Abstract
The intra- and intermolecular translocation processes whereby the bi-domain transcription factor Oct1 searches for its specific DNA target site have been investigated by residual dipolar coupling (RDC) and paramagnetic relaxation enhancement (PRE) measurements. The RDC data show that the orientation of the POUS and POUHD domains of Oct1 relative to the long axis of the DNA is the same for specific and nonspecific complexes with DNA. In the context of the specific Oct1-DNA complex, sparsely-populated, spectroscopically “invisible” states reveal their footprints on the PRE profiles observed for the specific complex. Analysis of the PRE data indicates that the POUHD domain searches the DNA primarily by rotation-coupled sliding (intramolecular translocation), while the POUS domain functions as an antenna to promote intersegment transfer via intermolecular translocation. The latter involves the formation of a bridged intermediate in which the POUHD domain is located on the first DNA molecule and the POUS domain on the second. The formation of the bridge intermediate promotes the completion of intermolecular translocation of Oct1 via a first order process involving dissociation and association of the POUHD domain from one DNA molecule to another. Thus cross-talk between the POUS and POUHD domains, each fulfilling different and complementary components of the search process ensures efficient sampling of DNA, thereby facilitating the location of specific Oct1 target sites.
Keywords: multidomain protein-DNA recognition, direct transfer, one-dimensional diffusion, nuclear magnetic resonance spectroscopy
The target search process whereby transcription factors locate their specific DNA binding sites within a sea of nonspecific DNA generally proceeds via three mechanisms: jumping, sliding, and intersegment or direct transfer (1–5). Jumping involves dissociation of the protein from the DNA into free solution followed by reassociation at another site on the DNA. In sliding, nonspecific DNA binding is followed by intramolecular translocation via 1D diffusion along the DNA. Finally, intersegment transfer or intermolecular translocation involves direct transfer of the protein from one DNA site to another either on a distant site (> 150 bp away) on the same DNA molecule or on another DNA molecule via a bridged complex without dissociation into free solution. We have previously made use of NMR paramagnetic relaxation enhancement (PRE) and z-exchange spectroscopy to directly demonstrate the occurrence of both sliding and intersegment transfer in a single domain DNA binding protein, namely the homeodomain transcription factor HoxD9 (6, 7). Both processes manifest themselves as sparsely-populated (< 1%), spectroscopically invisible states whose footprints are observed on the PRE profiles of the predominant (> 99%) species, namely the specific complex (6). Further, residual dipolar coupling (RDC) data on nonspecific complexes demonstrated that the orientation of HoxD9 on the DNA is maintained during the sliding process (8); that is HoxD9 tracks the grooves on the DNA in what can be termed rotation-coupled sliding (9). Many transcription factors, however, comprise two or more DNA binding domains connected by linkers of variable length, and the question arises as to the nature of the interplay between the DNA binding domains and intra- and intermolecular translocation processes.
A classic example of a bi-domain DNA binding protein is the transcription factor Oct1 (10) that plays a critical role in neural development, cell growth, and differentiation (11, 12). Oct1 comprises two helix-turn-helix DNA binding domains, namely the specific domain POUS and the homeodomain POUHD connected by a flexible (∼23 residue) linker (13, 14). Recent experiments employing z-exchange NMR spectroscopy have shown that, in addition to jumping, the two domains undergo direct transfer between specific sites located on different DNA molecules independently of one another, with POUS undergoing exchange between specific sites a factor of about 1.5 times faster than POUHD (15). Here we make use of PRE and RDC measurements to characterize the mechanism of Oct1 translocation on DNA. We show that translocation of Oct1 on the DNA predominantly involves sliding of POUHD and intersegment transfer of POUS. The data directly demonstrate the existence of transient, sparsely-populated bridging complexes involving two molecules of DNA, and suggest that POUS (whose affinity for DNA is less than that of POUHD) serves as an antenna to capture the second molecule of DNA followed by intersegment transfer of POUHD from the first to the second DNA molecule.
Results and Discussion
Interaction Between Oct1 and 24-bp DNA Duplexes.
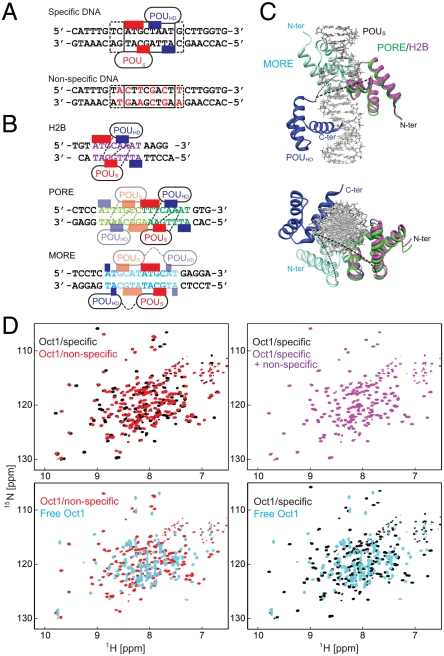
The interaction between Oct1 and 24-bp DNA duplexes with and without the specific binding site were studied by fluorescence anisotropy and heteronuclear NMR spectroscopy. The sequences of the two DNA duplexes are shown in Fig. 1A. The sequence of the specific site (boxed in Fig. 1A, top) is that from the Hoxb1 regulatory element (11, 16). The nonspecific DNA duplex was derived from the specific DNA duplex by introducing six mutations in the specific binding site (displayed in red in Fig. 1A, bottom). The KD for the binding of Oct1 to the 24-bp specific DNA duplex is 16 nM at a salt concentration of 150 mM NaCl (15). The six DNA mutations reduce the affinity by two orders of magnitude, and the KD for the nonspecific 24-bp DNA duplex determined by fluorescence anisotropy is 1.8 ± 0.5 μM at 150 mM NaCl (Fig. S1). The  chemical shifts of the specific and nonspecific complexes are distinct from one another (Fig. 1D, top left) and from the spectrum of free Oct1 (Fig. 1D, bottom). (Assignments of the 1H-15N cross-peaks for the nonspecific complex are shown in Fig. S2A). The largest
chemical shifts of the specific and nonspecific complexes are distinct from one another (Fig. 1D, top left) and from the spectrum of free Oct1 (Fig. 1D, bottom). (Assignments of the 1H-15N cross-peaks for the nonspecific complex are shown in Fig. S2A). The largest 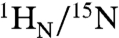 chemical shift differences between specific and nonspecific complexes at low salt involve regions in close contact with the DNA (14, 16), in particular helices 2–4 of the POUS domain, the linker connecting POUS and POUHD, and helices 1′ and 3′ of the POUHD domain (Fig. S3), and reflect the different base sequences in contact with Oct1 (i.e., a specific set of base pairs for the specific complex vs. multiple binding sites for the nonspecific DNA). When both specific and nonspecific duplexes are present at a 1.5-fold excess over Oct1, the spectrum is identical to that of the specific complex (Fig. 1D, top right), indicating, as expected, that Oct1 is located predominantly (> 99%) on the specific DNA. On the chemical shift time scale, exchange between free and bound Oct1 is in the slow exchange regime for the specific complex and in the fast exchange regime for the nonspecific complex.
chemical shift differences between specific and nonspecific complexes at low salt involve regions in close contact with the DNA (14, 16), in particular helices 2–4 of the POUS domain, the linker connecting POUS and POUHD, and helices 1′ and 3′ of the POUHD domain (Fig. S3), and reflect the different base sequences in contact with Oct1 (i.e., a specific set of base pairs for the specific complex vs. multiple binding sites for the nonspecific DNA). When both specific and nonspecific duplexes are present at a 1.5-fold excess over Oct1, the spectrum is identical to that of the specific complex (Fig. 1D, top right), indicating, as expected, that Oct1 is located predominantly (> 99%) on the specific DNA. On the chemical shift time scale, exchange between free and bound Oct1 is in the slow exchange regime for the specific complex and in the fast exchange regime for the nonspecific complex.
Fig. 1.
