution would produce a line as shown. The result implies that, at least in the scale relevant for DNA sequencing, the target site selection can be described as a random or near-random process.
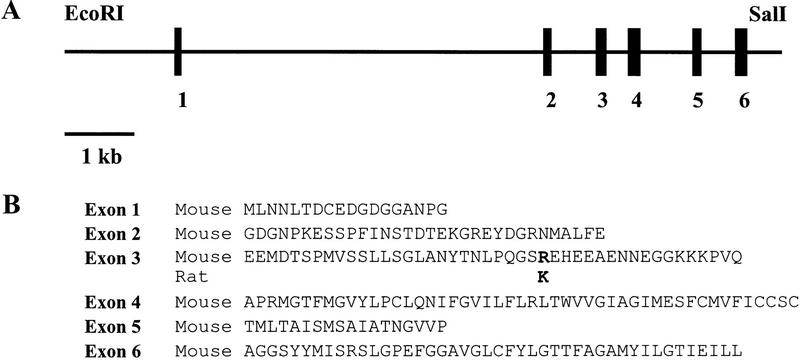
The exon–intron structure of the mouse genomic 10,288-bp Kcc2 fragment was compared with that of rat (Fig. 4). Six (putative) exons were identified that matched the sequence of rat Kcc2 cDNA (nucleotides 1–728, GenBank accession no. U55816). Nucleotide identity between the mouse exons and rat Kcc2 cDNA was 94%. Only a single amino acid difference between the mouse and rat KCC2 sequence was observed (mouse Arg-76 to rat Lys-76 in exon 3).
Figure 4.
(A) Exon/intron structure of mouse Kcc2 gene region. (Solid rectangles) Exons. (B). Predicted amino acid sequences encoded by the mouse Kcc2 exons. The only amino acid difference between mouse and rat sequence is shown in boldface type in exon 3.
DISCUSSION
The development of an efficient DNA sequencing strategy that is based on the bacteriophage Mu in vitro DNA transposition reaction allowed us to rapidly sequence >10 kb of mouse genomic DNA (Tables 1 and 2). The system has many attractive features that allow the production of reliable sequence information effectively and accurately (Table 3).
Table 3.
Comparison of the Characteristics of Mu and Ty1 Sequencing Strategies
| Mu | Ty1 | |
|---|---|---|
| Reaction catalyzed by | MuA transposase | Ty1 virus-like particles |
| Donor DNA length (kb) | 1.2 | 0.9 |
| Introduction of reaction products into cells | transformation/electroporation | electroporation |
| Reaction efficiency | ||
| maximum reported (insertions per target) | 1/10 | 1/2.5 × 103 |
| Target site duplication (bp) | 5a | 5b |
| Standard reaction | ||
| donor used (ng) | 18 | 100–500 |
| target used (ng) | 500 | 1000 |
| molar ratio of transposon to target | 1:2 | 10:1 |
| Reported recovery of clones | ||
| minipreps studied (no., %) | 95, 100 | 153, 100c |
| double insertions/cotransformants (%) | 2 | 8 |
| unknown plasmid map (%) | 0 | 3 |
| no transposon (%) | 0 | <1 |
| single insertion in one plasmid (%) | 98 | 88 |
Mu characteristics (this study); Ty1 characteristics (Devine and Boeke 1994).
Allet (1979); Kahmann and Kamp (1979); Mizuuchi and Mizuuchi (1993); this study.
Eichinger and Boeke (1988, 1990); Ji et al. (1993); Braiterman and Boeke (1994); Primer Island Transposition Kit Protocol (Applied Biosystems).
The numbers for the Ty1 system were calculated from Table 2 of Devine and Boeke (1994). For comparison the number of minipreps studied was adjusted to 100%
The Mu in vitro reaction is relatively simple and easy to perform. The critical macromolecular components in the reaction are the MuA transposase, the donor DNA, and the target DNA. In addition, only a simple buffer with standard components including BSA, glycerol, Triton X-100, and MgCl2 is required. Because no purified protein complexes or protein–DNA complexes are needed, the preparation of the reaction components by standard molecular techniques is relatively straightforward. We note that the amount of artificial transposon fragment required for the reaction is on a nanogram scale. In practice, it follows that substrate donor DNA for a large number of reactions can be prepared economically and with minimum effort. We have also used PCR for donor DNA preparation and found the method satisfactory and quick (data not shown).
