Abstract
Detailed understanding of protein function and malfunction hinges on the ability to characterize transiently populated states and the transitions between them. Here, we use 15N,  , and 13CO NMR R2 relaxation dispersion to investigate spontaneous unfolding and refolding events of native apomyoglobin. Above pH 5.0, dispersion is dominated by processes involving fluctuations of the F-helix region, which is invisible in NMR spectra. Measurements of R2 dispersion for residues contacted by the F-helix region in the native (N) structure reveal a transient state formed by local unfolding of helix F and undocking from the protein core. A similar state was detected at pH 4.75–4.95 and determined to be an on-pathway intermediate (I1) in a linear three-state unfolding scheme (N⇆I1⇆MG) leading to a transiently populated molten globule (MG) state. The slowest steps in unfolding and refolding are N → I1 (36 s-1) and MG → I1 (26 s-1), respectively. Differences in chemical shift between N and I1 are very small, except in regions adjacent to helix F, showing that their core structures are similar. Chemical shift changes between the N and MG states, obtained from R2 dispersion, reveal that the transient MG state is structurally similar to the equilibrium MG observed previously at high temperature and low pH. Analysis of MG state chemical shifts shows the location of residual helical structure in the transient intermediate and identifies regions that unfold or rearrange into nonnative structure during the N → MG transition. The experiments also identify regions of energetic frustration that “crack” during unfolding and impede the refolding process.
, and 13CO NMR R2 relaxation dispersion to investigate spontaneous unfolding and refolding events of native apomyoglobin. Above pH 5.0, dispersion is dominated by processes involving fluctuations of the F-helix region, which is invisible in NMR spectra. Measurements of R2 dispersion for residues contacted by the F-helix region in the native (N) structure reveal a transient state formed by local unfolding of helix F and undocking from the protein core. A similar state was detected at pH 4.75–4.95 and determined to be an on-pathway intermediate (I1) in a linear three-state unfolding scheme (N⇆I1⇆MG) leading to a transiently populated molten globule (MG) state. The slowest steps in unfolding and refolding are N → I1 (36 s-1) and MG → I1 (26 s-1), respectively. Differences in chemical shift between N and I1 are very small, except in regions adjacent to helix F, showing that their core structures are similar. Chemical shift changes between the N and MG states, obtained from R2 dispersion, reveal that the transient MG state is structurally similar to the equilibrium MG observed previously at high temperature and low pH. Analysis of MG state chemical shifts shows the location of residual helical structure in the transient intermediate and identifies regions that unfold or rearrange into nonnative structure during the N → MG transition. The experiments also identify regions of energetic frustration that “crack” during unfolding and impede the refolding process.
Keywords: transient protein unfolding, protein folding intermediate, protein folding mechanism
Within the cell, unfolding and refolding of proteins occurs constantly, and spontaneous unfolding and misfolding processes play a central role in the formation of amyloid fibrils (1). In addition, fluctuations between native protein structure and partially or fully unfolded states have functional significance for binding, allosteric regulation, translocation across membranes, protein trafficking, secretion, and degradation (2). Despite the importance of spontaneous unfolding for protein function and cellular proteostasis, little is known about the transient, partially unfolded states that are formed. Detailed structural characterization of such states is difficult because of their inherently low populations and the conformational heterogeneity present when both native and partially unfolded states simultaneously exist in solution.
Carr–Purcell–Meiboom–Gill (CPMG)-based R2 relaxation dispersion experiments are unique in their ability to measure the kinetics of microsecond–millisecond exchange processes involving transient states with populations as sparse as 1%, and, in addition, allow determination of the associated NMR chemical shift changes (Δω) (3–5). In these experiments, the effective transverse relaxation rate  is deconvoluted into the contribution from the exchange process, Rex, which varies with the pulsing frequency in the CPMG refocusing element, and the intrinsic relaxation rate
is deconvoluted into the contribution from the exchange process, Rex, which varies with the pulsing frequency in the CPMG refocusing element, and the intrinsic relaxation rate  . Here, we apply a series of amide and carbonyl R2 dispersion experiments to investigate transient unfolding and refolding events within the native (N) state free-energy landscape of apomyoglobin (apoMb).
. Here, we apply a series of amide and carbonyl R2 dispersion experiments to investigate transient unfolding and refolding events within the native (N) state free-energy landscape of apomyoglobin (apoMb).
ApoMb is an ideal model system for studies of protein folding/unfolding equilibria. The folded state is marginally stable at neutral pH, adopting a similar tertiary structure to the holoprotein except for disorder in the EF loop, F helix, FG loop, and N-terminal region of the G helix (residues 82–104), which appear to fluctuate between a folded native-like conformational substate and an ensemble of locally unfolded states (6, 7). ApoMb forms an equilibrium molten globule (MG) state at pH 4.1, amenable to direct investigation by NMR, which contains a significant number of native contacts and an overall compactness close to that of the N state (8–10). Kinetic refolding experiments show rapid formation of a burst-phase intermediate that resembles the equilibrium MG and contains native-like topology in the most structured regions (10–12). Folding to the N state from the MG is the overall rate-limiting event; although the structural changes that occur across this transition are well established, it is still unclear why folding is slow and which interactions are frustrated and responsible for the kinetic trap.
In the present work, we characterize the mechanism of transient unfolding of apoMb, identify the structural changes that occur, and measure the kinetics of the unfolding and refolding transitions. Chemical shift changes, obtained from analysis of R2 dispersion experiments, provide insights into the structures of the two intermediates that are formed and identify regions of energetic frustration that impede protein folding.