Interaction of Oct1 with 24-bp specific and nonspecific DNA duplexes. (A) Specific (top) and nonspecific (bottom) duplexes used in the current study. The red and blue bars along the specific DNA duplex indicate the bases involved in specific interactions with the POUS and POUHD domains, respectively, of Oct1 within the Hoxb1 promoter (16); the specific interactions are limited to an 8 bp segment of DNA (boxed) but interactions with the phosphate backbone extend a little further (dashed box). The bases marked in red in the nonspecific sequence indicate sites that were mutated relative to the specific duplex. (B) Mode of interaction of Oct1 observed crystallographically in the H2B (1OCT), PORE (1HF0), and MORE (1E3O) complexes (14, 21). The H2B site comprises a single octameric binding site similar to that in the Hoxb1 promoter, whereas PORE and MORE comprise two adjacent Oct1 binding sites in 15 and 12 bp sequences, respectively. (C) The relative orientation of the POUS and POUHD domains on the H2B and PORE sites is the same but different from that on the MORE site. The structures are superimposed on the POUHD domain (blue) with the POUS domain of H2B, PORE, and MORE in purple, green, and cyan, respectively. Note that the orientation of the POUS domain of MORE relative to the long axis of the DNA is approximately the same as that in the H2B/PORE complexes. (D) Comparisons of the 1H-15N TROSY correlation spectra of free and complexed Oct1: top left, specific (black) and nonspecific (red) complexes; top right, Oct1 in the presence of the specific duplex (black) and an equimixture of specific and nonspecific duplexes (purple); bottom left, nonspecific complex vs. free Oct1; bottom right, specific complex vs. free Oct1. The concentrations of protein and DNA are 0.2 and 0.3 mM, respectively.
Structure and Orientation of POUS and POUHD Domains on the Specific and Nonspecific DNA Duplexes.
The structure and orientation of the POUS and POUHD domains of Oct1 bound to the specific and nonspecific DNA duplexes were assessed by means of backbone amide (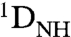 ) RDCs measured in a liquid crystalline medium of phage pf1 [11 mg/mL; (17, 18)]. The RDCs are dependent on the orientation of the N-H bond vectors relative to the alignment tensor and therefore provide an extremely sensitive indicator of structural perturbations (19).
) RDCs measured in a liquid crystalline medium of phage pf1 [11 mg/mL; (17, 18)]. The RDCs are dependent on the orientation of the N-H bond vectors relative to the alignment tensor and therefore provide an extremely sensitive indicator of structural perturbations (19).
Singular value decomposition (SVD) fitting of the RDCs (20) to the coordinates of the individual POUS and POUHD domains of Oct1 (21) yield RDC R-factors (defined in Table 1; ref. 22) of less than 22% for both the specific and nonspecific complexes (Table 1), indicating that the structures of the individual domains in both complexes are unchanged relative to those in the crystal structure. These conclusions are confirmed by the similarity of the backbone ϕ/ψ torsion angles for the two complexes derived from backbone chemical shifts using the program TALOS+ (23) (Fig. S2B).
Table 1.
SVD analysis of backbone 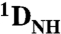 RDCs for the specific and nonspecific Oct1-DNA complexes at 0 and 150 mM NaCl *
RDCs for the specific and nonspecific Oct1-DNA complexes at 0 and 150 mM NaCl *
| 0 mM NaCl / 150 mM NaCl† | |||||||
| Number of RDCs | Euler angles (°)‡ |
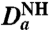 (Hz) (Hz) |
Rhombicity | R-factor (%)§ | |||
| ϕ | θ | ψ | |||||
| Oct1-DNA specific complex | |||||||
| Individual domain SVD fits | |||||||
| POUS | 37/35 | 98/98 | 82/80 | 119/116 | -16.9/-14.6 | 0.29/0.33 | 20.9/21.9 |
| POUHD | 37/35 | 99/96 | 86/80 | 125/121 | -17.3/-14.7 | 0.29/0.34 | 13.0/15.7 |
| Oct1 global SVD fits | |||||||
| H2B | 74/70 | 99/97 | 84/80 | 123/121 | -16.8/-14.7 | 0.30/0.35 | 17.0/18.4 |
| PORE | 74/70 | 99/97 | 87/84 | 121/116 | -17.3/-14.9 | 0.30/0.35 | 16.8/18.7 |
| MORE | 74/70 | 94/94 | 85/84 | 120/107 | -17.9/-15.6 | 0.10/0.18 | 21.9/24.3 |
| Oct1-DNA nonspecific complex | |||||||
| Individual domain SVD fits | |||||||
| POUS | 46/43 | 99/119 | 77/74 | 119/13 | -13.0/-12.1 | 0.30/0.38 | 19.5/19.0 |
| POUHD | 39/30 | 101/90 | 88/79 | 115/101 | -12.4/-12.1 | 0.37/0.21 | 21.8/20.6 |
| Oct1 global SVD fits | |||||||
| H2B | 85/73 | 99/100 | 76/70 | 121/18 | -11.8/-11.9 | 0.37/0.37 | 24.9/33.6 |
| PORE | 85/73 | 99/101 | 81/74 | 120/163 | -12.1/-11.9 | 0.35/0.35 | 22.7/37.8 |
| MORE | 85/73 | 97/106 | 94/100 | 271/222 | -13.5/-11.7 | 0.17/0.15 | 27.5/35.1 |
*The coordinates of the POUS and POUHD domains from the 1.9 Å resolution MORE structure [1E3O; (21)], with protons added in standard stereochemistry and geometry, are best-fitted onto the coordinates of the lower resolution H2B (1OCT; 3 Å resolution) and PORE (1HF0; 2.7 Å resolution) complexes (14, 21), and used for SVD analysis. Using this approach the differences in RDC R-factors and parameters of the alignment tensor reflect only differences in relative orientation of the POUS and POUHD domains. The R-factors for the individual domains using the MORE coordinates fall within the range expected for 1.5–2 Å resolution crystal structures (16, 19). (Note the R-factors for the individual domains using the coordinates of the two lower resolution crystal structures are somewhat higher, reflecting larger errors and uncertainties in the atomic positions of the N-H bond vectors).
†The first number lists the values obtained from the 0 mM NaCl; data, the second number the values obtained for the 150 mM NaCl data.
‡The POUHD domain is positioned in the same molecular frame throughout. In the fits to the individual domains, the Euler angles are calculated with the orientation of POUS relative to POUHD set to that in the H2B/PORE configuration.
§The RDC R-factor is defined as the ratio of the rms difference between observed and calculated RDCs to the expected rms if the N-H bond vectors were oriented completely randomly. The latter is given by 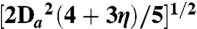 where Da is the magnitude of the alignment tensor and η the rhombicity (22).
where Da is the magnitude of the alignment tensor and η the rhombicity (22).
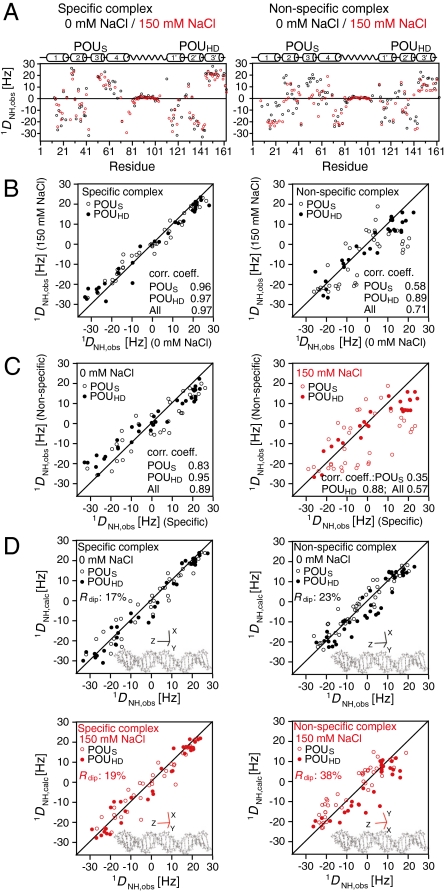
In the case of protein-DNA complexes involving unbent DNA, the alignment tensor in a charged liquid crystalline medium is dominated by the shape and charge distribution of the DNA, and the principal axis of the alignment tensor lies approximately parallel to the long axis of the DNA (8). For the specific Oct1-DNA complex the RDCs at low (0 mM NaCl) and high (150 mM NaCl) salt are highly correlated to one another (correlation coefficient r = 0.97 for the POUS and POUHD domains combined; Fig. 2B, left) indicating that the relative orientation of the POUS and POUHD domains to the long axis of the DNA is unperturbed by salt. For the nonspecific Oct1-DNA complex (Fig. 2B, right), however, the RDCs for the POUHD domain at low and high salt are highly correlated (r = 0.89), while those for the POUS domain are poorly correlated (r = 0.58). Further, while the RDCs for the specific and nonspecific Oct1-DNA complexes are highly correlated (r = 0.95 for POUHD and 0.83 for POUS) at low salt (Fig. 2C, left), only the RDCs for the POUHD domain are well correlated at high salt (r = 0.88 for POUHD vs. 0.35 for POUS; Fig. 2C, right). Thus, at low salt, the orientation of the POUS and POUHD domains relative to the long axis of the DNA is very similar in the specific and nonspecific Oct1-DNA complexes. At high salt, however, only the orientation of the POUHD domain with respect to the long axis of the DNA is unperturbed in the nonspecific Oct1-DNA complex relative to that in the specific Oct1-DNA complex; the POUS domain, on the other hand, must undergo some type of conformational rearrangement, for example partial dissociation from the DNA (see below).
