Abstract
Phospholamban (PLN) is a type II membrane protein that inhibits the sarcoplasmic reticulum Ca2+-ATPase (SERCA), thereby regulating calcium homeostasis in cardiac muscle. In membranes, PLN forms pentamers that have been proposed to function either as a storage for active monomers or as ion channels. Here, we report the T-state structure of pentameric PLN solved by a hybrid solution and solid-state NMR method. In lipid bilayers, PLN adopts a pinwheel topology with a narrow hydrophobic pore, which excludes ion transport. In the T state, the cytoplasmic amphipathic helices (domains Ia) are absorbed into the lipid bilayer with the transmembrane domains arranged in a left-handed coiled-coil configuration, crossing the bilayer with a tilt angle of approximately 11° with respect to the membrane normal. The tilt angle difference between the monomer and pentamer is approximately 13°, showing that intramembrane helix–helix association forces dominate over the hydrophobic mismatch, driving the overall topology of the transmembrane assembly. Our data reveal that both topology and function of PLN are shaped by the interactions with lipids, which fine-tune the regulation of SERCA.
Keywords: hybrid NMR method, PISEMA, calcium regulation, oligomeric protein, dipolar assisted rotational resonance recoupling
The membrane protein complex formed by Ca2+-ATPase (SERCA) and phospholamban (PLN) regulates Ca2+ concentration within the sarcoplasmic reticulum (SR), thereby controlling muscle excitation–contraction coupling (1, 2). PLN is a 52-residue transmembrane (TM) protein highly conserved across mammals (2). Its helix–loop–helix secondary structure is further subdivided into four dynamic domains: domain Ia (1–16), loop (17–22), domain Ib (23–30), and domain II (31–52) (3, 4). The hydrophobic TM domain II is the most conserved and responsible for SERCA inhibition, whereas the cytoplasmic domain harbors two phosphorylation sites that reverse PLN inhibitory function (2). PLN has a direct role in the pathophysiology of the heart muscle, with three lethal mutations linked to dilated cardiomyopathy in humans (R9C-PLN, R14del, and L39-truncated-PLN) (5). In both synthetic and cell membranes, PLN forms pentamers that dissociate into monomers upon interacting with SERCA (1, 6). Although the stoichiometry of the SERCA/PLN complex has been assessed (1, 6), both the role and the structure of the PLN pentamer remain a matter of active debate. Because PLN expression in both atria and ventricles is higher than SERCA, it is likely that oligomerization may participate in SERCA regulation (7). Insights into PLN organization in the membrane have come from biochemical and biophysical data (2, 6, 8). Initial electrophysiological measurements indicated that PLN formed Ca2+ channels (9). However, more recent electrochemical studies concluded that PLN does not conduct Cl- or Ca2+ ions (10).
Divergent structural models for the PLN pentamer have been proposed in the literature (8). Although very similar in the secondary structure content, these models differ in the membrane topology. The two most recent models are a solution NMR structure of PLN in detergent micelles (dodecylphosphocholine, DPC) with an unusual bellflower topology (11), and a low-resolution pinwheel model derived from fluorescence anisotropy data (12). The bellflower model, with its funnel-shaped TM pore, would support an active role of the pentamer in transporting Ca2+ or Cl- ions, contributing to the ion homeostasis in the SR. In contrast, the absence of a pore in the pinwheel topology would support the role of the pentamer assembly as a reservoir for active monomers and would constitute a supplementary regulatory mechanism for SERCA via monomer/pentamer equilibrium (13).
In this article, we present the high-resolution structure and topology of the PLN pentamer in its T state (14) as determined by a hybrid solution and solid-state NMR method (15, 16). In its T state, PLN adopts a pinwheel topology with the cytoplasmic helices making extensive contacts with the lipid bilayer surface. The TM helices traverse the membrane at an angle of approximately 11° with respect to the bilayer normal and have a narrow hydrophobic pore (average radius approximately 2 Å), which tapers to approximately 1 Å at the juxtamembrane region (residues 23–30).
Results
Structure and Topology of PLN in Detergent Micelles and Lipid Bilayers.
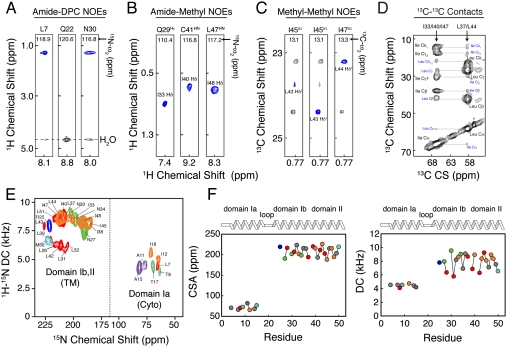
The overall strategy for the hybrid NMR method consists of combining secondary structure information from solution NMR with orientational restraints obtained from solid–state NMR in lipid membranes (15, 16). Here, we selected micelle and lipid conditions where PLN predominantly adopts a T-state conformation as probed by EPR and NMR (17–19). For solution NMR studies, we refolded bacterially expressed PLN (rabbit primary sequence) into DPC micelles. In DPC, PLN forms stable pentamers that have a melting temperature of approximately 70 °C (20), giving a homogenous and well-dispersed 2D [1H, 15N]-transverse relaxation optimized spectroscopy (21) heteronuclear single quantum coherence (HSQC) spectrum (Fig. S1A). To assign the PLN backbone and side-chain resonances, we utilized standard triple-resonance experiments (22, 23). The chemical shift assignments are in complete agreement with those reported for the human PLN pentamer (11), with the exception of Asn-27, which is Lys-27 in the rabbit sequence. Spatial correlations and other long-range distance restraints between nuclear spins were obtained from the quantification of a series of 3D [1H, 1H, 15N]-NOESY-HSQC experiments. We detected a total of 1945 intraprotomer NOEs (389 NOEs per protomer) (Fig. S1B). The chemical shift indices and the NOE patterns confirmed the existence of the helix–loop–helix structural motifs (Fig. S1 B and C) also present in monomeric PLN (24). Although micelle studies give useful insights into the topology of PLN, they may not reproduce the architecture of this protein in cell membranes (25). A more rigorous approach is to supplement micelle studies with solid-state NMR techniques in lipid bilayers (26–30). Therefore, we carried out solid-state NMR experiments in mechanically aligned lipid bilayers (31, 53, 54). PLN was reconstituted into 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)/1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) lipid bilayers at 4/1 (mol/mol) ratio, which maximizes the activity of SERCA (32) and closely mimics the SR membrane composition (33). To avoid nonspecific PLN aggregation, we used a lipid-to-protein molar ratio of 200/1. Orientational restraints were derived from PISEMA (34) and SAMPI4 (35) experiments, measuring dipolar couplings (DC) and anisotropic chemical shifts (CSA) on a series of selectively 15N labeled samples (see SI Appendix). Fig. 1E shows the overlay of several oriented solid-state NMR spectra located in two distinct regions: (i) resonances clustered at 200 ppm that correspond to the TM domains and (ii) resonances at 65–100 ppm, assigned to the cytoplasmic domain Ia. We measured a total of 29 DC and 31 CSA values (Table S2) and converted them into restraints for the hybrid structure calculations, where both solution and solid-state NMR restraints are implemented into the target energy function (15, 16).