Results
pH Dependence of N-State Structure and Dynamics:
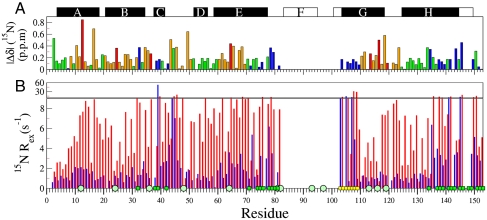
15N and 13CO chemical shifts were used to probe potential changes in the structure of the N state over the pH range of interest. Most amide 15N resonances shift by less than 0.2 ppm between pH 5.90 and 4.95 (Fig. 1A), showing that the native apoMb structure is retained over this pH range. These small shifts are in marked contrast to those associated with apoMb unfolding, which moves 15N resonances by an average of 3 ppm. Resonances with 15N chemical shift changes larger than 0.2 ppm largely belong to histidines, most of which titrate over this pH range (13), and residues that contact histidines. In addition, small 15N chemical shift changes are observed for residues in the N terminus of the G helix and in sites that contact the F helix in the holoMb structure, suggesting a small pH-dependent shift in the F-helix conformational ensemble. These effects can be monitored only indirectly, because exchange broadening prevents direct observation of amide resonances of the disordered F helix in native apoMb (6). 13CO chemical shift changes between pH 5.9 and 4.75 (Fig. S1A) corroborate these results.
Fig. 1.
Sensitivity of N-state 15N chemical shifts and Rex relaxation rates to changes in pH. (A) Magnitude of 15N chemical shift changes between pH 5.9 and 4.95, color-coded to highlight His (red); residues contacting His (orange); and residues contacting the disordered F helix, FG loop, and N-terminal region of helix G (blue); other residues are colored green. The solid black rectangles at the top of the figure depict helices in apoMb based on a TALOS+ (30) analysis of 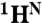 , 15N, 13CO,
, 15N, 13CO, 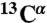 , and
, and 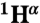 chemical shifts (6), whereas the boundaries of the F, G, and H helices in holoMb are shown as open rectangles. (B) 15N Rex (at a static magnetic field of 11.7 T) of deuterated apoMb at 35 °C; pH 5.9 (blue), pH 4.9 (red). Rex was estimated from the difference in R2eff at the lowest and highest 1/τcp values. The green squares on the horizontal axis indicate contact sites with helix F in holoMb, and the circles identify His residues. Yellow squares indicate the N-terminal region of helix G.
chemical shifts (6), whereas the boundaries of the F, G, and H helices in holoMb are shown as open rectangles. (B) 15N Rex (at a static magnetic field of 11.7 T) of deuterated apoMb at 35 °C; pH 5.9 (blue), pH 4.9 (red). Rex was estimated from the difference in R2eff at the lowest and highest 1/τcp values. The green squares on the horizontal axis indicate contact sites with helix F in holoMb, and the circles identify His residues. Yellow squares indicate the N-terminal region of helix G.
Single-quantum (SQ) 15N and 13CO R2 dispersion experiments were used to detect the formation of transiently populated states within the apoMb N-state ensemble in the pH range 5.2–5.9. Localized groups of residues display R2 dispersion with 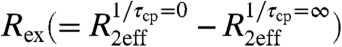 greater than 2 s-1 (Fig. 1B). These residues are located at or near sites where the F helix contacts helices C, E, and H in the native structure, suggesting that dispersion arises from millisecond timescale fluctuations in structure and/or packing of the F-helix region. Large Rex values are also observed for residues at the N terminus of the G helix, immediately following the FG turn. The pattern of 13CO Rex contributions largely matches that of 15N (Fig. S1B), with greater than average values in regions associated with helix F.
greater than 2 s-1 (Fig. 1B). These residues are located at or near sites where the F helix contacts helices C, E, and H in the native structure, suggesting that dispersion arises from millisecond timescale fluctuations in structure and/or packing of the F-helix region. Large Rex values are also observed for residues at the N terminus of the G helix, immediately following the FG turn. The pattern of 13CO Rex contributions largely matches that of 15N (Fig. S1B), with greater than average values in regions associated with helix F.
As the pH is lowered to 4.95, an additional exchange process becomes evident (Fig. 1B and Fig. S1C), associated with a large increase in Rex for residues throughout the structure. The dramatic pH dependence of Rex, together with the relatively small chemical shift perturbations between pH 5.9 and 4.95, is attributed to a shift in the N-state conformational ensemble such that there is an increased population of partially unfolded states at lower pH. At pH 5.9, R2 dispersion arises predominantly from localized fluctuations of the disordered F helix, whereas global conformational fluctuations dominate at pH values below 5.
Transient Unfolding at pH Greater than 5.0.
15N and 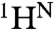 R2 dispersion curves for perdeuterated apoMb acquired at two static magnetic fields (11.7 and 18.8 T) at pH 5.5 fitted well to a global two-site exchange model (Fig. S2) with a reduced χ2 of 1.5. The small Rex contributions in regions outside of the sites that are contacted by residues 82–104 (Fig. 1B) most likely come from the exchange process that occurs below pH 5, although long-range effects of the F-helix fluctuations cannot be entirely ruled out. The kinetics of the fluctuations in the F-helix region were determined from a clustered fit of the pH 5.5 1H/15N R2 dispersion curves for residues within the contact sites of the F helix and FG loop in the native Mb structure. The fits reveal an exchange process at a rate of 533 ± 7 s-1 between the N state and a state with 7% population (Table 1).
R2 dispersion curves for perdeuterated apoMb acquired at two static magnetic fields (11.7 and 18.8 T) at pH 5.5 fitted well to a global two-site exchange model (Fig. S2) with a reduced χ2 of 1.5. The small Rex contributions in regions outside of the sites that are contacted by residues 82–104 (Fig. 1B) most likely come from the exchange process that occurs below pH 5, although long-range effects of the F-helix fluctuations cannot be entirely ruled out. The kinetics of the fluctuations in the F-helix region were determined from a clustered fit of the pH 5.5 1H/15N R2 dispersion curves for residues within the contact sites of the F helix and FG loop in the native Mb structure. The fits reveal an exchange process at a rate of 533 ± 7 s-1 between the N state and a state with 7% population (Table 1).
Table 1.