Fig. 2.
RDC analysis of the specific and nonspecific Oct1-DNA complexes in a liquid crystalline medium of phage pf1. (A) 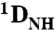 RDC profiles as a function of residue at 0 (black) and 150 (red) mM NaCl for the specific (left) and nonspecific (right) complexes. Note that the values of
RDC profiles as a function of residue at 0 (black) and 150 (red) mM NaCl for the specific (left) and nonspecific (right) complexes. Note that the values of 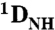 for the linker residues are close to zero suggesting that the linker is disordered. (B) Correlation of measured RDCs at 0 and 150 mM NaCl for the specific (left) and nonspecific (right) complexes. The data for the POUS and POUHD domains are displayed as open and filled-in circles, respectively. (C) Correlation of measured RDCs for the specific and nonspecific complexes at 0 (left) and 150 (right) mM NaCl. (D) SVD analysis of the specific (left) and nonspecific (right) complexes at 0 (top) and 150 (bottom) mM NaCl. The coordinates of the individual POUS and POUHD domains are taken from the 1.9 Å resolution structure of the MORE complex (1E3O) and placed in the orientation found in the H2B (1OCT)/PORE (HFO) complexes. Protons were added and SVD analysis carried out using Xplor-NIH (40). The axes of the alignment tensor relative to the DNA are displayed in each case; with the exception of the nonspecific complex at 150 mM NaCl, the principal axis (z) of the alignment tensor lies parallel to the long axis of the DNA. At 150 mM NaCl, a small deviation between the principal axis of the alignment tensor and the long axis of the DNA is observed for the nonspecific complex owing to the fact that the POUS domain is partially dissociated from the nonspecific DNA.
for the linker residues are close to zero suggesting that the linker is disordered. (B) Correlation of measured RDCs at 0 and 150 mM NaCl for the specific (left) and nonspecific (right) complexes. The data for the POUS and POUHD domains are displayed as open and filled-in circles, respectively. (C) Correlation of measured RDCs for the specific and nonspecific complexes at 0 (left) and 150 (right) mM NaCl. (D) SVD analysis of the specific (left) and nonspecific (right) complexes at 0 (top) and 150 (bottom) mM NaCl. The coordinates of the individual POUS and POUHD domains are taken from the 1.9 Å resolution structure of the MORE complex (1E3O) and placed in the orientation found in the H2B (1OCT)/PORE (HFO) complexes. Protons were added and SVD analysis carried out using Xplor-NIH (40). The axes of the alignment tensor relative to the DNA are displayed in each case; with the exception of the nonspecific complex at 150 mM NaCl, the principal axis (z) of the alignment tensor lies parallel to the long axis of the DNA. At 150 mM NaCl, a small deviation between the principal axis of the alignment tensor and the long axis of the DNA is observed for the nonspecific complex owing to the fact that the POUS domain is partially dissociated from the nonspecific DNA.
There are three crystal structures of Oct1-DNA complexes: the H2B complex involves an octamer binding site occupied by a single molecule of Oct1 (14), while the PORE and MORE complexes involve longer DNA target sites (15 and 12 bp, respectively) occupied by two molecules of Oct1 (21) (Fig. 1 B and C). The relative orientation of the POUS and POUHD domains within a single molecule of Oct1 is the same in the H2B and PORE complexes (Fig. 1C). In the case of the MORE complex, however, a very different intramolecular orientation of POUS to POUHD is found (Fig. 1C), but interestingly the intermolecular orientation of POUHD of one molecule of Oct1 and the POUS domain of the second molecule is similar (but not identical) to the intramolecular orientation observed in the H2B and PORE complexes. SVD fitting of the RDCs to the three crystal structures indicates that the orientation of the POUS and POUHD domains in the specific Oct1-DNA complex is the same as that found in the H2B and PORE complexes (RDC R-factors of 17–19%; see Table 1 and Fig. 2D left, top, and bottom).
Likewise, the orientation of the POUS and POUHD domains relative to the long axis of the DNA in the nonspecific oct1-DNA complex at low salt is also very similar to that in the H2B and PORE complexes with a RDC R-factor of 23–25% (see Table 1 and Fig. 2D, top right). Further the Euler angles obtained for the fits to the individual domains and the fits to both domains simultaneously in the H2B/PORE configuration are the same within error. This result indicates that (i) for each nonspecific site, the binding orientation of the POUS and POUHD domains relative to the long axis of the DNA is preserved, (ii) end-effects are minimal, and (iii) the negatively charged DNA dominates pf1-induced alignment such that the alignment tensor (which is close to axially symmetric) is minimally sensitive to the location of the POUS and POUHD domains along the DNA. Similar observations were previously made for the single homeodomain transcription factor HoxD9 (8). The distance, however, between the POUS and POUHD domains must be free to vary because there are no interdomain contacts to fix one domain relative to the other, the only constraint being the length of the 23 residue linker which can accommodate a wide range of spacing. Thus, these data indicate that each domain of Oct1 translates along the DNA grooves independently of each other by rotation-coupled sliding.
At high salt, however, the SVD fit of the RDCs for the nonspecific Oct1-DNA complex agree poorly with the X-ray structures (RDC R-factor of 33–38%; see Table 1 and Fig. 2D, bottom right). This finding is as expected given that the RDCs for the POUS domain in the nonspecific complex at high salt are so poorly correlated with those of either the specific complex or the nonspecific complex at low salt (Fig. 2 B and C, right). It should be noted that the poor agreement between observed and calculated RDCs for the nonspecific Oct1-DNA complex at high salt cannot be attributed to the presence of free Oct1 as the population of unbound Oct1 is less than 2% under the experimental conditions employed, and therefore will not contribute in any significant way to the measured RDCs (24).
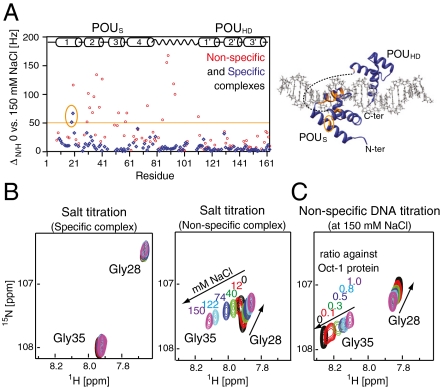
To further understand the binding of Oct1 to nonspecific DNA under high salt conditions we monitored the perturbation in backbone 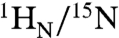 chemical shifts (
chemical shifts (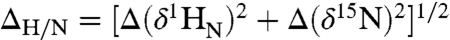 in Hz) as a function of added salt and nonspecific DNA. Negligible ΔH/N perturbations are observed for the specific Oct1-DNA complex upon addition of NaCl (Fig. 3 A and B, left , and Fig. S4A). For the nonspecific Oct1-DNA complex, however, large salt-induced ΔH/N perturbations are observed for the POUS domain and linker, but not the POUHD domain which is largely unaffected by salt (Fig. 3 A and B, right, and Fig. S4B). Because the affinity of the isolated POUS domain for its cognate DNA target site is less than that for the isolated POUHD domain (25), these data, together with the RDC data presented above, suggest that at high salt the POUS domain is partially dissociated from nonspecific DNA while the POUHD domain remains anchored to the DNA. This conclusion was further addressed by titrating nonspecific DNA into an Oct1 solution at 150 mM NaCl. The shifts induced by addition of salt (Fig. 3B, right) and nonspecific DNA (Fig. 3C) display a colinear relationship, thereby enabling one to assess the fraction of POUS that is locally dissociated from the DNA in the high salt nonspecific Oct1-DNA complex. Assuming that the POUS domain in the context of Oct1 is 100% bound to the nonspecific DNA duplex at 0 mM NaCl, the fraction of POUS that is locally dissociated at 150 mM NaCl is calculated to be 58%.