Fig. 1.
Multidimensional solution and solid-state NMR spectra defining the structural topology of PLN in DPC micelles, planar lipid bilayers, and lipid vesicles. (A) Two-dimensional planes from a 3D [1H, 1H,15N]-NOESY-HSQC spectrum (300-ms mixing time) showing NOEs between amide protons of PLN and methyl and methylene groups of DPC. Note the peaks at approximately 4.5 ppm are exchange cross-peaks between the protein amide resonances and the water signal. (B) Two-dimensional planes from 3D [1H, 1H,15N]-NOESY-HSQC (400-ms mixing time) on a mixed PLN sample with 1/1 of 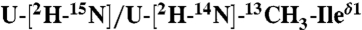 PLN. (C) Two-dimensional planes from 3D [1H, 13C, 13C]-HSQC-NOESY-HSQC experiment performed on a sample containing 1/1 of
PLN. (C) Two-dimensional planes from 3D [1H, 13C, 13C]-HSQC-NOESY-HSQC experiment performed on a sample containing 1/1 of 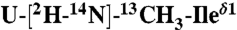 and
and 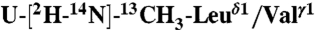 PLN. (D) Two-dimensional-DARR experiment (200-ms mixing time) on a 1/1 sample of U-13C-Leu/U-13C-Ile PLN. Intraresidue and interprotomer cross-peaks are labeled in black and blue, respectively. (E) Overlay of 2D [1H, 15N]-PISEMA spectra of selectively labeled PLN in oriented DOPC/DOPE (4/1) lipid bilayers. (F) Plot of the CSA (Left) and DC (Right) values versus residue.
PLN. (D) Two-dimensional-DARR experiment (200-ms mixing time) on a 1/1 sample of U-13C-Leu/U-13C-Ile PLN. Intraresidue and interprotomer cross-peaks are labeled in black and blue, respectively. (E) Overlay of 2D [1H, 15N]-PISEMA spectra of selectively labeled PLN in oriented DOPC/DOPE (4/1) lipid bilayers. (F) Plot of the CSA (Left) and DC (Right) values versus residue.
Interprotomer Distance Restraints.
Interprotomer distance restraints were derived from NOESY experiments using an asymmetric isotopic labeling scheme that allowed for the unambiguous assignment of interprotomer methyl–methyl NOEs, defining the quaternary structure (24). We prepared two different mixed pentamer samples. The first was a 1/1 molar mixture of 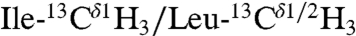 ,
, 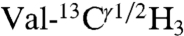 in a U-[2H,12C] background, where interprotomer NOEs were measured between methyl protons (24). The second sample consisted of a 1/1 mixture of
in a U-[2H,12C] background, where interprotomer NOEs were measured between methyl protons (24). The second sample consisted of a 1/1 mixture of 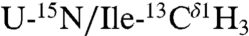 in a U-[2H,12C] background. This sample gave quaternary restraints between Ile methyl protons and backbone amide protons. We used a total of 75 interprotomer NOEs (15 NOEs per protomer) in the structure calculation (Fig. 1 and Table S3). Although several other NOEs are present in the spectral region corresponding to the Leu residues in the Ile/Leu zipper (e.g., Leu37, Leu44, and Leu51), these peaks are highly overlapped and we did not use them in the calculations. To validate the interprotomer distance restraints obtained in micelles, we carried out dipolar assisted rotational resonance recoupling (DARR) magic-angle-spinning (MAS) experiments in 4/1 DOPC/DOPE lipid bilayers (see SI Methods). These experiments (Fig. 1B and Fig. S5) utilized a Leu/Ile asymmetric labeling method and confirmed the dipolar connectivities measured by NOESY experiments (Fig. S5C). The DARR cross-peaks are also in good agreement with the Ile/Leu zipper present in the pentamer. To define the quaternary arrangement in the juxtamembrane domain Ib, we carried out two sets of calculations with and without the following two restraints: (i) an MAS Rotational Echo Double Resonance (REDOR) contact between the nitrogen side chain of Gln-29 of PLN found by Smith and co-workers (36) and (ii) an interprotomer NOE between Ile-33Hδ1 and Gln-29Hε we detected in micelles (Fig. 1B). The two structural ensembles calculated without the REDOR and NOE restraints are almost identical with an rmsd between the TM domains < 0.8 Å (Fig. S7), demonstrating that these contacts were not crucial to define the overall fold of the pentamer. In addition, the agreement between the distances obtained by solution and solid-state NMR is further evidence that the T-state structures in micelles and bilayers are very similar and supports using the hybrid restraints for structure calculations. Finally, an EPR-derived restraint between spin labels at Lys-3 in each monomer (31) was used in order to better define the arrangement of the cytoplasmic helices.