Kinetic parameters of apoMb transient unfolding and refolding measured by R2 relaxation dispersion
| Probes | pH | kN-I1 , s-1 | kI1-N , s-1 | kI1-MG, s-1 | kMG-I1, s-1 | PN | PI1 | PMG |
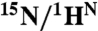 * *
|
5.50 | 38 ± 1 | 495 ± 7 | — | — | 0.93 ± 0.02 | 0.07 ± 0.00 | — |
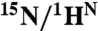 † †
|
4.95 | 31.7 ± 0.5 | 240 ± 12 | 53 ± 1 | 26 ± 4 | 0.71 ± 0.03 | 0.09 ± 0.00 | 0.20 ± 0.03 |
| 3CO/15N‡ | 4.75 | 78 ± 18 | 650 ± 140 | 113 ± 16 | 17 ± 1 | 0.52 ± 0.01 | 0.06 ± 0.01 | 0.42 ± 0.01 |
*Parameters from a two-state fit to amide datasets 1H-SQ and 15N-SQ using the cluster of residues in contact with the F helix and the first helical turn of G (listed in the caption of Fig. S2). Error values were obtained using Monte Carlo sampling.
Equilibrium Unfolding Process at pH Less than 5.
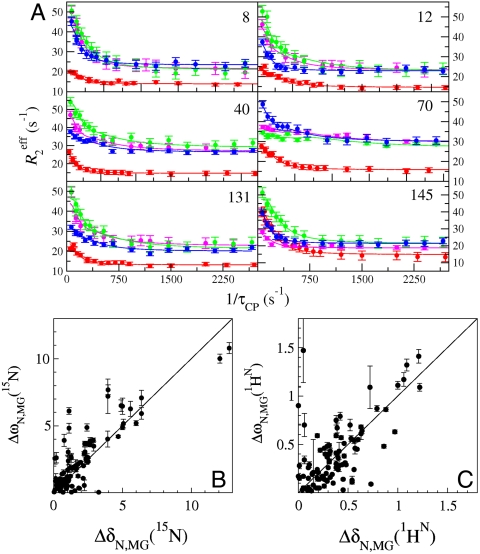
Analysis of SQ 15N R2 dispersion experiments at pH 4.75 (SI Text) showed clearly that more than two states are involved in the transient unfolding process at pH < 5. To robustly fit more complex exchange models, dispersion data were acquired on 2H/15N-labeled apoMb at pH 4.95 using 1H SQ (4), 15N SQ (3, 4), 1H/15N double-quantum (DQ) and 1H/15N zero-quantum (ZQ) coherences (14), while incorporating the respective chemical shift differences ΔωH, ΔωN, (ΔωN + ΔωH), and (ΔωN - ΔωH) into a global analysis (15, 16) (Fig. 2). Because of the destabilizing effect of aliphatic deuteration (17), 15N Rex values for perdeuterated samples at pH 4.95 were of similar amplitude to those of protonated 15N-labeled apoMb at pH 4.75.
Fig. 2.
Chemical shift differences obtained from R2 dispersion show that apoMb populates the MG state below pH 5. (A) Representative R2 dispersion curves for apoMb at pH 4.95, 35 °C at a static magnetic field of 18.8 T: 15N-SQ (red), 1H-SQ (purple), 1H15N-ZQ (green), and 1H15N-DQ (blue), showing fits to a three-state exchange model. Data acquired at 11.7 T were included in the global fit, but are omitted from the plots for clarity. Correlation of amide 15N (B) and 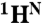 (C) equilibrium chemical shift differences (ppm) between the N (pH 4.95) and MG (pH 4.1) states, |ΔδN,MG|, and the chemical shift differences |ΔωN,MG| obtained from a global fit of the dispersion curves to the linear model N⇆I1⇆MG. A line of slope 1 is shown as a visual guide. Some scatter in the correlation plots for amide 15N and
(C) equilibrium chemical shift differences (ppm) between the N (pH 4.95) and MG (pH 4.1) states, |ΔδN,MG|, and the chemical shift differences |ΔωN,MG| obtained from a global fit of the dispersion curves to the linear model N⇆I1⇆MG. A line of slope 1 is shown as a visual guide. Some scatter in the correlation plots for amide 15N and 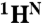 resonances is to be expected because of unavoidable differences in sample conditions, required to obtain spectra of the equilibrium MG (pH 4.1, 50 °C, 10% ethanol) (8).
resonances is to be expected because of unavoidable differences in sample conditions, required to obtain spectra of the equilibrium MG (pH 4.1, 50 °C, 10% ethanol) (8).
Dispersion curves for 89 residues were fitted using a global three-state exchange model, which gave significantly better fits than two-state exchange models according to reduced χ2 values and the Akaike information criterion (SI Text). All possible three-state exchange models were evaluated, including triangular connectivity between states and linear schemes with N in the center or on the periphery. A linear exchange model (A⇆B⇆C) yielded the best global fit and physically meaningful Δω values (SI Text). The kinetics and populations are summarized in Table 1. A linear correlation is observed between the 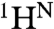 and 15N chemical shift differences (ΔωA,C) derived by fitting the dispersion curves and the equilibrium chemical shift differences (Δδ) between native apoMb (N) and the pH 4.1 MG, thus identifying states A and C as N and MG, respectively (Figs. 2 B and C). The previously unknown I1 state is therefore an on-pathway intermediate between the N and MG states (Scheme 1).
and 15N chemical shift differences (ΔωA,C) derived by fitting the dispersion curves and the equilibrium chemical shift differences (Δδ) between native apoMb (N) and the pH 4.1 MG, thus identifying states A and C as N and MG, respectively (Figs. 2 B and C). The previously unknown I1 state is therefore an on-pathway intermediate between the N and MG states (Scheme 1).
Scheme 1.
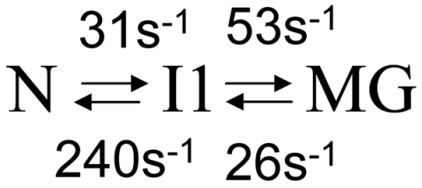
The 15N chemical shift differences between the N and I1 states (ΔωN,I1) are very small, < 0.4 ppm for most residues; residues with larger Δω values are predominantly associated with F-helix contact sites (Figs. S3 A and B and S4), suggesting that the transition to I1 involves fluctuations in the F helix or dissociation of the F helix from the protein core. In contrast, much larger 15N chemical shift differences occur in the transition between the N and MG states, with Δω(15N) > 1 ppm for the majority of resonances that exhibit dispersion (Fig. 2). Based on the small amplitude of ΔωN,I1 for both 15N and 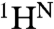 (Fig. S3A), I1 is clearly similar in overall structure to N and is formed through a local, partial unfolding process involving the NMR-invisible residues of the F helix.