in Hz) as a function of added salt and nonspecific DNA. Negligible ΔH/N perturbations are observed for the specific Oct1-DNA complex upon addition of NaCl (Fig. 3 A and B, left , and Fig. S4A). For the nonspecific Oct1-DNA complex, however, large salt-induced ΔH/N perturbations are observed for the POUS domain and linker, but not the POUHD domain which is largely unaffected by salt (Fig. 3 A and B, right, and Fig. S4B). Because the affinity of the isolated POUS domain for its cognate DNA target site is less than that for the isolated POUHD domain (25), these data, together with the RDC data presented above, suggest that at high salt the POUS domain is partially dissociated from nonspecific DNA while the POUHD domain remains anchored to the DNA. This conclusion was further addressed by titrating nonspecific DNA into an Oct1 solution at 150 mM NaCl. The shifts induced by addition of salt (Fig. 3B, right) and nonspecific DNA (Fig. 3C) display a colinear relationship, thereby enabling one to assess the fraction of POUS that is locally dissociated from the DNA in the high salt nonspecific Oct1-DNA complex. Assuming that the POUS domain in the context of Oct1 is 100% bound to the nonspecific DNA duplex at 0 mM NaCl, the fraction of POUS that is locally dissociated at 150 mM NaCl is calculated to be 58%.
Fig. 3.
Results of salt and DNA titration experiments. (A) Backbone 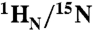 amide chemical shift perturbation (ΔN/H = [(ΔδH)2 + (ΔδN)2]1/2 in Hz at a spectrometer frequency of 500 MHz) upon addition of salt (150 vs. 0 mM NaCl) for the specific (blue) and nonspecific (red) complexes with regions exhibiting significant (ΔN/H > 50 Hz) salt-induced shifts for the nonspecific complex color-coded in orange on the structure of the specific complex. The salt-induced perturbations for the nonspecific complex are predominantly located in the POUS domain and linker. (B) Selected region of the 1H-15N TROSY correlation spectrum illustrating the behavior of the cross-peaks of Gly28 and Gly35 upon titration of salt (0, 12, 40, 74, 122, and 150 mM NaCl) into the specific (left) and nonspecific (right) complexes. (C) The same region of the 1H-15N correlation spectrum illustrating the effects of titration of nonspecific DNA into Oct1 (0.2 mM) at 150 mM NaCl. The DNA:Oct1 ratios shown are 0∶1, 0.1∶1, 0.3∶1, 0.5∶1, 0.8∶1 and 1∶1; the spectra at ratios of 1.5∶1, 2.5∶1 and 4.5∶1 are the same as that at a ratio of 1∶1. Note that purple cross-peaks in the right side of (B) and in (C) are identical and the chemical shift perturbations in the two boxes follow a colinear relationship.
amide chemical shift perturbation (ΔN/H = [(ΔδH)2 + (ΔδN)2]1/2 in Hz at a spectrometer frequency of 500 MHz) upon addition of salt (150 vs. 0 mM NaCl) for the specific (blue) and nonspecific (red) complexes with regions exhibiting significant (ΔN/H > 50 Hz) salt-induced shifts for the nonspecific complex color-coded in orange on the structure of the specific complex. The salt-induced perturbations for the nonspecific complex are predominantly located in the POUS domain and linker. (B) Selected region of the 1H-15N TROSY correlation spectrum illustrating the behavior of the cross-peaks of Gly28 and Gly35 upon titration of salt (0, 12, 40, 74, 122, and 150 mM NaCl) into the specific (left) and nonspecific (right) complexes. (C) The same region of the 1H-15N correlation spectrum illustrating the effects of titration of nonspecific DNA into Oct1 (0.2 mM) at 150 mM NaCl. The DNA:Oct1 ratios shown are 0∶1, 0.1∶1, 0.3∶1, 0.5∶1, 0.8∶1 and 1∶1; the spectra at ratios of 1.5∶1, 2.5∶1 and 4.5∶1 are the same as that at a ratio of 1∶1. Note that purple cross-peaks in the right side of (B) and in (C) are identical and the chemical shift perturbations in the two boxes follow a colinear relationship.
Translocation of Oct1 on DNA.
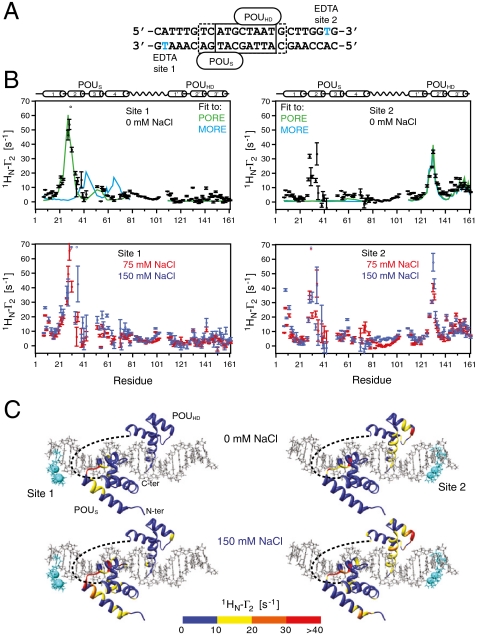
Intra- and intermolecular translocation processes, (corresponding to rotation-coupled sliding and intersegment transfer, respectively) of Oct1 bound to its specific DNA target site involve short-lived, sparsely-populated (≤ 1%) states that sample nonspecific DNA binding sites. These phenomena are readily probed by intermolecular PRE measurements (6, 24, 26–28) in which the DNA is paramagnetically labeled with dT-EDTA chelated to Mn2+ and PREs are observed on the protein bound to the DNA (29) (Fig. 4A). The PRE effect is proportional to the 〈r-6〉 distance between the paramagnetic label and the protons of interest (in this instance the backbone amide protons). In a fast exchanging system, the PRE observed on the resonances of specifically bound Oct1 (the major species) will be a population weighted average of the PREs for the specific complex and the alternate states located outside the specific DNA binding site. If the distances between the paramagnetic label and the backbone amide protons in the sparsely-populated alternate species are shorter than the corresponding distances in the specific complex, the PRE profiles will bear the footprint of the minor species (6, 28, 30). The transverse  PRE relaxation rates are obtained by taking the difference in transverse
PRE relaxation rates are obtained by taking the difference in transverse  relaxation rates between paramagnetic (Mn2+) and diamagnetic (Ca2+) states of the system (31). The 24-bp specific DNA duplex was paramagnetically labeled at two sites separately, on either end of the DNA (Fig. 4A), and PREs recorded at 0 mM NaCl (Fig. 4B, top) and higher salt (75 and 150 mM NaCl; Fig. 4B, bottom). The PRE profiles mapped onto the structure of the specific Oct1-DNA complex in the H2B/PORE configuration are shown in Fig. 4C, and a summary of PRE Q-factors (32) for the agreement between observed and calculated PRE profiles is provided in Table 2 (see Materials and Methods for the definition of the PRE Q-factor).
relaxation rates between paramagnetic (Mn2+) and diamagnetic (Ca2+) states of the system (31). The 24-bp specific DNA duplex was paramagnetically labeled at two sites separately, on either end of the DNA (Fig. 4A), and PREs recorded at 0 mM NaCl (Fig. 4B, top) and higher salt (75 and 150 mM NaCl; Fig. 4B, bottom). The PRE profiles mapped onto the structure of the specific Oct1-DNA complex in the H2B/PORE configuration are shown in Fig. 4C, and a summary of PRE Q-factors (32) for the agreement between observed and calculated PRE profiles is provided in Table 2 (see Materials and Methods for the definition of the PRE Q-factor).
Fig. 4.
Intermolecular PRE profiles for the specific Oct1-DNA complex. (A) The 24-bp DNA duplexes employed with the paramagnetic label (dT-EDTA-Mn2+) located either at the POUS (site 1) or POUHD (site 2) sides of the duplex. (B) Intermolecular PRE profiles for the paramagnetic label at sites 1 (left) and 2 (right) at 0 mM NaCl (top, black circles) and 75 (red circles) and 150 (cyan circles) mM NaCl (bottom). Error bars, 1 SD. The Oct1 and DNA concentrations are 0.4 and 0.44 mM, respectively. The back-calculated PREs for Oct1 in the H2B/PORE (green) and MORE (cyan) configurations are displayed as solid lines. Note the location of the POUHD domain in the two configurations is the same. The PREs are back-calculated in Xplor-NIH (40) using an ensemble representation for the paramagnetic label (32). (C) Experimental PRE profiles at 0 (top) and 150 (bottom) mM NaCl mapped onto the structure of the specific complex in the H2B/PORE configuration (14, 21). The color scale for the PRE 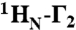 rates is shown at the bottom of the figure.
rates is shown at the bottom of the figure.