in a U-[2H,12C] background. This sample gave quaternary restraints between Ile methyl protons and backbone amide protons. We used a total of 75 interprotomer NOEs (15 NOEs per protomer) in the structure calculation (Fig. 1 and Table S3). Although several other NOEs are present in the spectral region corresponding to the Leu residues in the Ile/Leu zipper (e.g., Leu37, Leu44, and Leu51), these peaks are highly overlapped and we did not use them in the calculations. To validate the interprotomer distance restraints obtained in micelles, we carried out dipolar assisted rotational resonance recoupling (DARR) magic-angle-spinning (MAS) experiments in 4/1 DOPC/DOPE lipid bilayers (see SI Methods). These experiments (Fig. 1B and Fig. S5) utilized a Leu/Ile asymmetric labeling method and confirmed the dipolar connectivities measured by NOESY experiments (Fig. S5C). The DARR cross-peaks are also in good agreement with the Ile/Leu zipper present in the pentamer. To define the quaternary arrangement in the juxtamembrane domain Ib, we carried out two sets of calculations with and without the following two restraints: (i) an MAS Rotational Echo Double Resonance (REDOR) contact between the nitrogen side chain of Gln-29 of PLN found by Smith and co-workers (36) and (ii) an interprotomer NOE between Ile-33Hδ1 and Gln-29Hε we detected in micelles (Fig. 1B). The two structural ensembles calculated without the REDOR and NOE restraints are almost identical with an rmsd between the TM domains < 0.8 Å (Fig. S7), demonstrating that these contacts were not crucial to define the overall fold of the pentamer. In addition, the agreement between the distances obtained by solution and solid-state NMR is further evidence that the T-state structures in micelles and bilayers are very similar and supports using the hybrid restraints for structure calculations. Finally, an EPR-derived restraint between spin labels at Lys-3 in each monomer (31) was used in order to better define the arrangement of the cytoplasmic helices.
Positioning of PLN in Micelle and Lipid Bilayers.
To position the two helical segments (1–16 and 23–52), we used planar restraints (see SI Methods) derived by measuring NOEs between the amide backbone of PLN and the detergent hydrocarbon tails in a 3D [1H, 1H, 15N]-NOESY spectrum (Fig. 1A and Fig. S4). Because PLN was fully deuterated (except the back-exchanged amide protons), the presence of cross-peaks at 1–2 ppm in the 1H dimension indicated a close proximity between the amide hydrogen and the methylene and methyl protons of the DPC micelle. These contacts between PLN and the micelle were observed throughout the pentamer with the exception of residues 21 to 23 within the dynamic and solvent-exposed loop (Fig. 1A and Fig. S4). Residues in the cytoplasmic domains Ia and Ib displayed NOE contacts with the micelle hydrocarbon tails, indicating a strong association with the DPC micelle. These results confirmed unambiguously that the amphipathic domain Ia is absorbed on the micelle surface, with the most polar residues facing the bulk water and the hydrophobic residues pointing toward the inner core of the micelle, mirroring previous paramagnetic mapping studies (31).
Hybrid PLN Structural Ensemble.
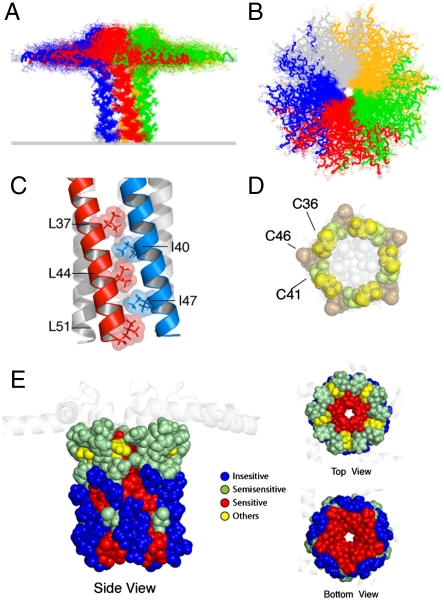
The calculation protocol is illustrated in Fig. S2. Symmetry distance restraints (37) were used to maintain the pentamer geometry during the simulated annealing calculations. The relative position of the two domains (depth of insertion) was constrained with planar restraints derived from protein/micelle NOEs (38) (see SI Methods). Fig. 2 A and B displays the final conformational ensemble of PLN (Protein Data Bank ID code 2KYV), with a well-defined pinwheel topology. The low energy conformers show the convergence of the TM domains with an rmsd of approximately 0.5 Å (Table S1). We assessed the quality of our structural ensemble with the MolProbity software (39). Our 2KYV ensemble obtained a total score of 1.8 (94th percentile), showing an excellent van der Waals packing and local covalent geometry (Table S1). The PLN protomers are held together by a tight Leu/Ile zipper, which is the driving force for PLN oligomerization that confers high thermostability (2). The interlocking of alternating Leu and Ile residues gives rise to a left-handed coiled-coil assembly (Fig. 2C) (8). The interior of the TM domain is lined with residues that are sensitive to mutations (Fig. 2E). Single mutations of these residues lead to PLN deoligomerization and enhanced inhibitory function (Fig. 2E) (1). Other crucial residues for PLN oligomerization are the three cysteines (Fig. 2D and Fig. S3), which participate in quaternary contacts (8, 40). Indeed, we detected long-range interprotomer NOEs between the amide hydrogen of Cys-41 and the methyl protons of Ile-40 (Fig. 1). We also observed short-range intraprotomer NOEs between the sulfhydryl hydrogen of Cys-36 and the amide hydrogens of Phe-35 and Leu-37, as well as the sulfhydryl hydrogen of Cys-41 and the amide hydrogen of Ile-40 (Fig. S3B). These experimental observables confirmed that Cys-36 and Cys-41 are positioned at the interface between the protomers, which is consistent with their chemical reactivity (40). The intraprotomer NOEs resulting in hydrogen bonding distances between the Cys backbone amides and the (i - 4) carbonyl oxygens (Fig. S3B) are in agreement with mutagenesis experiments and computational modeling (8, 40). Overall, the hybrid ensemble agrees with a wealth of structural data from different techniques and various membrane mimicking systems. The only inconsistency is relative to the dynamic loop region (Table S4).