(Fig. S3A), I1 is clearly similar in overall structure to N and is formed through a local, partial unfolding process involving the NMR-invisible residues of the F helix.
A linear correlation (slope = 1.0) is observed between ΔωN,I1 and Δω values for residues in F-helix contact sites derived from the two-state clustered fit of dispersion curves at pH 5.5 (Fig. S3). This confirms that I1, formed transiently at pH < 5, corresponds to the state associated with F-helix fluctuations at pH 5.5. The unfolding rates at pH 5.5 and 4.95 are similar (Table 1), whereas the refolding rate at pH 5.5 is twice that at pH 4.95, resulting in a smaller population of the transiently unfolded F helix at pH 5.5 (7%) than for I1 at pH 4.95 (9% population).
Probing Backbone Conformation at pH 4.75 Using 13CO R2 Dispersion.
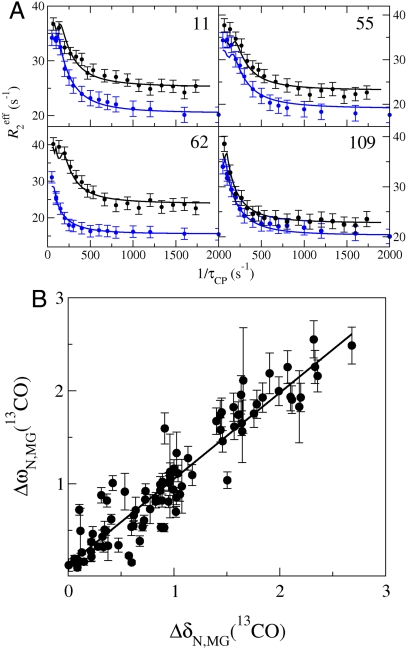
Transient unfolding of secondary structure at pH 4.75 was monitored using 13CO R2 dispersion (18) (Fig. 3). To provide an internal control to account for small and unavoidable differences in sample conditions, 13CO and 15N data were acquired on the same 15N/13C/1H-labeled sample. The dispersion curves yield 15N Δω values that match those determined with solely 15N-labeled samples. Dispersion curves were analyzed for 117 residues located throughout the structure, except for the F-helix region and C terminus of the H helix where amide resonances are broadened beyond detection by conformational exchange. The 15N and 13CO dispersion data were fit simultaneously to a three-state model using a single set of kinetic parameters (Table 1 and Fig. 3). The ability to simultaneously fit both datasets provides evidence that the two probes report on the same three-state exchange process and supports a model of cooperative partial unfolding of tertiary and secondary structure. The exchange rates between N and I1 and the population of the MG state derived from fits of the 13CO/15N dataset are slightly higher than those extracted from the 1H/15N data (Table 1), probably due to differences in sample conditions (SI Text and Fig. S1C).
Fig. 3.
Probing backbone conformational changes using 13CO R2 dispersion. (A) Representative 13CO (black) and 15N (blue) R2 dispersion curves collected on a single sample of 13C, 15N-apoMb, pH 4.75, at a static magnetic field of 18.8 T. Data acquired at a static field strength of 14.1 T were included in the global fit but are omitted from the figure for clarity. The solid curves represent three-state global fits to the exchange model N⇆I1⇆MG. (B) Correlation between ΔωN,MG (ppm) obtained from a global three-state fit to 13CO and 15N R2 dispersion data at pH 4.75, and |ΔδN,MG| determined from 3D NMR data under equilibrium conditions (pH 4.75 and 4.1 for N and MG, respectively). The line shows a linear fit to the data (R2 = 0.88, slope 0.93). A description of parameter constraints and uncertainty analysis is in the SI Text.
Discussion
Exchange Rates.
Fast kinetics experiments have shown that apoMb folds by a sequential pathway (U↔Ia↔Ib↔N) involving two intermediates separated by an energy barrier (19–21). The first intermediate (Ia) is helical in the A, G, and H regions, whereas Ib contains additional structure in the B, C, and E regions (22). The kMG,I1 folding rate from three-state fits of the dispersion curves (26 s-1 at 35 °C; Table 1) agrees remarkably well with the folding rate for the Ib to N transition (20 s-1 at 26 °C) determined from fast kinetics (21). The rate of apoMb unfolding to the MG state at 4.5 °C ranges from 13 s-1 at pH 4.2 to ∼50 s-1 at pH 3.7 (23). Given the differences in temperature and pH, these rates appear consistent with that determined here for the I1 → MG transition (53 s-1 at pH 4.95, 35 °C).
Unfolding to the MG Occurs Through the I1 State.
The dispersion data at pH 4.75 fit best to a linear three-state exchange model, in which I1 is an on-pathway intermediate between N and MG; both unfolding and refolding transitions proceed by way of I1. Native apoMb contains a compact globular core, with secondary and tertiary structure similar to that of the holoprotein (7). However, the F helix and neighboring regions are dynamically disordered and fluctuate between a compact ordered state, in which the empty heme-binding pocket is partially closed (24), and a partially unfolded, solvent-exposed state (6). In the present work, we attribute the process that dominates R2 dispersion at pH 5.5, and also contributes to dispersion at lower pH values, to the intrinsic fluctuations of the F helix to form state I1. The correlation between Δω, determined from a two-site exchange model at pH 5.5, and ΔωN,I1, obtained by fitting a three-site model at pH 4.75 (Fig. S3B), strongly supports the interpretation of N⇆I1 exchange as a local unfolding process. The dispersion measurements report indirectly on fluctuations of the F helix, the amide resonances of which are broadened beyond detection in the N state, through exchange contributions at F-helix contact sites. Transient unfolding of helix F does not significantly perturb the structure of the apoMb core; chemical shift changes between N and I1 (Figs. S3 and S5) are very small for the majority of residues showing that the secondary and tertiary structure of the apoMb core is preserved. Amides with the largest values of ΔωN,I1(15N) are located in F-helix contact sites (Figs. S3 and S4). Changes in 15N shifts in the transition to I1 are also observed for several residues in helix E; these likely reflect undocking of the F-helix region, which probably occupies partially the empty heme pocket of the native apoMb, where it would make direct contact with helix E (24). Some residues that directly flank helix F have ΔωN,I1(13CO) between 0.4–0.9 ppm, suggesting partial fraying at the C terminus of E and the N terminus of helix G in I1 (Figs. S4 and S5). Clusters of residues with ΔωN,I1(13CO) > 0.2 ppm are also found at F-helix contact sites in the CD region and in the H helix.