Table 2.
Agreement between observed and calculated intermolecular PREs for the specific Oct1-DNA complex
| NaCl (mM) | PRE Q-factor | |||||
| H2B/PORE * | MORE * | |||||
| All | POUS | POUHD | All | POUS | POUHD | |
| Site 1 (POUS side) | ||||||
| 0 | 0.35 | 0.29 | 0.95 | 0.96 | 0.96 | 0.95 |
| 75 | 0.42 | 0.37 | 0.97 | 0.98 | 0.98 | 0.97 |
| 150 | 0.72 | 0.69 | 0.97 | 0.96 | 0.96 | 0.97 |
| Site 2 (POUHD side) | ||||||
| 0 | 0.71 | 0.98 | 0.31 | 0.72 | 0.99 | 0.31 |
| 75 | 0.71 | 0.97 | 0.36 | 0.72 | 0.99 | 0.36 |
| 150 | 0.69 | 0.98 | 0.33 | 0.70 | 0.99 | 0.33 |
*The POUHD domain in the H2B/PORE (14) and MORE (21) configurations is located at the same position on the DNA. The position of Oct1 in the H2B/PORE configuration on the 24-bp specific DNA duplex bearing the specific octameric target site from the Hoxb1 regulatory element is taken from the NMR structure of the Oct1-Sox2-Hoxb1 DNA ternary complex (16). The entire experimental PRE profiles are best-fitted by optimizing the position of the paramagnetic labels, represented by a three-member ensemble, using simulated annealing (32). The definition of the PRE Q-factor is provided in Materials and Methods.
We first consider the PRE profiles at 0 mM NaCl (Fig. 4B, top). When the label is located on the POUS side of the DNA (site 1, Fig. 4B, left top), the overall experimental PRE profile agrees well with that back-calculated for the specific Oct1-DNA complex in the H2B/PORE geometry (PRE Q-factor = 0.35, green line) but very poorly with that for the MORE geometry (Q-factor = 0.96, blue line). A more detailed examination, however, shows that while the PREs for the POUS domain agree extremely well (Q-factor = 0.29) with those calculated for the H2B/PORE geometry, the agreement for the POUHD domain is actually poor in terms of Q-factor (Q = 0.95), although not unreasonable in terms of rms difference between observed and calculated PRE rates (3.2 s-1). This result is because no PREs are predicted for the POUHD domain from the structure of the specific complex, whereas some small PRE effects (ranging from 5–10 s-1) are in fact observed at the N terminus of helix 1′ (residues 110–113) and the N and C termini of helix 2′ (residues 131 and 142) of the POUHD domain (Fig. 4B, top left). Thus, although the POUHD domain clearly spends the vast majority of its time bound to its cognate DNA target site, a small population (≪ 1%) must sample alternate configurations that come close to the paramagnetic label located at site 1. When the paramagnetic label is placed on the POUHD side of the DNA (Fig. 4B, top right), excellent agreement (Q-factor = 0.31) is obtained for the POUHD domain, but very poor agreement is apparent for the POUS domain (Q-factor = 0.98) where large PREs ranging from 10–40 s-1 are observed within helix 2 of POUS (residues 28–36) but no PREs are predicted. Thus, the POUS domain must sample states in close proximity to the paramagnetic label at site 2, with a population that is significantly larger than the population of POUHD states in close proximity to the paramagnetic label at site 1.
When the salt concentration is increased to 75 or 150 mM NaCl (Fig. 4B, bottom), the magnitude of all the PREs is progressively increased with the paramagnetic label positioned at either sites 1 or 2, but the overall PRE profiles remain qualitatively similar to those obtained at 0 mM NaCl. These findings reflect screening of electrostatic interactions by salt resulting in enhanced sliding and intersegment transfer (i.e., the addition of salt increases the exchange rate between Oct1 located at specific and nonspecific DNA sites and/or increases the population of sparsely-populated nonspecific sites).
In the case of the paramagnetic label at site 1, the quality of agreement for the POUS domain is progressively decreased upon raising the salt concentration (Q-factor = 0.37 and 0.69 at 75 and 150 mM NaCl, respectively) and new PRE effects are observed at the N terminus (residues 8–12) of helix 1 of the POUS domain (Fig. 4B, bottom left). These observations imply the existence of a low population of states in which the orientation of the POUS domain is flipped by 180° relative to that in the specific Oct1-DNA complex. The PRE profile for the POUHD domain is essentially the same as that at 0 mM NaCl but a few PRE rates are now increased in the range of 10–20 s-1. Indeed, the PRE effects observed at the C terminus of helix 2 of POUHD also imply the existence of states in which the orientation of POUHD is flipped by 180° relative to that in the specific Oct1-DNA complex.
With the paramagnetic label at site 2, the agreement with the POUHD domain remains essentially unchanged as the salt concentration is increased (Q-factor = 0.33 at 150 mM NaCl; Fig. 4B, bottom right) but closer examination would suggest that the slight increase in PRE rates observed for the N-terminal residues of helix 1 of POUHD confirms the existence of states with a 180° reorientation relative to that in the specific Oct1-DNA complex. For the POUS domain not only are the PREs within helix 2 increased relative to those at 0 mM NaCl, but large PREs are now also observed at the N terminus of helix 1. These PREs must arise from states in the same orientation as the specific Oct1-DNA complex but translated along the DNA in the direction of site 2.
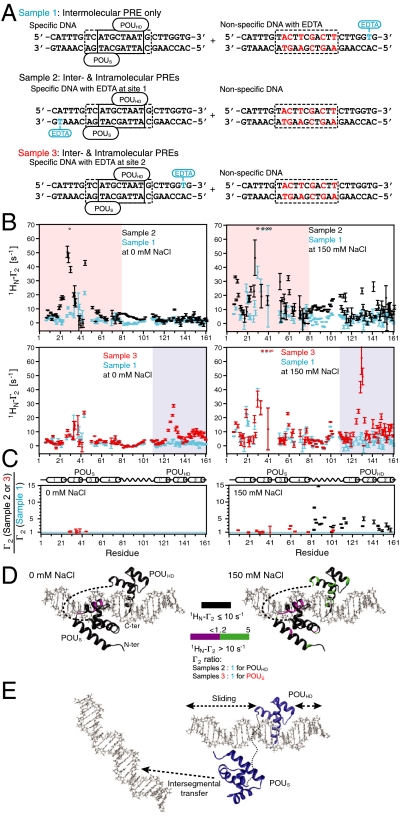
To analyze the contributions to the PRE profiles from intra- (sliding) and intermolecular translocation we carried out a series of experiments involving samples comprising an equal mixture of specific and nonspecific DNA duplexes (6) (Fig. 5). In the first set of PRE experiments (sample 1), the paramagnetic label was placed on the nonspecific DNA (Fig. 5A, top), and therefore any PREs observed in the spectrum of the specific complex can only arise from intermolecular translocation involving transient excursions from the specific DNA to the nonspecific DNA and back to the specific DNA. In the second set of experiments, the paramagnetic label was placed on the specific DNA duplex, either on the POUS side (Sample 2; Fig. 5A, middle) or the POUHD side (sample 3; Fig. 5A, bottom) with the nonspecific DNA unlabeled. The PREs observed in samples 2 and 3 will therefore comprise effects arising from both intramolecular (i.e., sliding) and intermolecular translocation (arising from direct hopping of Oct1 between DNA molecules bearing the specific site), as well as PREs arising directly from the specific complex itself. The latter will be manifested on the POUS domain for sample 2 and the POUHD domain for sample 3. Thus, if the ratio of a given PRE for samples 2 or 3 to sample 1 has a value greater than 1 outside regions directly attributable to the specific complex, that PRE must arise from both intra- and intermolecular translocation. The results of these experiments are summarized in Fig. 5.
Fig. 5.