Fig. 2.
Pinwheel architecture of pentameric PLN in lipid bilayers. (A and B) PLN hybrid conformational ensemble. Overlay of the 20 lowest energy structures. The conformers were aligned using heavy atoms from residues 24 to 48. See Table S1 for structural statistics. (C) Domain II Leu/Ile zipper motif. Residues are shown using space filling model with atomic radii. (D) Spatial arrangement of the three cysteines in the PLN TM domain. The hydrophobic residues lining the inner pore are shown in light gray. Cys-41 and Cys-36 are at the interface between protomers. (E) Mapping of the mutagenesis studies on the PLN pentamer. Residues sensitive to mutation and oligomerization (1) are indicated in red. (Left) Side view of the PLN pentamer with residues from 24 to 52 shown in space filling model.
Comparison Between Structural Models.
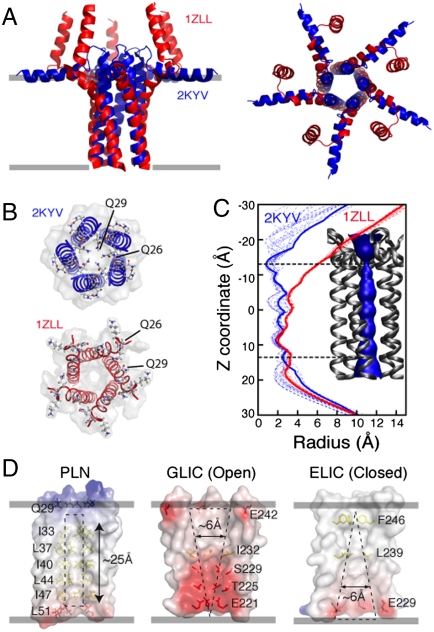
The hybrid PLN ensemble differs significantly from the model proposed by Chou and Oxenoid (11). In the bellflower model, the five domain-Ia helices are oriented almost perpendicularly to the bilayer normal, which probably resulted from the selection of one of the ghost orientations derived from the analytical solutions of the orientationally dependent residual dipolar couplings (RDCs) (41). In the hybrid ensemble, domains Ia are embedded within the top leaflet of the lipid bilayer (Figs. 3A and 4C), with the juxtamembrane domains Ib more tightly packed, resulting in straight α-helices crossing the membrane at an angle of approximately 11°. Conversely, the bellflower model (Molprobity score 3.5, 9th percentile) presents a peculiar bending in the TM helices, diverging from one another at the juxtamembrane region (Fig. 3B). The distorted α-helical domain Ib obtained in the bellflower arrangement contrasts with the hydrogen bonds found in this region (36) and is probably due to dynamics (42) in this domain that averages the RDC values (41). Also, the torsion angle predictions from TALOS (43) agree remarkably well with our conformational ensemble, whereas they deviate substantially in the bellflower ensemble (Fig. S6). The most striking difference for the two models can be appreciated in the back-calculation of the PISEMA spectra (Fig. S8). Although the structural ensemble for the bellflower model differs considerably from the spectra determined in lipid membranes, the pinwheel structure agrees well with the experimental data. The CSA values and the scaling of the DC (Fig. 1 E and F) in domain Ia are strong evidence that this domain has a perpendicular orientation with respect to the bilayer normal.
Fig. 3.
Comparison between 2KYV and 1ZLL structural models. (A) Overlay of the TM regions of the 1ZLL (red) and 2KYV (blue) models. Protein backbones were aligned using heavy atoms for residues 24 to 48. (B) Top view of the 1ZLL and 2KYV models. (C) Width of the central pore calculated with the program HOLE2 (68) for the 2KYV (blue dashed traces) and 1ZLL (red dashed traces) ensembles. Thick solid lines refer to the structures shown in A. (Inset) Surface rendering of the pore for the 2KYV model. (D) Electrostatic surface potentials of two pentameric ligand-gated ion channels (ELIC and GLIC) (44, 45) and PLN (2KYV).
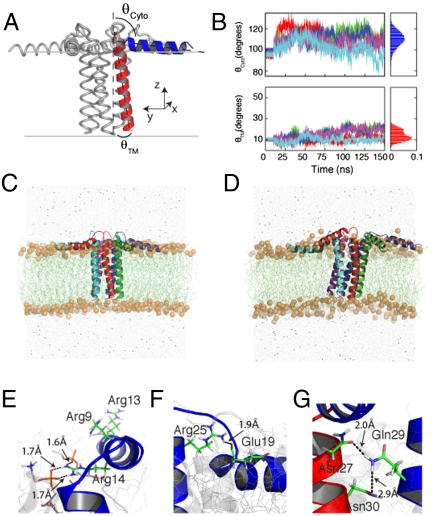
Fig. 4.
MD simulations of PLN in explicit DOPC lipids. (A) Average structure of PLN from the hybrid NMR conformational ensemble with the TM domain (red) tilted by 11°. (B) Time course of the tilt angles of both the TM and cytoplasmic helices of PLN pentamer. (C and D) Snapshots of PLN from the MD trajectories taken at 10 and 150 ns. (E) Orientation of the three Arg residues in domain Ia. The guanidinium group of Arg-14 forms transient hydrogen bonds with several DOPC lipid molecules. (F) Arg-25 and Glu-19 salt bridge persistent throughout the MD trajectory. (G) Interprotomer contacts between the side chain of Gln-29, Asn-27, and Asn-30.