Disruption of the buried His24–His119 hydrogen bond is the primary trigger for pH-dependent unfolding of native apoMb (13). His24 has an abnormally low pKa (< 4) in the N state and a normal pKa in the transition state and MG (25). The His24 chemical shifts are unchanged in I1 (ΔωN,I1 ≈ 0 ppm), indicating that His24 remains in a native-like environment (with low pKa) in the I1 intermediate. Protonation of His24, which drives unfolding (13, 25) and causes large changes in His24 amide chemical shifts, must therefore occur before the transition state between I1 and MG is reached (Fig. 4), In support, ΔωI1,MG is large for both 15N and 1H (1.0 and 0.5 ppm, respectively).
Fig. 4.
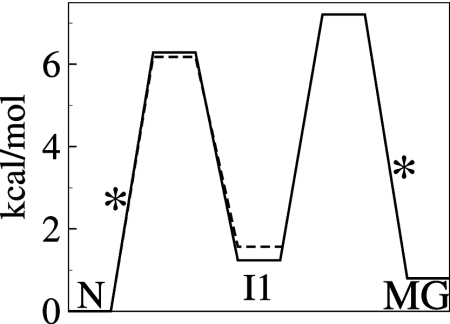
Schematic free energy diagram for three-state transient unfolding (N⇆I1⇆MG) of apoMb at pH 4.95 (solid) and two-state unfolding (N⇆I1) at pH 5.5 (dashed). Barrier heights were calculated from the rates in Table 1 and Table S1 using transition-state theory and a standard assumption for the prefactor for protein folding (35). The rate-limiting barriers are marked with asterisks. The free energies of the I1 and MG states are 1.2 and 0.9 kcal/mol, respectively, above the N state.
Recent studies of apoMb folding and unfolding kinetics (26, 27) suggest that the initial event during unfolding and the final event during refolding involve solvation and desolvation, respectively. Indeed, it has been suggested that desolvation of backbone amides may contribute to the energy barrier in the final rate-limiting step of the folding pathway (27). Although our data do not directly probe solvation effects, it is likely that the N → I1 transition, driven by the undocking of the F helix, is accompanied by substantial hydration of the empty heme-binding pocket.
Although the secondary and tertiary structure of the compact globular core of N and I1 is very similar, I1 is thermodynamically less stable than either the native or MG states (Fig. 4). The difference in free energy between I1 and MG is small (0.3 kcal/mol) yet striking, because it suggests structural frustration in the formation of the less stable, highly native-like I1 state from the MG.
Structure of the Transient MG.
The Δω values for 15N, 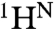 , and 13CO (Tables S2–S4) yield information about the structural changes that occur between the N and MG states during the process of folding and unfolding, at equilibrium under a single set of experimental conditions (temperature, pH, and buffer). Whereas 13CO chemical shifts are determined primarily by backbone conformation, and hence Δω(13CO) reports directly on changes in backbone dihedral angles and secondary structure, amide 15N chemical shifts are in addition highly sensitive to hydrogen bonding interactions and local environment (28, 29). Large values of ΔωN,MG(15N) for residues in the AB loop (residues 17–21), in the CD loop (residues 41–47), at the C terminus of helix D and beginning of helix E (residues 56–61), and in the GH loop (residues 117–122) indicate substantial conformational rearrangements or unfolding/folding transitions between the N and MG states (Fig. 5A). Changes in structure or packing are also evident in the helix B/helix G interface, in the middle of helix E (residues 67–70) where it contacts the AB loop in the native fold, and in helix H (residues 134, 137, 138) where it packs against the EF turn.
, and 13CO (Tables S2–S4) yield information about the structural changes that occur between the N and MG states during the process of folding and unfolding, at equilibrium under a single set of experimental conditions (temperature, pH, and buffer). Whereas 13CO chemical shifts are determined primarily by backbone conformation, and hence Δω(13CO) reports directly on changes in backbone dihedral angles and secondary structure, amide 15N chemical shifts are in addition highly sensitive to hydrogen bonding interactions and local environment (28, 29). Large values of ΔωN,MG(15N) for residues in the AB loop (residues 17–21), in the CD loop (residues 41–47), at the C terminus of helix D and beginning of helix E (residues 56–61), and in the GH loop (residues 117–122) indicate substantial conformational rearrangements or unfolding/folding transitions between the N and MG states (Fig. 5A). Changes in structure or packing are also evident in the helix B/helix G interface, in the middle of helix E (residues 67–70) where it contacts the AB loop in the native fold, and in helix H (residues 134, 137, 138) where it packs against the EF turn.
Fig. 5.