Intermolecular PREs arising from inter- and intramolecular translocation. (A) DNA samples comprise an equimixture of specific and nonspecific 24-bp DNA duplexes. In sample 1, the specific DNA duplex is unlabeled and the nonspecific DNA duplex bears the paramagnetic label. In samples 2 and 3, the nonspecific DNA duplex is unlabeled, and the specific DNA duplex is paramagnetically labeled at either the POUS or POUHD sides of the DNA, respectively. The PRE effects observed for sample 1 arise entirely from intermolecular translocation, while those for samples 2 and 3 are given by the sum of PREs arising from the specific complex and sparsely-populated states sampled by intra- and intermolecular translocation processes. The concentrations of Oct1, specific DNA, and nonspecific DNA are 0.40, 0.44, and 0.44 mM, respectively. (B) Intermolecular PRE profiles at 0 (left) and 150 (right) mM NaCl, with intermolecular PREs from samples 1, 2, and 3 shown as cyan, black, and red circles respectively (error bars, 1 SD). The veiled-orange and blue shading indicates the regions in samples 2 (POUS domain) and 3 (POUHD domain), respectively, arising from PREs to the Oct1 domains bound to their specific target site. (C) Ratio of PRE rates for samples 2 or 3 to sample 1 at 0 (left) and 150 (right) mM NaCl for residues with 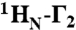 rates > 10 s-1 located in regions where the PREs arise from sampling of sparsely-populated states (i.e., residues 1–108 comprising POUS and the linker for sample 3; and residues 80–163 comprising the linker plus POUHD for sample 2). For Γ2(samples 2 or 3)/Γ2(sample 1) ∼ 1, the PRE effects arise predominantly from intermolecular translocation; Γ2(samples 2 or 3)/Γ2(sample 1) ratios larger than 1 indicate contributions from both intramolecular and intermolecular translocation where the contribution from the latter is provided by the PRE profile for sample 1. (D) Mapping of Γ2 ratios at 0 (left) and 150 (right) mM NaCl shown in (C) mapped onto the structure of the specific complex in the H2B/PORE configuration (14, 21). (E) Model for the predominant translocation processes involving the POUS and POUHD domains of Oct1.
rates > 10 s-1 located in regions where the PREs arise from sampling of sparsely-populated states (i.e., residues 1–108 comprising POUS and the linker for sample 3; and residues 80–163 comprising the linker plus POUHD for sample 2). For Γ2(samples 2 or 3)/Γ2(sample 1) ∼ 1, the PRE effects arise predominantly from intermolecular translocation; Γ2(samples 2 or 3)/Γ2(sample 1) ratios larger than 1 indicate contributions from both intramolecular and intermolecular translocation where the contribution from the latter is provided by the PRE profile for sample 1. (D) Mapping of Γ2 ratios at 0 (left) and 150 (right) mM NaCl shown in (C) mapped onto the structure of the specific complex in the H2B/PORE configuration (14, 21). (E) Model for the predominant translocation processes involving the POUS and POUHD domains of Oct1.
At both 0 and 150 mM NaCl, no significant intermolecular PRE effects within the POUHD domain are observed for sample 1 with the exception of some small effects (Γ2 = 5–10 s-1) at the N terminus of helix 1′ and within helix 2′ (Fig. 5B). Similar results are also seen for a sample comprising a paramagnetically labeled 11-bp nonspecific DNA duplex and an unlabeled 16-bp specific DNA duplex (Fig. S5). Thus, the POUHD domain remains largely bound to the specific DNA duplex and transient intermolecular translocation to the nonspecific DNA duplex occur only very rarely. As noted above, in the experiments conducted with only the specific DNA duplex, a few PREs are observed on the POUHD domain when the paramagnetic label is placed on the POUS side of the duplex. The ratio of the sample 2 to sample 1 PREs within the POUHD domain as well as the linker are greater than 1 (Fig. 5B, top left) and these effects are increased substantially at 150 mM NaCl (Fig. 5 B, top right, and C, bottom right). Thus, the POUHD domain largely undergoes intramolecular translocation.
In contrast sizeable PREs within the POUS domain are observed for sample 1 (Fig. 5B, as well as Fig. S5), and are increased in magnitude upon increasing the salt concentration: at 0 mM NaCl significant PREs are located within helix 2 of POUS (Fig. 5B, left) but extend to helix 1 and the C terminus of helix 3 of POUS at high salt (Fig. 5B, right). At both low and high salt, the ratio of the sample 3 to sample 1 PREs observed for the POUS domain are equal to 1 within experimental error (Fig. 5C), indicating that the PRE effects observed on the POUS domain when the paramagnetic label is placed on the POUHD side of the specific DNA duplex are largely due to intermolecular translocation.
Observation of Nonspecific Binding Site Sampling by PRE.
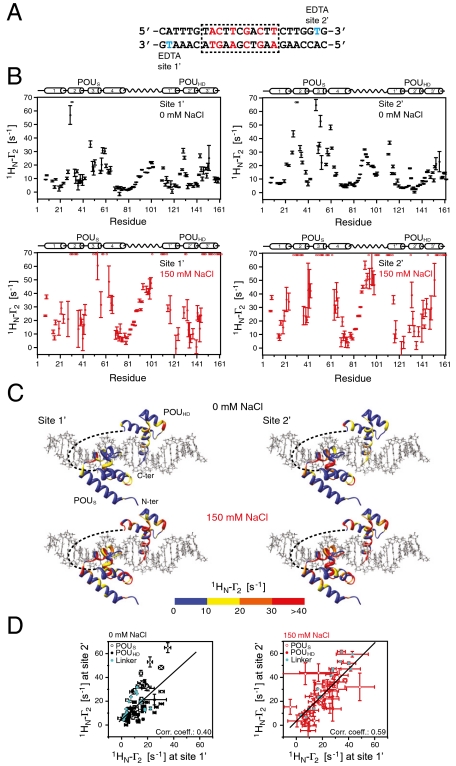
Intermolecular PRE experiments were also conducted using a nonspecific 24-bp DNA duplex with the paramagnetic label located at either end, denoted as sites 1′ and 2′ (Fig. 6A). The observed PRE profiles obtained at sites 1′ and 2′ (Fig. 6 B and C) are qualitatively similar with correlation coefficients of 0.40 and 0.59 at 0 and 150 mM NaCl, respectively (Fig. 6D). The effect of salt (i.e., electrostatic screening) is largely to increase the magnitude of the PREs, reflecting increased exchange rates between nonspecific sites close and far from the paramagnetic labels. The regions that exhibit PREs are all located in structural elements that are in close proximity to the DNA (Fig. 6C), as expected from the RDC data that indicate that the orientation of the domains relative to the long axis of the DNA is essentially the same for specific and nonspecific DNA binding (Fig. 3C, left). These data indicate that the POUS and POUHD domains sample numerous binding sites on the DNA in two approximately equally populated orientations related by a 180° rotation with respect to the long axis of the DNA.
Fig. 6.
PRE profiles for the nonspecific Oct1-DNA complex. (A) 24-bp nonspecific DNA duplex. The two sites chosen to covalently attach EDTA to thymine (one site at a time) are shown in cyan. (B) PRE profiles at 0 (black, top) and 150 (red, bottom) mM NaCl. Oct1 and nonspecific DNA concentrations are 0.40 and 0.44 mM, respectively. (C) PRE profiles at 0 and 150 mM NaCl mapped onto the structure of the specific Oct1-DNA complex in the H2B/PORE configuration (14, 21). The color scale for the PRE Γ2 rates is shown below the structures. (D) Correlation of site 1′ and site 2′ PREs at 0 (left) and 150 (right) mM NaCl.
Concluding Remarks
The chemical shift perturbation, RDC and PRE data indicate that the orientation of the POUS and POUHD domains relative to the long axis of the DNA is the same in both specific and nonspecific complexes of Oct1 with DNA. The intermolecular PRE data indicate that both the POUS and POUHD domains in the context of the specific Oct1-DNA complex sample sparsely-populated, transient states that are spectroscopically “invisible” and occupy nonspecific DNA binding sites. From the ratio of the equilibrium dissociation constants for specific and nonspecific DNA binding of Oct1, the population of the sparsely-populated states is estimated as < 1%. For both domains, orientations parallel and antiparallel to that found in the specific Oct1-DNA complex are sampled. The key finding is that the transition from the specific Oct1-DNA complex that is observed in the NMR spectrum to the spectroscopically “invisible” states involve predominantly intramolecular translocation by rotation-coupled sliding for the POUHD domain but intermolecular translocation in the form of intersegment transfer for the POUS domain. This finding is consistent with the following observations: (i) in the nonspecific Oct1-DNA complex the POUS domain at high salt exists as an approximately equal mixture of associated and dissociated states while the POUHD domain remains fully associated; (ii) the affinity of the isolated POUS domain for DNA is less than that of the POUHD domain (25); and (iii) the direct intersegment transfer rate between specific DNA binding sites located on different DNA molecules is ∼1.5-fold faster for the POUS domain than the POUHD domain (15).