Our hybrid structural ensemble has an average width of the pentameric pore that is too narrow to fit a fully hydrated ion (approximately 3 Å, Fig. 3C). Unlike other pentameric ligand-gated ion channels, whose inner pores are interspersed with hydrophilic residues (44, 45), the PLN inner pore, which encompasses domain II (approximately 20 Å), is coated with hydrophobic residues, making it energetically improbable for a hydrated ion to diffuse through it (Fig. 3D). Theoretical calculations support our findings, showing that the free energy for crossing the PLN pore for chloride ions is approximately 20 kcal/mol, whereas for calcium ions it is more than 40 kcal/mol (10). In addition, the three cysteine residues of the TM α-helices (Fig. 2C) do not face the interior of the pore (8), precluding their participation in the ion transport.
Pentamer to Monomer Topological Changes.
Overall, the structure of each protomer within the pentameric assembly of PLN is similar to that of the monomeric PLN (AFA-PLN, 2KB7) (16). However, the two forms differ noticeably in the membrane topology (Fig. 4A). In monomeric PLN, the tilt angle is approximately 24°, whereas in the pentamer it is approximately 11°. This difference is probably due to the driving forces for folding and stability of the two species. For the monomer, the driving force is the hydrophobic mismatch between protein and bilayer to optimize lipid–protein interactions, causing the TM domain to adopt a more pronounced tilt. In contrast, the orientation of each monomer within the pentamer is driven by interprotomer interactions (Leu/Ile zipper in domain II and hydrogen bonds in the domain Ib), which dominate over the hydrophobic mismatch, giving rise to a smaller tilt angle.
Molecular Dynamic (MD) Simulations of PLN in DOPC Bilayers.
The lowest energy conformer from the final conformational ensemble was embedded in a DOPC lipid bilayer (see SI Appendix for details) and the pentamer dynamics were analyzed for 150 ns. Unlike previous MD simulations that used the bellflower model as starting conformations (46, 47), our data show that the pinwheel topology of PLN does not significantly deviate from the initial configuration (Fig. 4B). During the simulations, the tilt angles of the TM domains fluctuate around 13° in agreement with the solid-state NMR data (Fig. 4B). The cytoplasmic domains, however, undergo larger fluctuations without detaching from the bilayer surface (Fig. 4B), in agreement with the dynamics determined for these domains (48). The simulations reveal several interesting protein/protein and protein/lipid interactions that contribute to the stability of the pinwheel topology. Specifically, the guanidinium group of Arg-14 snorkeling on the surface of the membrane, makes several hydrogen bonds with the oxygens of the lipid’s glycerol moiety (Fig. 4E). These interactions are functionally important because the deletion of this residue generates a loss-of-function species that is unable to properly regulate SERCA (5). Additionally, the side chain of Arg-25 in domain Ib forms a salt bridge with the side chain of Glu-19, which is persistent throughout most of the simulations (Fig. 4F). Several other interprotomer interactions take place in the juxtamembrane portion of PLN, with Gln-29 forming a hydrogen bond with the backbone carbonyl oxygen of Asn-27 of the neighboring monomer (Fig. 4G). In contrast, MD studies showed that the bellflower topology (11) is highly unstable, with domains Ia (constituting the putative vestibule of the bellflower channel) bending after a few nanoseconds and interacting readily with the lipid bilayers (10, 46, 47). Finally, MD trajectories show that the bellflower pore closes after a few nanoseconds, excluding water and ions and preventing transport across the bilayer (10).
Discussion
The T state of PLN is the most populated in living cells (13). It has been hypothesized that its interactions with lipid membranes are important for both oligomerization (49) and SERCA regulation (50). The T-state structure of PLN reported here reveals that both interprotomer interactions and lipid/protein interactions contribute to the overall folding and stability. The cytoplasmic domains contain well-defined helices, with the hydrophobic faces absorbed onto the membrane surface (Fig. 4C). This topology agrees with rotational dynamics measurements carried out by EPR (51) and solid-state NMR (4). MD calculations show that the pinwheel structure does not deviate significantly from the initial topology, with the domains Ia maintaining a tight association with the lipid bilayer (Fig. 4 C and D). The pinwheel topology dismisses the previous models and agrees with a plethora of structural, biophysical, biochemical, and mutagenesis data (31, 36, 50, 52, 55). The narrow pore within the TM domain is not consistent with the proposed ion-channel function of PLN (9).
What is the function of the pentameric assembly? Are the interactions with lipids significant for PLN biological function? Lipid-mediated clustering and oligomerization are emerging aspects that define the biological function of membrane proteins. These mechanisms are responsible for allosteric signaling, ion-channel formation, and enzyme regulation (56). Here, we propose that the ground state of PLN is in a quiescent T state, with the amphipathic helices absorbed into the membrane. Upon membrane detachment, the cytoplasmic domains unfold (42) into a higher energy R state, potentially acting as a mechanism by which the PLN pentamer dissociates one or more subunits to interact with SERCA (14).
Although in vivo and in vitro data show that promoting PLN deoligomerization increases SERCA inhibition (1, 6, 57), we propose that the PLN pentamer actively participates in the regulatory mechanism, tuning the extent of monomeric species available for SERCA inhibition. The latter is supported by cardiac-specific simultaneous overexpression studies of pentameric and monomeric PLN (58). It was found that the PLN monomer (C41F) is able to reduce Ca2+ affinity of SERCA to the same extent as the PLN pentamer, but it was not able to reduce the rate of myocardial relaxation (58). Moreover, PLN phosphorylation enhances the population of the pentamer (6), whereas an increase in SERCA expression shifts the equilibrium toward the monomeric species (59). Importantly, this equilibrium plays a crucial role in muscle pathophysiology. Our recent studies show that an increased stability of the pentamer via the R9C mutation results in decreased inhibition of SERCA and hampers PLN phosphorylation at Ser-16 by protein kinase A, which may lead to dilated cardiomyopathy (20).