The Mu in vitro transposition reaction is highly efficient. With just one 25-μl reaction, which contained only 18 ng of donor DNA and 500 ng of target DNA, and of which only one-tenth was electrotransformed into E. coli cells, >500 bacterial clones containing plasmids with transposon inserts were produced. We used electrocompetent E. coli cells that, in control electrotransformation, yielded ∼107 cfu/μg supercoiled pUC19 DNA. The result indicated that if cells of extremely high competence (e.g., competence of ∼109 cfu/μg DNA) were to be used, the maximum number of recoverable integrants from one 25-μl reaction would be 500,000. Such collection of transposon-containing plasmids is expected to be appropriate even for the most demanding sequencing projects and, in principle, all of the clones needed for a project can easily be generated in a matter of one day. The result also indicated that transformation frequencies that are easily obtained with conventionally prepared competent cells (i.e., 106–107 cfu/μg DNA) are efficient enough for obtaining a sufficient number of clones for sequencing of lengthy inserts. In fact, we have used the system in several sequencing projects with competent cells prepared by the standard CaCl2 method (Sambrook et al. 1989) and easily obtained several hundred clones for sequencing purposes. Highly productive recovery of plasmids with transposon insertions is the result of efficient in vitro reaction as well as favorable primary reaction product profile. By adjusting donor DNA concentration in the reaction, up to 10% of targets can be converted to integrants, and essentially all primary reaction products are derived from two-ended integrations of one donor molecule into the target (data not shown). For comparison, the reported efficiency of Ty1 in vitro reaction was 1 insertion per 2.5 × 103 targets (Devine and Boeke 1994) and the system utilized electroporation as a means to introduce transposon-containing plasmids into the host cells (see Table 3).
The system used appears to recapitulate the genuine Mu transposition reaction in that the hallmark target site duplication (Allet 1979; Kahmann and Kamp 1979) is faithfully generated in the process. All of the 71 sequenced clones contained a duplicated 5-bp target site flanking the transposon. Furthermore, in several other projects we have mapped by sequencing >200 integration sites and found the accurate 5-bp duplication in each of them. In favorable conditions, one sequencing run can produce 500–800 nucleotides worth of sequence information. Consequently, one minicontig can span a region of 1000–1600 nucleotides. Relatively few clones are therefore required to sequence a typical cloned insert in a plasmid vector. We mapped the transposon insertions with restriction enzyme cuts that distinguish between insertions into the vector portion and into the few restriction fragments of the insert portion of the plasmid. In some other cases, however, and especially if most of the sequence has already been retrieved, more accurate mapping or prescreen might be pertinent. An efficient strategy would utilize an enzyme that cuts the transposon sequence once or more but the rest of the plasmid only once.
One of the important aspects of artificial transposon is the selectable marker that should provide a reliable identification for transposon insertions. In the artificial cat–Mu transposon, we used a cat gene that has been one of the most widely used selectable markers in bacterial genetics over the years (Sambrook et al. 1989; Miller 1992). In this project, this gene also proved to be convenient and easy to use for selection; we found no background colonies among the clones studied (for comparison to the Ty1 system, see recovery of clones in Table 3).