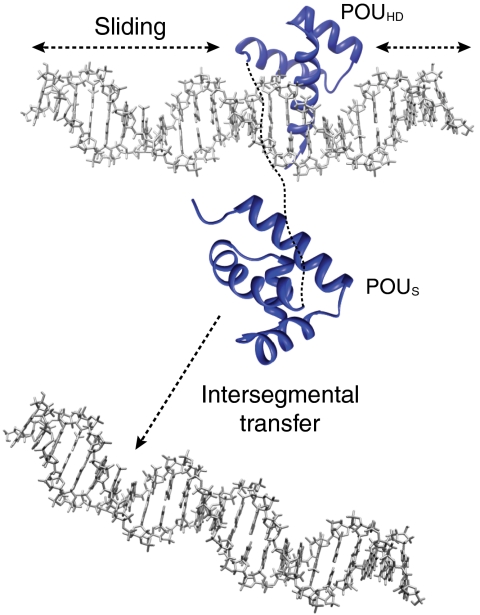
Taken together, the data presented here suggest the following model whereby Oct1 efficiently explores the DNA landscape to locate its specific target site within an overwhelming sea of nonspecific sites. The POUHD domain largely searches the DNA by sliding (i.e., intramolecular translocation), while the POUS domain acts as an antenna to promote intersegment transfer via intermolecular translocation (Fig. 5E), in a manner that is analogous to flycasting provided by the tails of single domain transcription factors (33). Transfer from one DNA molecule (or distantly located DNA segment) to another occurs via a bridged intermediate that is formed by the POUS domain latching on to the second DNA molecule. Once this intermediate is formed, the probability of completing intermolecular translocation of Oct1 by dissociation of the POUHD domain from the first DNA molecule followed by association to the second DNA molecule is significantly enhanced as this involves a first order process. In essence, the affinity of the POUHD domain for DNA is significantly larger than that of the POUS, (25) and the two domains undergo dissociation/reassociation events from DNA essentially independently of one another. The rate at which the POUS domain can hop from one DNA molecule to another is higher than that of the more tightly binding POUHD domain, but once a ternary complex is formed with POUHD bound to the original DNA molecule and POUS to the second DNA molecule, the probability of POUHD hopping to the second DNA molecule is increased significantly relative to the situation in which the POUS domain would be absent. Thus, cross-talk between the POUS and POUHD domains of Oct1, each fulfilling distinct and mutually complementary components of the search process, is essential for efficient sampling. It is gratifying that the present experimental results fully confirm the theoretical findings of recent coarse-grained simulations on facilitated DNA searching by multidomain DNA binding proteins in which intersegment transfer was promoted by the weaker DNA binding domain (34).
Materials and Methods
Sample Preparation.
The POU region (POUS + POUHD) of human Oct1 (residues 280–442) was expressed and purified as described previously (16). Uniform 2H/15N or 2H/13C/15N isotopic labeling was achieved by growing Escherichia coli BL21-CodonPlus(DE3)-RIPL cells in minimal medium with 99.9% D2O, d7-glucose or 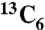 , d7-glucose, and
, d7-glucose, and 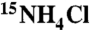 . Single-stranded unmodified and EDTA- or rhodamine-conjugated DNA oligonucleotides were purchased from Invitrogen Inc. and Midland Certified Reagents, respectively, and purified by ion-exchange chromatography on a Mono-Q (GE Healthcare Bioscience) column with a NaCl gradient in a buffer of 50 mM Tris-HCl, pH 7.5 and 1 mM EDTA. After annealing, DNA duplexes were further purified by ion-exchange chromatography to remove any residual single-stranded DNA (29). NMR samples were prepared in 10 mM Hepes, 94% H2O/6%D2O, pH 6.5.
. Single-stranded unmodified and EDTA- or rhodamine-conjugated DNA oligonucleotides were purchased from Invitrogen Inc. and Midland Certified Reagents, respectively, and purified by ion-exchange chromatography on a Mono-Q (GE Healthcare Bioscience) column with a NaCl gradient in a buffer of 50 mM Tris-HCl, pH 7.5 and 1 mM EDTA. After annealing, DNA duplexes were further purified by ion-exchange chromatography to remove any residual single-stranded DNA (29). NMR samples were prepared in 10 mM Hepes, 94% H2O/6%D2O, pH 6.5.
NMR Spectroscopy.
All NMR experiments were carried out at 303 K on Bruker 500 and 600 MHz spectrometers equipped with z gradient triple resonance cryoprobes. Spectra were processed using NMRPipe (35), and analyzed using the programs XIPP (in-house software written by D.S. Garrett) and NMRView (36). 1H, 15N, and 13C backbone resonances of Oct1 bound nonspecifically to DNA were assigned using transverse relaxation optimized (TROSY) versions (37, 38) of conventional 3D triple resonance correlation experiments (39).
PRE 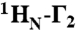 data were acquired on samples comprising 0.4 mM 2H/15N-labeled Oct1 and 0.44 mM DNA-EDTA chelated to either Mn2+ (paramagnetic) or Ca2+ (diamagnetic) using a two-time point measurement (with a ΔT difference of 16 ms) as described previously (31). Three samples comprising 0.4 mM 2H/15N-labeled Oct1, 0.44 mM specific DNA and 0.44 mM nonspecific DNA were employed to analyze the relative contribution of intra- and intermolecular translocation processes: in the first sample, the paramagnetic label was attached to the nonspecific DNA duplex and therefore reports exclusively on intermolecular translocation; in the other two samples, the paramagnetic label was attached to either end of the specific DNA duplex, and therefore report on both intra- and intermolecular translocation. PRE
data were acquired on samples comprising 0.4 mM 2H/15N-labeled Oct1 and 0.44 mM DNA-EDTA chelated to either Mn2+ (paramagnetic) or Ca2+ (diamagnetic) using a two-time point measurement (with a ΔT difference of 16 ms) as described previously (31). Three samples comprising 0.4 mM 2H/15N-labeled Oct1, 0.44 mM specific DNA and 0.44 mM nonspecific DNA were employed to analyze the relative contribution of intra- and intermolecular translocation processes: in the first sample, the paramagnetic label was attached to the nonspecific DNA duplex and therefore reports exclusively on intermolecular translocation; in the other two samples, the paramagnetic label was attached to either end of the specific DNA duplex, and therefore report on both intra- and intermolecular translocation. PRE 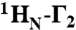 rates were back-calculated from structural models of the Oct1-DNA specific complex using a three-conformer ensemble representation for the EDTA-Mn2+ groups in conjunction with the model-free extension of the Solomon-Bloemberg PRE equations to account for flexibility of the paramagnetic label as well as local protein motions (32). The coordinates of the EDTA-Mn2+ moieties were optimized by simulated annealing in Xplor-NIH (40), and agreement between observed and calculated values is expressed as a PRE Q-factor given by
rates were back-calculated from structural models of the Oct1-DNA specific complex using a three-conformer ensemble representation for the EDTA-Mn2+ groups in conjunction with the model-free extension of the Solomon-Bloemberg PRE equations to account for flexibility of the paramagnetic label as well as local protein motions (32). The coordinates of the EDTA-Mn2+ moieties were optimized by simulated annealing in Xplor-NIH (40), and agreement between observed and calculated values is expressed as a PRE Q-factor given by 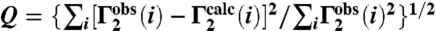 (32).
(32).
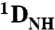 RDCs were measured on samples comprising 0.2 mM 2H/15N-labeled Oct1 and 0.3 mM specific or nonspecific DNA by taking the difference in one-bond 1H-15N splittings in aligned [11 mg/mL phage pf1; (17, 18)] and isotropic media using transverse relaxation optimized (TROSY)-based 1H-15N correlation spectroscopy as described (41). Fitting of observed RDCs to structure coordinates by SVD was carried out in Xplor-NIH (40).