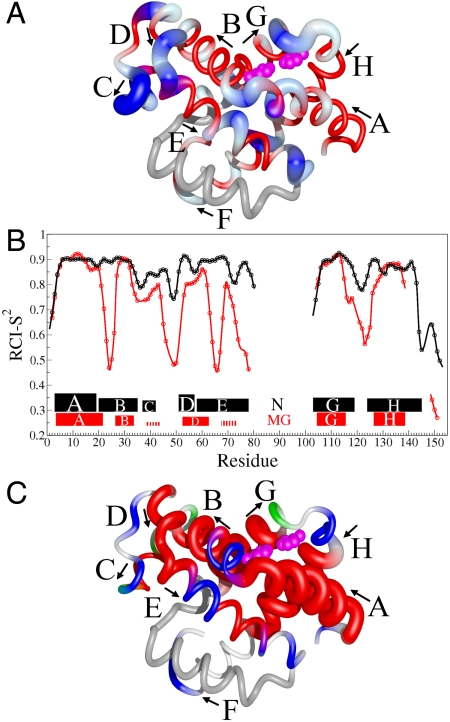
Sites of local unfolding and structural rearrangement in the transient MG. (A) Mapping of 15N ΔωN,MG chemical shift differences onto the structure of holoMb. The protein backbone is represented as a tube. Residues with smaller than average ΔωN,MG are colored red, whereas those with above-average shifts are colored blue and vary in thickness and color saturation according to the magnitude of ΔωN,MG. (B) Secondary structure and RCI-S2 parameter (32) for the transient MG state and the N state. The secondary structure of the MG state was predicted by TALOS+ (30) from 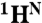 , 15N, and 13CO chemical shifts (derived from ΔωN,MG as described in SI Text). A more complete dataset (
, 15N, and 13CO chemical shifts (derived from ΔωN,MG as described in SI Text). A more complete dataset (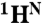 , 15N, 13CO,
, 15N, 13CO, 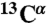 ,
, 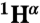 shifts) was available for the N state; however, the helical boundaries and RCI-S2 for N were not significantly altered when predicted from a limited set of
shifts) was available for the N state; however, the helical boundaries and RCI-S2 for N were not significantly altered when predicted from a limited set of 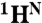 , 15N, 13CO shifts. The rectangles depict the location of helical structure in each state; the thickness of each rectangle is proportional to the population of helix. The hatched lines indicate the small population of transient helical structure in the C- and E-helix regions of MG. (C) Changes in secondary structure accompanying the N⇆MG transition, mapped to the structure of holoMb. Residues predicted to be helical by TALOS+ are red. The population of helix in the MG ensemble is indicated by the tube radius, with a larger radius indicating higher population. Flexible regions with RCI S2 < 0.7 are blue, and coil regions with S2 > 0.7 are green. A white backbone trace indicates regions for which no predictions are available. The His24 and His119 side chains are shown as pink spheres in A and C. The figure was prepared using the program MolMol (36) from the coordinates of holoMb (Protein Data Bank ID code 1MBC).
, 15N, 13CO shifts. The rectangles depict the location of helical structure in each state; the thickness of each rectangle is proportional to the population of helix. The hatched lines indicate the small population of transient helical structure in the C- and E-helix regions of MG. (C) Changes in secondary structure accompanying the N⇆MG transition, mapped to the structure of holoMb. Residues predicted to be helical by TALOS+ are red. The population of helix in the MG ensemble is indicated by the tube radius, with a larger radius indicating higher population. Flexible regions with RCI S2 < 0.7 are blue, and coil regions with S2 > 0.7 are green. A white backbone trace indicates regions for which no predictions are available. The His24 and His119 side chains are shown as pink spheres in A and C. The figure was prepared using the program MolMol (36) from the coordinates of holoMb (Protein Data Bank ID code 1MBC).
To obtain direct insights into changes in secondary structure, backbone dihedral angles for the transient MG intermediate were determined from 15N, 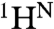 , and 13CO chemical shifts (derived from ΔωN,MG values) using TALOS+ (30). Helical structure is predicted with high confidence in several regions (Fig. 5 B and C); estimates of helix population were obtained from the average deviations of the 13CO shifts from sequence-corrected random coil values (31). Highly populated helices are indicated for residues 4–21 (80% population), 27–33 (65%), 105–115 (80%), and 127–138 (85%), corresponding respectively to the A helix, C terminus of B, and central regions of the G and H helices of native apoMb (Fig. 5B). Whereas the B helix is shortened in the transient MG, the A helix extends to form nonnative helix between residues 19–21. Nonnative helical structure (population 55%) is also predicted for residues 53–62, extending from helix D into the first turn of helix E. Finally, a small helical propensity (≤ 30%) is found for residues 39–43 (C helix in native apoMb) and 69–73, in the middle of helix E. The transient MG is similar in structure to that formed at pH 4.1 (see SI Text for details).
, and 13CO chemical shifts (derived from ΔωN,MG values) using TALOS+ (30). Helical structure is predicted with high confidence in several regions (Fig. 5 B and C); estimates of helix population were obtained from the average deviations of the 13CO shifts from sequence-corrected random coil values (31). Highly populated helices are indicated for residues 4–21 (80% population), 27–33 (65%), 105–115 (80%), and 127–138 (85%), corresponding respectively to the A helix, C terminus of B, and central regions of the G and H helices of native apoMb (Fig. 5B). Whereas the B helix is shortened in the transient MG, the A helix extends to form nonnative helix between residues 19–21. Nonnative helical structure (population 55%) is also predicted for residues 53–62, extending from helix D into the first turn of helix E. Finally, a small helical propensity (≤ 30%) is found for residues 39–43 (C helix in native apoMb) and 69–73, in the middle of helix E. The transient MG is similar in structure to that formed at pH 4.1 (see SI Text for details).
The random coil chemical shift index (RCI) order parameter (S2) (32) shows that the A, B, G, and H helical regions in the transient MG are as well ordered as in the N state (Fig. 5B), suggesting that they pack together to form a compact hydrophobic core. The other helical regions, including the weakly populated C helix, the D–E region, and the central part of helix E, have lower S2 than in N, consistent with the disorder implied by their fractional helical populations. It is likely that these regions are highly dynamic and dock only transiently against the ABGH core of the MG. The RCI order parameters in the N-terminal region of helix B, in the CD region, throughout most of the E helix, at the C terminus of helix G, and in the GH turn also indicate greatly increased flexibility in the transient MG and loss of structure relative to the N state. The S2 decreases slightly for Val21, suggesting dynamic fraying at the end of the nonnative A helix.
The observed changes in secondary structure and local regions of unfolding indicated by the RCI S2 parameters suggest a plausible model for the unfolding process. Residues surrounding His24 at the N terminus of helix B and His 119 at the C terminus of helix G unfold during the I1 to MG transition and are disordered in the MG state. These changes reflect loss of the stabilizing His24–His119 hydrogen bond and provide graphic evidence that “cracking” (33) of the structure in this region and protonation of His24 acts as a trigger (13, 25) for apoMb unfolding. Loss of structure at the N terminus of helix B disrupts critical contacts with the N-terminal part of helix E, which also unfolds and becomes dynamically disordered. Unfolding of helix E, in turn, disrupts the packing interface for the CD loop, leading to unfolding of this entire region and formation of the nonnative D–E helix. This nonnative structure forms spontaneously (20% population) in acid-unfolded apoMb at pH 2.1 (31); the increased population in MG probably reflects stabilizing tertiary contacts in the compact MG state. Apart from a small propensity for helix between residues 69–72, most likely stabilized by transient packing of this strongly hydrophobic region onto the MG core, and the nonnative structure at the helix D/helix E junction, the entire E helix is dynamically disordered.