The cat-Mu insertions were recovered in all areas of the region to be sequenced (Fig. 2 and Table 2) and their distribution was relatively even (Fig. 3). The selection of target sequences by the MuA transposase, therefore, appears to be random enough to be useful in DNA sequencing. This situation contrasts with Mu reaction in vivo and with plasmid substrates in vitro when MuB is included in the reaction. Regional target preference was observed for which the MuB protein was implicated (Mizuuchi and Mizuuchi 1993). Recently, we have used the Mu system also in other DNA sequencing projects and reproducibly found similar, relatively even distribution of transposon insertions (data not shown). In this study (see Table 2) the mean interval distance of sequenced insertions needed to obtain the final sequence was ∼150 bp and thus well below the length of a typical sequencing run. Using an up to date DNA sequencer, we experienced no difficulty in producing reliable final sequence with these 71 insertions even though the maximum insertion distance was as long as 570 bp. However, in some cases, it may be desirable to lower the mean interval distance of insertions; this can be done by simply sequencing more insertions. Our data indicate that for obtaining reliable final sequence for a 2-kb insert (with typical sequencing parameters used in this study, see Table 1) ∼10 clones need to be sequenced (see the 2322-bp and 2239-bp fragments in Fig. 2). However, when longer inserts (without prescreening) are to be sequenced, on the basis of statistics of random insertions, relatively more clones are progressively required (see also the 5727-bp fragment in Fig. 2).
The described in vitro system is easily amenable to modifications to meet the needs of various sequencing projects. For example, different selectable marker genes may be desirable instead of the cat gene; any selectable or screenable marker is expected to be usable in minitransposons between the Mu ends. For screening purposes, a transposon containing a rare restriction site such as NotI might be desirable. PCR-based screening strategies will probably be very useful for mapping of individual transposon insertion sites.
The Mu in vitro transposition reaction-based DNA sequencing strategy is simple and, therefore, amenable to automatization. Consequently, high-throughput sequencing projects are expected to benefit substantially from the described Mu strategy. The system is currently in routine use in the sequencing facility of the Institute of Biotechnology and it is regarded as the method of choice for miscellaneous DNA sequencing projects.
METHODS
Proteins, Plasmids, and Reagents
MuA was overexpressed and purified in collaboration with Finnzymes (Espoo, Finland) essentially as described (Baker et al. 1993) except that the chromatographic steps were performed on heparin–agarose (Kemotex, Tallinn, Estonia) and Mono S (Pharmacia). The target for transposition-aided sequencing was plasmid pSR11 containing a 10.3-kb mouse genomic EcoRI–SalI insert. The insert was screened from a 129/Sv mouse library (Stratagene) with a rat Kcc2 cDNA (Payne et al. 1996) fragment as a probe and subcloned into pBluescript II SK+ (Stratagene) vector backbone between EcoRI and SalI sites. The plasmid was purified by use of a Qiagen plasmid purification kit. Restriction endonucleases were from New England Biolabs, T4 DNA ligase from Life Technologies, BSA from Sigma, Triton X-100 from Boehringer Mannheim, and agarose from Promega.
cat–Mu Artificial Transposon
Construction of the cat–Mu artificial transposon-containing plasmid will be described in detail elsewhere. The cat–Mu transposon (Fig. 1A) contains the CAT gene from plasmid pBC SK+ (Stratagene) that is flanked by a pair of 50-bp Mu right-end segments (Savilahti et al. 1995) in inverted orientation relative to each other. From the vector plasmid, the transposon fragment was released by BglII digestion that leaves four nucleotide 5′-overhangs flanking the exposed transposon ends. The cat–Mu transposon is therefore in precut configuration that ensures efficient transpososome assembly and stability (Savilahti et al. 1995; Savilahti and Mizuuchi 1996). The cat–Mu fragment was purified chromatographically with a TSK-gel DNA-NPR (Tosohaas, Stuttgart, Germany) anion exchange column.
In Vitro Transposition Reactions
In vitro transposition reaction (Fig. 1B) was performed with the cat–Mu artificial transposon as a donor DNA and plasmid pSR11 as a target DNA. Transposition reactions (25 μl) contained 0.02 pmole (18 ng) donor, 0.06 pmole (500 ng) target, 2.7 pmoles (0.2 μg) MuA, 25 mm Tris-HCl at pH 8.0, 100 μg/ml BSA, 15% (wt/vol) glycerol, 0.05% (wt/vol) Triton X-100, 126 mm NaCl, and 10 mm MgCl2. Reactions were carried out at 30°C for 1 hr and stopped by freezing in liquid nitrogen.