RDCs were measured on samples comprising 0.2 mM 2H/15N-labeled Oct1 and 0.3 mM specific or nonspecific DNA by taking the difference in one-bond 1H-15N splittings in aligned [11 mg/mL phage pf1; (17, 18)] and isotropic media using transverse relaxation optimized (TROSY)-based 1H-15N correlation spectroscopy as described (41). Fitting of observed RDCs to structure coordinates by SVD was carried out in Xplor-NIH (40).
Oct1 binds specifically to the octamer recognition site within the Hoxb1 promoter (see Fig. 1A) in the H2B/PORE geometry (14, 21), as determined by NMR (16). PREs and RDCs were back-calculated for this structure as well as for a structure in which the position of the POUS domain was placed in the same position as that found in the MORE complex (21). Note that the actual coordinates of the individual POUS and POUHD domains are taken from the 1.9 Å resolution X-ray structure of the PORE complex (21) because these are found to fit the RDCs the best [as expected given the higher resolution of these coordinates relative to the 3 Å resolution H2B complex and the 2.7 Å resolution PORE complex; (14, 21)].
Fluorescence Anisotropy.
The KD for the binding of Oct1 to nonspecific DNA at 25 °C was determined by fluorescence anisotropy using a Jobin-Yvon FluoroMax-3 spectrometer as described previously (8). The wavelengths for excitation and emission were 550 and 580 nm, respectively. Oct1 was titrated (0–13.2 μM final concentration) into a rhodamine-conjugated nonspecific 24-bp DNA duplex (1.7 μM) dissolved in 10 mM Tris-HCl and 150 mM NaCl at pH 7.4. The KD was calculated from the titration data as described previously (42).
Supplementary Material
Acknowledgments.
We thank Dr. M. Doucleff for initial help at the start of this work, and Drs. Garrett, Baber and Ying for technical support. This work was supported by the intramural program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, and by the AIDS Targeted Antiviral Program of the Office of the Director of the NIH (G.M.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 8929.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100050108/-/DCSupplemental.
References
- 1.Berg OG, von Hippel PH. Diffusion-controlled macromolecular interactions. Annu Rev Biophys Biophys Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 2.von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 3.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vuzman D, Azia A, Levy Y. Searching DNA via a “Monkey Bar” mechanism: the significance of disordered tails. J Mol Biol. 2010;396:674–684. doi: 10.1016/j.jmb.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 5.Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17:932–938. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 7.Iwahara J, Clore GM. Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy. J Am Chem Soc. 2006;128:404–405. doi: 10.1021/ja056786o. [DOI] [PubMed] [Google Scholar]
- 8.Iwahara J, Zweckstetter M, Clore GM. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc Natl Acad Sci USA. 2006;103:15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blainey PC, et al. Nonspecifically bound proteins spin while diffusing along DNA. Nat Struct Mol Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sturm RA, Herr W. The POU domain is a bipartite DNA-binding structure. Nature. 1988;336:601–604. doi: 10.1038/336601a0. [DOI] [PubMed] [Google Scholar]
- 11.Di Rocco G, et al. The recruitment of SOX/OCT complexes and the differential activity of HOXA1 and HOXB1 modulate the Hoxb1 auto-regulatory enhancer function. J Biol Chem. 2001;276:20506–20515. doi: 10.1074/jbc.M011175200. [DOI] [PubMed] [Google Scholar]
- 12.Andersen B, Rosenfeld MG. POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr Rev. 2001;22:2–35. doi: 10.1210/edrv.22.1.0421. [DOI] [PubMed] [Google Scholar]
- 13.Herr W, et al. The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- 14.Klemm JD, Rould MA, Aurora R, Herr W, Pabo CO. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 15.Doucleff M, Clore GM. Global jumping and domain-specific intersegment transfer between DNA cognate sites of the multidomain transcription factor Oct-1. Proc Natl Acad Sci USA. 2008;105:13871–13876. doi: 10.1073/pnas.0805050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams DC, Jr, Cai M, Clore GM. Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1•Sox2•Hoxb1 DNA ternary transcription factor complex. J Biol Chem. 2004;279:1449–1457. doi: 10.1074/jbc.M309790200. [DOI] [PubMed] [Google Scholar]
- 17.Clore GM, Starich MR, Gronenborn AM. Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod-shaped viruses. J Am Chem Soc. 1998;120:10571–10572. [Google Scholar]
- 18.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat Struct Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 19.Bax A, Kontaxis G, Tjandra N. Dipolar couplings in macromolecular structure determination. Methods Enzymol. 2001;339:127–174. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 20.Losonczi JA, Andrec M, Fischer MW, Prestegard JH. Order matrix analysis of residual dipolar couplings using singular value decomposition. J Magn Reson. 1999;138:334–342. doi: 10.1006/jmre.1999.1754. [DOI] [PubMed] [Google Scholar]
- 21.Remenyi A, et al. Differential dimer activities of the transcription factor Oct-1 by DNA-induced interface swapping. Mol Cell. 2001;8:569–580. doi: 10.1016/s1097-2765(01)00336-7. [DOI] [PubMed] [Google Scholar]
- 22.Clore GM, Garrett DS. R-factor,free R and complete cross-validation for dipolar coupling refinement of NMR structures. J Am Chem Soc. 1999;121:9008–9012. [Google Scholar]
- 23.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang C, Schwieters CD, Clore GM. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449:1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 25.Klemm JD, Pabo CO. Oct-1 POU domain-DNA interactions: cooperative binding of isolated subdomains and effects of covalent linkage. Genes Dev. 1996;10:27–36. doi: 10.1101/gad.10.1.27. [DOI] [PubMed] [Google Scholar]
- 26.Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 27.Volkov AN, Worrall JA, Holtzmann E, Ubbink M. Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR. Proc Natl Acad Sci USA. 2006;103:18945–18950. doi: 10.1073/pnas.0603551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwahara J, Anderson DE, Murphy EC, Clore GM. EDTA-derivatized deoxythymidine as a tool for rapid determination of protein binding polarity to DNA by intermolecular paramagnetic relaxation enhancement. J Am Chem Soc. 2003;125:6634–6635. doi: 10.1021/ja034488q. [DOI] [PubMed] [Google Scholar]
- 30.Clore GM. Exploring sparsely-populated states of macromolecules by diamagnetic and paramagnetic NMR relaxation. Protein Sci. 2010 doi: 10.1002/pro.576. epub ahead of print doi: 10.1002/pro.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwahara J, Tang C, Clore GM. Practical aspects of 1H transverse paramagnetic relaxation enhancement measurements on macromolecules. J Magn Reson. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwahara J, Schwieters CD, Clore GM. Ensemble approach for NMR structure refinement against 1H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J Am Chem Soc. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 33.Levy Y, Onuchic JN, Wolynes PG. Fly-casting in protein-DNA binding: frustration between protein folding and electrostatics facilitates target recognition. J Am Chem Soc. 2007;129:738–739. doi: 10.1021/ja065531n. [DOI] [PubMed] [Google Scholar]
- 34.Vuzman D, Polonsky M, Levy Y. Facilitated DNA search by multidomain transcription factors: cross talk via a flexible linker. Biophys J. 2010;99:1202–1211. doi: 10.1016/j.bpj.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 36.Johnson BA, Blevins RA. NMR View: a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 37.Tugarinov V, Muhandiram R, Ayed A, Kay LE. Four-dimensional NMR spectroscopy of a 723-residue protein: chemical shift assignments and secondary structure of malate synthase G. J Am Chem Soc. 2002;124:10025–10035. doi: 10.1021/ja0205636. [DOI] [PubMed] [Google Scholar]
- 38.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clore GM, Gronenborn AM. Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 1998;16:22–34. doi: 10.1016/S0167-7799(97)01135-9. [DOI] [PubMed] [Google Scholar]
- 40.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR structure determination. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;48:47–62. [Google Scholar]
- 41.Schwieters CD, et al. Solution structure of the 128 kDa enzyme I dimer from Escherichia coli and its 146 kDa complex with HPr using residual dipolar couplings and small- and wide-angle X-ray scattering. J Am Chem Soc. 2010;132:13026–13045. doi: 10.1021/ja105485b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iwahara J, Schwieters CD, Clore GM. Characterization of nonspecific protein-DNA interactions by 1H paramagnetic relaxation enhancement. J Am Chem Soc. 2004;126:12800–12808. doi: 10.1021/ja046246b. [DOI] [PubMed] [Google Scholar]