In addition to the pentamer/monomer equilibrium, the conformational interconversion from the more structured T state to a more dynamic, unfolded, and membrane-detached R state (14, 19, 60) is a salient aspect of PLN regulatory function. The T state is the most populated in both micelles and lipid membranes (19, 31). However, this equilibrium can be shifted toward the R state by phosphorylation (19, 61), binding partners (62), lipid composition (50, 63), or protein aggregation. The tendency of monomeric PLN domain Ia to unfold into the R state was also detected by solid-state NMR (64). These authors showed that in 1,2-dimyristoyl- sn-glycero-3-phosphocholine (20∶1 lipid-to-protein ratio), the TM domain of PLN is helical, whereas the domain Ia is mostly unfolded. Remarkably, we found a direct correlation between the extent of R state and SERCA regulatory function (60, 65). These studies indicate a potential avenue to design PLN analogs for gene therapy in heart failure (66).
From a biophysical standpoint, our study reveals a unique aspect of TM oligomerization. The packing of the PLN pentamer prevails over the hydrophobic mismatch of the lipid membrane, causing a change in the topology of the TM domain relative to the monomer. Given the dynamic nature of lipids, changes in TM topology within the greasy bilayer may be energetically favorable over folding and unfolding of secondary structure domains and may represent the way protein–protein interactions take place within lipid membranes.
Materials and Methods
PLN was expressed in Escherichia coli bacteria and purified as previously described (31). For solution NMR experiments, PLN was solubilized in 300 mM deuterated DPC, 120 mM NaCl, 20 mM NaH2PO4 (pH ∼ 6), and 50 mM 2-mercaptoethanol. For solid-state NMR spectroscopy, PLN was reconstituted in 4/1 DOPC/DOPE lipid bilayers and spread onto glass plates as previously described (31) or packed into a 3.2-mm MAS rotor. NMR experiments were performed at 1H frequencies of 600 and 700 MHz using Bruker (DMX) and Varian (VNMRS) spectrometers, respectively, equipped with low electric field flat-coil probes (67) and a 3.2-mm bioMAS probe. Structure calculations were carried out with XPLOR-NIH (v.2.23) software, using the hybrid solution and solid-state NMR method (15, 16). For details on MD simulations, see SI Appendix.
Supplementary Material
Acknowledgments.
The authors thank Dr. David Thomas for many helpful discussions. We also thank P. Gor’kov, W. Brey, R. Fu, and T. Cross at the National High Magnetic Field Laboratory, Tallahassee, FL (DMR-0084173) for solid-state NMR experiments, and the National Magnetic Resonance Facility at Madison (Dr. M. Tonelli) for solution NMR experiments. This work was supported by National Institutes of Health Grants GM64742 and HL80081 (to G.V.) and by the University of Minnesota Supercomputing Institute.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The NMR atomic coordinates, chemical shifts, and restraints have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 2KYV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016535108/-/DCSupplemental.
References
- 1.MacLennan DH, Kranias EG. Phospholamban: A crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. 2003;4:566–577. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 2.Simmerman HK, Jones LR. Phospholamban: Protein structure, mechanism of action, and role in cardiac function. Physiol Rev. 1998;78:921–47. doi: 10.1152/physrev.1998.78.4.921. [DOI] [PubMed] [Google Scholar]
- 3.Zamoon J, Mascioni A, Thomas DD, Veglia G. NMR solution structure and topological orientation of monomeric phospholamban in dodecylphosphocholine micelles. Biophys J. 2003;85:2589–2598. doi: 10.1016/s0006-3495(03)74681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Traaseth NJ, et al. Structural and dynamic basis of phospholamban and sarcolipin inhibition of ca(2+)-ATPase. Biochemistry. 2008;47:3–13. doi: 10.1021/bi701668v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haghighi K, et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc Natl Acad Sci USA. 2006;103:1388–1393. doi: 10.1073/pnas.0510519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornea RL, Jones LR, Autry JM, Thomas DD. Mutation and phosphorylation change the oligomeric structure of phospholamban in lipid bilayers. Biochemistry. 1997;36:2960–2967. doi: 10.1021/bi961955q. [DOI] [PubMed] [Google Scholar]
- 7.Boknik P, et al. Regional expression of phospholamban in the human heart. Cardiovasc Res. 1999;43:67–76. doi: 10.1016/s0008-6363(99)00053-x. [DOI] [PubMed] [Google Scholar]
- 8.Arkin IT, Adams PD, Brunger AT, Smith SO, Engelman DM. Structural perspectives of phospholamban, a helical transmembrane pentamer. Annu Rev Biophys Biomol Struct. 1997;26:157–179. doi: 10.1146/annurev.biophys.26.1.157. [DOI] [PubMed] [Google Scholar]
- 9.Kovacs RJ, Nelson MT, Simmerman HK, Jones LR. Phospholamban forms Ca2+-selective channels in lipid bilayers. J Biol Chem. 1988;263:18364–18368. [PubMed] [Google Scholar]
- 10.Becucci L, et al. On the function of pentameric phospholamban: Ion channel or storage form? Biophys J. 2009;96:L60–L62. doi: 10.1016/j.bpj.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oxenoid K, Chou JJ. The structure of phospholamban pentamer reveals a channel-like architecture in membranes. Proc Natl Acad Sci USA. 2005;102:10870–10875. doi: 10.1073/pnas.0504920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robia SL, Flohr NC, Thomas DD. Phospholamban pentamer quaternary conformation determined by in-gel fluorescence anisotropy. Biochemistry. 2005;44:4302–4311. doi: 10.1021/bi0478446. [DOI] [PubMed] [Google Scholar]