Recovery of Plasmids Carrying Inserted CAT–Mu Transposon and Insertion Screen
In vitro transposition reaction was sequentially phenol and chloroform extracted, ethanol precipitated, washed with 75% ethanol, dried, and resuspended in 5 μl of TEN buffer (10 mm Tris-HCl at pH 7.5, 0.5 mm EDTA, 50 mm NaCl). From the reaction, 0.5 μl was then electrotransformed into 25 μl of competent E. coli DH5α (Life Technologies) cells prepared as described (Ausubel et al. 1989). Electrotransformation was done with 1-mm cuvettes in a Bio-Rad Genepulser II with the following settings: capacitance, 25 μF; voltage 1.8 kV; and resistance, 200 ohms. One milliliter of Luria Broth (LB) was then added, and the bacteria were grown for 1 hr at 37°C, collected by centrifugation, and plated on LB plates containing chloramphenicol (5 μg/ml) and ampicillin (100 μg/ml). Plasmid preparations were made from colonies obtained by a quick boiling method (Holmes and Quigley 1981). To distinguish between transposon insertions into the vector portion of the plasmid and those into the cloned mouse DNA insert, a restriction analysis was performed by KpnI and EcoRI double digestion (Fig. 2) in CA buffer (20 mm Tris-HCl at pH 7.5, 7 mm MgCl2, 100 mm KCl, 2 mm β-mercaptoethanol).
DNA Techniques, Sequencing, and Data Analysis
Standard DNA techniques were performed as described (Sambrook et al. 1989). Plasmids for sequencing were prepared by use of a QIAprep Spin Miniprep kit (Qiagen). The transposon specific primers were as follows: Muc1 (5′-GCTCTCCCCGTGGAGGTAAT-3′) and Muc2 (5′-TTCCGTCACAGGTATTTATTCGGT-3′). The vector specific primers for sequencing the ends of cloned mouse insert were as follows: RP (5′- TTTCACACAGGAAACAGCTATGAC-3′) and UP (5′-CGACGTTGTAAAACGACGGCCAGT-3′). Sequencing reactions were performed with a BigDye terminator cycle sequencing kit (Perkin Elmer) and analyzed by an ABI 377 XL automated DNA sequencer (Perkin Elmer). The sequence assembly and data analysis were done by the xgap program in the Staden Package (Bonfield et al. 1995) on a SUN workstation. The sequence has been deposited in the European Molecular Biology Laboratory GenBank under accession number AJ011033.
Acknowledgments
Sari Nieminen, Pirjo Rahkola, and Noora Salovuori are acknowledged for excellent technical assistance. Dr. John Payne kindly provided the rat Kcc2 cDNA. We thank Erkki Koivunen, Arja Lamberg, and Pekka Lappalainen for critical comments on the manuscript and Hannu Rita for valuable suggestions concerning statistical analysis of the data. Financial support was obtained from Biocentrum Helsinki (to M.A., L.P., and H.S.), Academy of Finland (to M.A. and H.S.), Technology Development Centre (to H.S.), and Oskar Öflund Foundation (to S.S.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
NOTE ADDED IN PROOF
More information about the function of the K+/Cl− cotransporter KCC2 is now available (Rivera et al. 1998, Nature 397: 251–255).
Footnotes
E-MAIL harri.savilahti@helsinki.fi; FAX 358-9-708 59366.