The MG state observed in the present experiments appears to correspond to the kinetic Ib intermediate (21, 22), containing stable helical structure in the B region and transient structure in helix C and part of helix E that would not be formed in Ia. The R2 dispersion data provide additional structural insights into Ib, which complement those from hydrogen exchange pulse labeling (10, 22). Firstly, dispersion analysis provides direct and unambiguous evidence that the high degree of exchange protection of amide protons in the A, B, G, and H regions in the kinetic burst-phase intermediate is due to formation of stable helical structure. Second, residues that exhibit partial proton occupancies in hydrogen exchange experiments (10) are seen to be located in disordered or transiently helical regions in MG. Finally, information is available from dispersion for residues whose amides are poorly protected and are therefore not observed in hydrogen exchange experiments (10, 22). For example, dispersion reveals unfolding at the N terminus of helix B and C terminus of helix G and an extension of helical structure at the N terminus of helix H, where no hydrogen exchange probes are available. Conversely, hydrogen exchange experiments suggest formation of stable helical structure between residues 139 and 144, for which dispersion data are missing because of the broadness of the N-state resonances. Differences are observed at the C terminus of the E helix; this region is predicted to behave as dynamic coil in the transient MG state, yet displays mild H–D exchange protection in Ib. The amide closing rates involved in the formation of the kinetic intermediate indicate that the E helix is stabilized initially at its C terminus.
The R2 dispersion methods described here should be generally applicable to study equilibrium unfolding/folding transitions in other proteins under mildly destabilizing conditions. This approach offers several advantages, including measurement of unfolding and refolding kinetics under identical conditions in a single experiment, and characterization of any intermediates formed. R2 dispersion methods could prove particularly powerful for characterization of the earliest protein unfolding events that lead to protein aggregation, amyloid formation, and disease.
Materials and Methods
Expression and Purification.
Protein expression, purification, and NMR sample preparation are described in SI Text.
NMR Data Acquisition.
The assignment of amide and 13CO resonances is described in SI Text. R2 dispersion data were acquired simultaneously on 500- and 800-, or 600- and 800-MHz Bruker spectrometers equipped with cryogenic probes, using constant time relaxation compensated CPMG pulse sequences (3). The sample temperatures were calibrated to 35 °C with perdeuterated methanol (34). SQ 15N and 13CO dispersion experiments (4, 18) were performed on 13C, 15N-labeled samples. Separate experiments were conducted on 2H, 15N-labeled apoMb using 1H SQ (4), 15N SQ (3, 4), 1H/15N DQ, and 1H/15N ZQ coherences (14).
Data Fitting.
Dispersion data were fit using the in-house program GLOVE (5). The fitting parameters included the microscopic kinetic rates connecting the ground and transient states, the populations of all states, the differences in chemical shifts between each of the states (Δω), and the relaxation rate in the absence of conformational exchange, 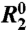 . The error in
. The error in 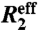 was defined as 10% of the amplitude of the R2 dispersion curve (Rex) because this adequately reflects the level of random noise in
was defined as 10% of the amplitude of the R2 dispersion curve (Rex) because this adequately reflects the level of random noise in 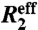 . A detailed description of the global optimization and model selection is given in SI Text.
. A detailed description of the global optimization and model selection is given in SI Text.
Supplementary Material
Acknowledgments.
We thank Jane Dyson for invaluable discussions, Gerard Kroon for assistance with NMR experiments, and Daniel Felitsky for advice about data fitting and optimization of sample conditions. This work was supported by the National Institutes of Health Grant DK34909 and the Skaggs Institute for Chemical Biology. D.W.M. was the recipient of Ruth L. Kirschstein National Research Service Award GM84594.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105682108/-/DCSupplemental.
References
- 1.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 2.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 3.Loria JP, Rance M, Palmer AG. A relaxation-compensated Carr–Purcell–Meiboom–Gill sequence for characterizing chemical exchange by NMR spectroscopy. J Am Chem Soc. 1999;121:2331–2332. [Google Scholar]
- 4.Tollinger M, Skrynnikov NR, Mulder FA, Forman-Kay JD, Kay LE. Slow dynamics in folded and unfolded states of an SH3 domain. J Am Chem Soc. 2001;123:11341–11352. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- 5.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 6.Eliezer D, Wright PE. Is apomyoglobin a molten globule? Structural characterization by NMR. J Mol Biol. 1996;263:531–538. doi: 10.1006/jmbi.1996.0596. [DOI] [PubMed] [Google Scholar]
- 7.Lecomte JT, Sukits SF, Bhattacharjya S, Falzone CJ. Conformational properties of native sperm whale apomyoglobin in solution. Protein Sci. 1999;8:1484–1491. doi: 10.1110/ps.8.7.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eliezer D, Chung J, Dyson HJ, Wright PE. Native and non-native structure and dynamics in the pH 4 intermediate of apomyoglobin. Biochemistry. 2000;39:2894–2901. doi: 10.1021/bi992545f. [DOI] [PubMed] [Google Scholar]
- 9.Eliezer D, et al. The radius of gyration of an apomyoglobin folding intermediate. Science. 1995;270:487–488. doi: 10.1126/science.270.5235.487. [DOI] [PubMed] [Google Scholar]