- 13.Robia SL, et al. Forster transfer recovery reveals that phospholamban exchanges slowly from pentamers but rapidly from the SERCA regulatory complex. Circ Res. 2007;101:1123–1129. doi: 10.1161/CIRCRESAHA.107.159947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zamoon J, Nitu F, Karim C, Thomas DD, Veglia G. Mapping the interaction surface of a membrane protein: Unveiling the conformational switch of phospholamban in calcium pump regulation. Proc Natl Acad Sci USA. 2005;102:4747–4752. doi: 10.1073/pnas.0406039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi L, et al. A refinement protocol to determine structure, topology, and depth of insertion of membrane proteins using hybrid solution and solid-state NMR restraints. J Biomol NMR. 2009;44:195–205. doi: 10.1007/s10858-009-9328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Traaseth NJ, et al. Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proc Natl Acad Sci USA. 2009;106:10165–10170. doi: 10.1073/pnas.0904290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Traaseth NJ, Buffy JJ, Zamoon J, Veglia G. Structural dynamics and topology of phospholamban in oriented lipid bilayers using multidimensional solid-state NMR. Biochemistry. 2006;45:13827–13834. doi: 10.1021/bi0607610. [DOI] [PubMed] [Google Scholar]
- 18.Mascioni A, Karim C, Zamoon J, Thomas DD, Veglia G. Solid-state NMR and rigid body molecular dynamics to determine domain orientations of monomeric phospholamban. J Am Chem Soc. 2002;124:9392–9393. doi: 10.1021/ja026507m. [DOI] [PubMed] [Google Scholar]
- 19.Karim CB, Zhang Z, Howard EC, Torgersen KD, Thomas DD. Phosphorylation-dependent conformational switch in spin-labeled phospholamban bound to SERCA. J Mol Biol. 2006;358:1032–1040. doi: 10.1016/j.jmb.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 20.Ha KN, et al. Lethal Arg9Cys phospholamban mutation hinders Ca2+-ATPase regulation and phosphorylation by protein kinase A. Proc Natl Acad Sci USA. 2011;108:2735–2740. doi: 10.1073/pnas.1013987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kay LE, Ikura M, Tschudin R, Bax A. Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. J Magn Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Grzesiek S, Bax A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 24.Traaseth NJ, Verardi R, Veglia G. Asymmetric methyl group labeling as a probe of membrane protein homo-oligomers by NMR spectroscopy. J Am Chem Soc. 2008;130:2400–2401. doi: 10.1021/ja711499r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cross TA, Sharma M, Yi M, Zhou HX. Influence of solubilizing environments on membrane protein structures. Trends Biochem Sci. 2010;36:117–125. doi: 10.1016/j.tibs.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Page RC, Kim S, Cross TA. Transmembrane helix uniformity examined by spectral mapping of torsion angles. Structure. 2008;16:787–797. doi: 10.1016/j.str.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chem Rev. 2004;104:3587–606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong M. Oligomeric structure, dynamics, and orientation of membrane proteins from solid-state NMR. Structure. 2006;14:1731–1740. doi: 10.1016/j.str.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Baldus M. Magnetic resonance in the solid state: Applications to protein folding, amyloid fibrils and membrane proteins. Eur Biophys J. 2007;36(Suppl 1):37–48. doi: 10.1007/s00249-007-0174-y. [DOI] [PubMed] [Google Scholar]
- 30.Abu-Baker S, et al. The structural topology of wild-type phospholamban in oriented lipid bilayers using 15N solid-state NMR spectroscopy. Protein Sci. 2007;16:2345–2349. doi: 10.1110/ps.072977707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Traaseth NJ, et al. Spectroscopic validation of the pentameric structure of phospholamban. Proc Natl Acad Sci USA. 2007;104:14676–14681. doi: 10.1073/pnas.0701016104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cornea RL, Thomas DD. Effects of membrane thickness on the molecular dynamics and enzymatic activity of reconstituted ca-ATPase. Biochemistry. 1994;33:2912–2920. doi: 10.1021/bi00176a022. [DOI] [PubMed] [Google Scholar]
- 33.Bick RJ, Buja LM, Van Winkle WB, Taffet GE. Membrane asymmetry in isolated canine cardiac sarcoplasmic reticulum: Comparison with skeletal muscle sarcoplasmic reticulum. J Membr Biol. 1998;164:169–175. doi: 10.1007/s002329900402. [DOI] [PubMed] [Google Scholar]
- 34.Wu CH, Ramamoorthy A, Opella SJ. High-resolution heteronuclear dipolar solid-state NMR spectroscopy. J Magn Reson. 1994;109:270–272. [Google Scholar]
- 35.Nevzorov AA, Opella SJ. Selective averaging for high-resolution solid-state NMR spectroscopy of aligned samples. J Magn Reson. 2007;185:59–70. doi: 10.1016/j.jmr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Liu W, Fei JZ, Kawakami T, Smith SO. Structural constraints on the transmembrane and juxtamembrane regions of the phospholamban pentamer in membrane bilayers: Gln29 and Leu52. Biochim Biophys Acta. 2007;1768:2971–2978. doi: 10.1016/j.bbamem.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Donoghue SI, Nilges M. Structure Computation and Dynamics in Protein NMR. New York: Kluwer Academic; 1999. pp. 131–158. [Google Scholar]
- 38.Teriete P, Franzin CM, Choi J, Marassi FM. Structure of the na,K-ATPase regulatory protein FXYD1 in micelles. Biochemistry. 2007;46:6774–6783. doi: 10.1021/bi700391b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karim CB, Stamm JD, Karim J, Jones LR, Thomas DD. Cysteine reactivity and oligomeric structure of phospholamban and its mutants. Biochemistry. 1998;37:12074–12081. doi: 10.1021/bi980642n. [DOI] [PubMed] [Google Scholar]