REFERENCES
- Adachi T, Mizuuchi M, Robinson EA, Appella E, O’Dea MH, Gellert M, Mizuuchi K. DNA sequence of the E. coli gyrB gene: Application of a new sequencing strategy. Nucleic Acids Res. 1987;15:771–784. doi: 10.1093/nar/15.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzuma K, Mizuuchi K. Target immunity of Mu transposition reflects a differential distribution of MuB protein. Cell. 1988;53:257–266. doi: 10.1016/0092-8674(88)90387-x. [DOI] [PubMed] [Google Scholar]
- ————— Interaction of proteins located at a distance along DNA: Mechanism of target immunity in the Mu DNA strand-transfer reaction. Cell. 1989;57:41–47. doi: 10.1016/0092-8674(89)90170-0. [DOI] [PubMed] [Google Scholar]
- Allet B. Mu insertion duplicates a 5 base pair sequence at the host inserted site. Cell. 1979;16:123–129. doi: 10.1016/0092-8674(79)90193-4. [DOI] [PubMed] [Google Scholar]
- Ansorge W, Voss H, Zimmermann J. DNA sequencing strategies: Automated and advanced approaches. New York, NY: John Wiley & Sons Inc.; 1997. [Google Scholar]
- Ash C. Year of the genome. Trends Microbiol. 1997;5:135–139. doi: 10.1016/S0966-842X(97)01031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Curr Prot Mol Biol. 1989;1:1.8.4.–1.8.10. [Google Scholar]
- Baker TA, Mizuuchi K. DNA-promoted assembly of the active tetramer of the Mu transposase. Genes & Dev. 1992;6:2221–2232. doi: 10.1101/gad.6.11.2221. [DOI] [PubMed] [Google Scholar]
- Baker TA, Mizuuchi M, Savilahti H, Mizuuchi K. Division of labor among monomers within the Mu transposase tetramer. Cell. 1993;74:723–733. doi: 10.1016/0092-8674(93)90519-v. [DOI] [PubMed] [Google Scholar]
- Beck S, Sterk P. Genome-scale DNA sequencing: Where are we? Curr Opin Biotechnol. 1998;9:116–120. doi: 10.1016/s0958-1669(98)80094-3. [DOI] [PubMed] [Google Scholar]
- Berg CM, Wang G, Straughbaugh LD, Berg DE. Transposon-facilitated sequencing of DNAs cloned in plasmids. Methods Enzymol. 1993;218:279–306. doi: 10.1016/0076-6879(93)18022-5. [DOI] [PubMed] [Google Scholar]
- Bonfield JK, Smith KF, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braiterman LT, Boeke JD. In vitro integration of retrotransposon Ty1: A direct physical assay. Mol Cell Biol. 1994;14:5719–5730. doi: 10.1128/mcb.14.9.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaconas G, Lavoie BD, Watson MA. DNA transposition: Jumping gene machine, some assembly required. Curr Biol. 1996;6:817–820. doi: 10.1016/s0960-9822(02)00603-6. [DOI] [PubMed] [Google Scholar]
- Craigie R, Mizuuchi K. Role of DNA topology in Mu transposition: Mechanism of sensing the relative orientation of two DNA segments. Cell. 1986;45:793–800. doi: 10.1016/0092-8674(86)90554-4. [DOI] [PubMed] [Google Scholar]
- ————— Transposition of Mu DNA: Joining of Mu to target DNA can be uncoupled from cleavage at the ends of Mu. Cell. 1987;51:493–501. doi: 10.1016/0092-8674(87)90645-3. [DOI] [PubMed] [Google Scholar]
- Devine SE, Boeke JD. Efficient integration of artificial transposons into plasmid targets in vitro: A useful tool for DNA mapping, sequencing and genetic analysis. Nucleic Acids Res. 1994;22:3765–3772. doi: 10.1093/nar/22.18.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine SE, Chissoe SL, Eby Y, Wilson RK, Boeke JD. A Transposon-based strategy for sequencing repetitive DNA in eukaryotic genomes. Genome Res. 1997;7:551–563. doi: 10.1101/gr.7.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger DJ, Boeke JD. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: Cell-free Ty1 transposition. Cell. 1988;54:955–966. doi: 10.1016/0092-8674(88)90110-9. [DOI] [PubMed] [Google Scholar]
- ————— A specific terminal structure is required for Ty1 transposition. Genes & Dev. 1990;4:324–330. doi: 10.1101/gad.4.3.324. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Fleischmann RD. Strategies for whole microbial genome sequencing and analysis. Electrophoresis. 1997;18:1207–1216. doi: 10.1002/elps.1150180803. [DOI] [PubMed] [Google Scholar]