- 10.Nishimura C, Dyson HJ, Wright PE. Enhanced picture of protein-folding intermediates using organic solvents in H/D exchange and quench-flow experiments. Proc Natl Acad Sci USA. 2005;102:4765–4770. doi: 10.1073/pnas.0409538102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jennings PA, Wright PE. Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Science. 1993;262:892–896. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- 12.Nishimura C, Dyson HJ, Wright PE. Identification of native and non-native structure in kinetic folding intermediates of apomyoglobin. J Mol Biol. 2006;355:139–156. doi: 10.1016/j.jmb.2005.10.047. [DOI] [PubMed] [Google Scholar]
- 13.Barrick D, Hughson FM, Baldwin RL. Molecular mechanisms of acid denaturation. The role of histidine residues in the partial unfolding of apomyoglobin. J Mol Biol. 1994;237:588–601. doi: 10.1006/jmbi.1994.1257. [DOI] [PubMed] [Google Scholar]
- 14.Orekhov VY, Korzhnev DM, Kay LE. Double- and zero-quantum NMR relaxation dispersion experiments sampling millisecond time scale dynamics in proteins. J Am Chem Soc. 2004;126:1886–1891. doi: 10.1021/ja038620y. [DOI] [PubMed] [Google Scholar]
- 15.Korzhnev DM, Kloiber K, Kay LE. Multiple-quantum relaxation dispersion NMR spectroscopy probing millisecond time-scale dynamics in proteins: Theory and application. J Am Chem Soc. 2004;126:7320–7329. doi: 10.1021/ja049968b. [DOI] [PubMed] [Google Scholar]
- 16.Korzhnev DM, Neudecker P, Mittermaier A, Orekhov VY, Kay LE. Multiple-site exchange in proteins studied with a suite of six NMR relaxation dispersion experiments: An application to the folding of a Fyn SH3 domain mutant. J Am Chem Soc. 2005;127:15602–15611. doi: 10.1021/ja054550e. [DOI] [PubMed] [Google Scholar]
- 17.Hattori A, Crespi HL, Katz JJ. Effect of side-chain deuteration on protein stability. Biochemistry. 1965;4:1213–1225. doi: 10.1021/bi00883a002. [DOI] [PubMed] [Google Scholar]
- 18.Lundstrom P, Hansen DF, Kay LE. Measurement of carbonyl chemical shifts of excited protein states by relaxation dispersion NMR spectroscopy: Comparison between uniformly and selectively (13)C labeled samples. J Biomol NMR. 2008;42:35–47. doi: 10.1007/s10858-008-9260-4. [DOI] [PubMed] [Google Scholar]
- 19.Jamin M, Baldwin RL. Two forms of the pH 4 folding intermediate of apomyoglobin. J Mol Biol. 1998;276:491–504. doi: 10.1006/jmbi.1997.1543. [DOI] [PubMed] [Google Scholar]
- 20.Weisbuch S, et al. Cooperative sub-millisecond folding kinetics of apomyoglobin pH 4 intermediate. Biochemistry. 2005;44:7013–7023. doi: 10.1021/bi047372v. [DOI] [PubMed] [Google Scholar]
- 21.Uzawa T, et al. Collapse and search dynamics of apomyoglobin folding revealed by submillisecond observations of α-helical content and compactness. Proc Natl Acad Sci USA. 2004;101:1171–1176. doi: 10.1073/pnas.0305376101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uzawa T, et al. Hierarchical folding mechanism of apomyoglobin revealed by ultra-fast H/D exchange coupled with 2D NMR. Proc Natl Acad Sci USA. 2008;105:13859–13864. doi: 10.1073/pnas.0804033105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamin M, Yeh SR, Rousseau DL, Baldwin RL. Submillisecond unfolding kinetics of apomyoglobin and its pH 4 intermediate. J Mol Biol. 1999;292:731–740. doi: 10.1006/jmbi.1999.3074. [DOI] [PubMed] [Google Scholar]
- 24.Hambly DM, Gross ML. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J Am Soc Mass Spectrom. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Jamin M, Geierstanger B, Baldwin RL. The pKa of His-24 in the folding transition state of apomyoglobin. Proc Natl Acad Sci USA. 2001;98:6127–6131. doi: 10.1073/pnas.111157998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng ZY, Ha JH, Loh SN. Identifying the site of initial tertiary structure disruption during apomyoglobin unfolding. Biochemistry. 1999;38:14433–14439. doi: 10.1021/bi991933e. [DOI] [PubMed] [Google Scholar]
- 27.Nishiguchi S, Goto Y, Takahashi S. Solvation and desolvation dynamics in apomyoglobin folding monitored by time-resolved infrared spectroscopy. J Mol Biol. 2007;373:491–502. doi: 10.1016/j.jmb.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Wishart DS, Case DA. Use of chemical shifts in macromolecular structure determination. Methods Enzymol. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]
-
29.Xu XP, Case DA. Probing multiple effects on 15N, 13C alpha, 13C beta, and
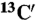 chemical shifts in peptides using density functional theory. Biopolymers. 2002;65:408–423. doi: 10.1002/bip.10276. [DOI] [PubMed] [Google Scholar]
chemical shifts in peptides using density functional theory. Biopolymers. 2002;65:408–423. doi: 10.1002/bip.10276. [DOI] [PubMed] [Google Scholar] - 30.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao J, Chung J, Eliezer D, Wright PE, Dyson HJ. NMR structural and dynamic characterization of the acid-unfolded state of apomyoglobin provides insights into the early events in protein folding. Biochemistry. 2001;40:3561–3571. doi: 10.1021/bi002776i. [DOI] [PubMed] [Google Scholar]
- 32.Berjanskii MV, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J Am Chem Soc. 2005;127:14970–14971. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- 33.Miyashita O, Onuchic JN, Wolynes PG. Nonlinear elasticity, proteinquakes, and the energy landscapes of functional transitions in proteins. Proc Natl Acad Sci USA. 2003;100:12570–12575. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Findeisen M, Brand T, Berger S. A 1H-NMR thermometer suitable for cryoprobes. Magn Reson Chem. 2007;45:175–178. doi: 10.1002/mrc.1941. [DOI] [PubMed] [Google Scholar]
- 35.Hagen SJ, Hofrichter J, Szabo A, Eaton WA. Diffusion-limited contact formation in unfolded cytochrome c: Estimating the maximum rate of protein folding. Proc Natl Acad Sci USA. 1996;93:11615–11617. doi: 10.1073/pnas.93.21.11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.