- 41.Shi L, et al. Paramagnetic-based NMR restraints lift residual dipolar coupling degeneracy in multidomain detergent-solubilized membrane proteins. J Am Chem Soc. 2011;133:2232–4221. doi: 10.1021/ja109080t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Traaseth NJ, Veglia G. Probing excited states and activation energy for the integral membrane protein phospholamban by NMR CPMG relaxation dispersion experiments. Biochim Biophys Acta. 2010;1798:77–81. doi: 10.1016/j.bbamem.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 44.Bocquet N, et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 45.Hilf RJ, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 46.Maffeo C, Aksimentiev A. Structure, dynamics, and ion conductance of the phospholamban pentamer. Biophys J. 2009;96:4853–4865. doi: 10.1016/j.bpj.2009.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim T, Lee J, Im W. Molecular dynamics studies on structure and dynamics of phospholamban monomer and pentamer in membranes. Proteins. 2009;76:86–98. doi: 10.1002/prot.22322. [DOI] [PubMed] [Google Scholar]
- 48.Metcalfe EE, Zamoon J, Thomas DD, Veglia G. (1)H/(15)N heteronuclear NMR spectroscopy shows four dynamic domains for phospholamban reconstituted in dodecylphosphocholine micelles. Biophys J. 2004;87:1205–1214. doi: 10.1529/biophysj.103.038844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang XM, Kimura Y, Inui M. Effects of phospholipids on the oligomeric state of phospholamban of the cardiac sarcoplasmic reticulum. Circ J. 2005;69:1116–1123. doi: 10.1253/circj.69.1116. [DOI] [PubMed] [Google Scholar]
- 50.Hughes E, Clayton JC, Middleton DA. Cytoplasmic residues of phospholamban interact with membrane surfaces in the presence of SERCA: A new role for phospholipids in the regulation of cardiac calcium cycling? Biochim Biophys Acta. 2009;1788:559–566. doi: 10.1016/j.bbamem.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 51.Nesmelov YE, Karim CB, Song L, Fajer PG, Thomas DD. Rotational dynamics of phospholamban determined by multifrequency electron paramagnetic resonance. Biophys J. 2007;93:2805–2812. doi: 10.1529/biophysj.107.108910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chu S, Abu-Baker S, Lu J, Lorigan GA. (15)N solid-state NMR spectroscopic studies on phospholamban at its phosphorylated form at ser-16 in aligned phospholipid bilayers. Biochim Biophys Acta. 2010;1798:312–317. doi: 10.1016/j.bbamem.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma M, et al. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marassi FM, Opella SJ. Simultaneous assignment and structure determination of a membrane protein from NMR orientational restraints. Protein Sci. 2003;12:403–411. doi: 10.1110/ps.0211503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slovic AM, Lear JD, DeGrado WF. De novo design of a pentameric coiled-coil: Decoding the motif for tetramer versus pentamer formation in water-soluble phospholamban. J Pept Res. 2005;65:312–321. doi: 10.1111/j.1399-3011.2005.00244.x. [DOI] [PubMed] [Google Scholar]
- 56.Bray D, Duke T. Conformational spread: The propagation of allosteric states in large multiprotein complexes. Annu Rev Biophys Biomol Struct. 2004;33:53–73. doi: 10.1146/annurev.biophys.33.110502.132703. [DOI] [PubMed] [Google Scholar]
- 57.Zhai J, et al. Cardiac-specific overexpression of a superinhibitory pentameric phospholamban mutant enhances inhibition of cardiac function in vivo. J Biol Chem. 2000;275:10538–10544. doi: 10.1074/jbc.275.14.10538. [DOI] [PubMed] [Google Scholar]
- 58.Chu G, et al. Pentameric assembly of phospholamban facilitates inhibition of cardiac function in vivo. J Biol Chem. 1998;273:33674–33680. doi: 10.1074/jbc.273.50.33674. [DOI] [PubMed] [Google Scholar]
- 59.Reddy LG, Jones LR, Thomas DD. Depolymerization of phospholamban in the presence of calcium pump: A fluorescence energy transfer study. Biochemistry. 1999;38:3954–3962. doi: 10.1021/bi981795d. [DOI] [PubMed] [Google Scholar]
- 60.Ha KN, et al. Controlling the inhibition of the sarcoplasmic Ca2+-ATPase by tuning phospholamban structural dynamics. J Biol Chem. 2007;282:37205–37214. doi: 10.1074/jbc.M704056200. [DOI] [PubMed] [Google Scholar]
- 61.Traaseth NJ, Thomas DD, Veglia G. Effects of Ser16 phosphorylation on the allosteric transitions of phospholamban/Ca(2+)-ATPase complex. J Mol Biol. 2006;358:1041–1050. doi: 10.1016/j.jmb.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 62.Masterson LR, et al. Dynamics connect substrate recognition to catalysis in protein kinase A. Nat Chem Biol. 2010;6:821–828. doi: 10.1038/nchembio.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abu-Baker S, Lorigan GA. Phospholamban and its phosphorylated form interact differently with lipid bilayers: A 31P, 2H, and 13C solid-state NMR spectroscopic study. Biochemistry. 2006;45:13312–13322. doi: 10.1021/bi0614028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andronesi OC, et al. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J Am Chem Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- 65.Gustavsson M, et al. Lipid-mediated folding/unfolding of phospholamban as a regulatory mechanism for the sarcoplasmic reticulum ca(2+)-ATPase. J Mol Biol. 2011;408:755–765. doi: 10.1016/j.jmb.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoshijima M, et al. Chronic suppression of heart-failure progression by a pseudophosphorylated mutant of phospholamban via in vivo cardiac rAAV gene delivery. Nat Med. 2002;8:864–871. doi: 10.1038/nm739. [DOI] [PubMed] [Google Scholar]
- 67.Gor’kov PL, et al. Using low-E resonators to reduce RF heating in biological samples for static solid-state NMR up to 900 MHz. J Magn Reson. 2007;185:77–93. doi: 10.1016/j.jmr.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 68.Smart OS, Goodfellow JM, Wallace BA. The pore dimensions of gramicidin A. Biophys J. 1993;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.