- Holmes DS, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hunkapiller T, Kaiser RJ, Koop BF, Hood L. Large-scale and automated DNA sequence determination. Science. 1991;254:59–67. doi: 10.1126/science.1925562. [DOI] [PubMed] [Google Scholar]
- Ji H, Moore DP, Blomberg MA, Braiterman LT, Voytas DF, Natsoulis G, Boeke JD. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- Kahmann R, Kamp D. Nucleotide sequences of the attachment sites of bacteriophage Mu DNA. Nature. 1979;280:247–250. doi: 10.1038/280247a0. [DOI] [PubMed] [Google Scholar]
- Kasai H, Isono S, Kitakawa M, Mineno J, Akiyama H, Kurnit DM, Berg DE, Isono K. Efficient large-scale sequencing of the Escherichia coli genome: Implementation of a transposon- and PCR-based strategy for the analysis of ordered lambda phage clones. Nucleic Acids Res. 1992;20:6509–6515. doi: 10.1093/nar/20.24.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Big time for small genomes. Genome Res. 1997;7:418–421. doi: 10.1101/gr.7.5.418. [DOI] [PubMed] [Google Scholar]
- Lavoie BD, Chaconas G. Transposition of phage Mu. Curr Top Microbiol Immunol. 1996;204:83–99. doi: 10.1007/978-3-642-79795-8_4. [DOI] [PubMed] [Google Scholar]
- Lavoie BD, Chan BS, Allison RG, Chaconas G. Structural aspects of a higher order nucleoprotein complex: Induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. EMBO J. 1991;10:3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Mizuuchi K. Transpositional recombination: Mechanistic insights from studies of Mu and other elements. Annu Rev Biochem. 1992a;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- ————— Polynucleotidyl transfer reactions in transpositional DNA recombination. J Biol Chem. 1992b;267:21273–21276. [PubMed] [Google Scholar]
- Mizuuchi M, Mizuuchi K. Efficient Mu transposition requires interaction of transposase with a DNA sequence at the Mu operator: Implications for regulation. Cell. 1989;58:399–408. doi: 10.1016/0092-8674(89)90854-4. [DOI] [PubMed] [Google Scholar]
- ————— Target site selection in transposition of phage Mu. Cold Spring Harbor Symp Quant Biol. 1993;58:515–523. doi: 10.1101/sqb.1993.058.01.058. [DOI] [PubMed] [Google Scholar]
- Payne JA, Stevenson TJ, Donaldson LF. Molecular characterization of a putative K-Cl cotransporter in rat brain. A neuronal-specific isoform. J Biol Chem. 1996;271:16245–16252. doi: 10.1074/jbc.271.27.16245. [DOI] [PubMed] [Google Scholar]
- Phadnis SH, Huang HV, Berg DE. Tn5supF, a 264-base-pair transposon derived from Tn5 for insertion mutagenesis and sequencing DNAs cloned in phage λ. Proc Natl Acad Sci. 1989;86:5908–5912. doi: 10.1073/pnas.86.15.5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A laboratory manual. 2nd edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Savilahti H, Mizuuchi K. Mu transpositional recombination: Donor DNA cleavage and strand transfer in trans by the Mu transposase. Cell. 1996;85:271–280. doi: 10.1016/s0092-8674(00)81103-4. [DOI] [PubMed] [Google Scholar]
- Savilahti H, Rice PA, Mizuuchi K. The phage Mu transpososome core: DNA requirements for assembly and function. EMBO J. 1995;14:4893–4903. doi: 10.1002/j.1460-2075.1995.tb00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathmann M, Hamilton BA, Mayeda CA, Simon MI, Meyerowitz EM, Palazzolo MJ. Transposon-facilitated DNA sequencing. Proc Natl Acad Sci. 1991;88:1247–1250. doi: 10.1073/pnas.88.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette MG, Buch SJ, Chaconas G. Transpososomes: Stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell. 1987;49:253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]