Abstract
Interferons and interleukin-10 are involved in key aspects of the host defence mechanisms. Human chromosome 21 harbors the interferon/interleukin-10 receptor gene cluster linked to the GART gene. This cluster includes both components of the interferon α/β-receptor (IFNAR1 and IFNAR2) and the second components of the interferon γ-receptor (IFNGR2) and of the IL-10 receptor (IL10R2). We report here the complete gene content of this GART–cytokine receptor gene cluster and the use of comparative genomic analysis to identify chicken IFNAR1, IFNAR2, and IL10R2. We show that the large-scale structure of this locus is conserved in human and chicken but not in the pufferfish Fugu rubripes. This establishes that the receptor components of these host defense mechanisms were fixed in an ancestor of the amniotes. The extraordinary diversification of the interferon ligand family during the evolution of birds and mammals has therefore occured in the context of a fixed receptor structure.
[The sequence data described in this paper have been submitted to GenBank under accession nos. AF039904, AF039905, AF039906, AF039907, AF045606, AF082664, AF082665, AF082666, AF082667, and AF083221.]
The cytokine receptor (CR) family is structurally defined by the presence in the extracellular domain of its members of a typical 200-amino-acid domain (D200). This domain is built of two subdomains of 100 amino acids (SD100A and SD100B) that are both structured like the constant domain of the immunoglobulins (Bazan 1990; Thoreau et al. 1991). Conserved cysteines allow the definition of class I and class II CR and clearly distinguish the CR family from the fibronectin type III family and the immunoglobulin superfamily. The CR family is also genetically defined by the intron/exon structures encoding the D200: All members of the family have introns conserved both in position and in phase (Lutfalla et al. 1992; Nakagawa et al. 1994). The genes encoding the members of the CR family are scattered all over the human genome. When clusters are observed, they encode proteins that are structurally the most similar, and where the orientation of transcription of the genes in the clusters is known, they are transcribed in the same orientation (Gorman et al. 1992; Kremer et al. 1993; Lutfalla et al. 1995). This suggests that the family has evolved through successive rounds of duplications, that the oldest duplications have been scattered all over the genome, but that the most recent ones are still conserved in tandem. This evolution is probably a vertebrate story because no CR has ever been reported in any invertebrate species. The CRs are part of the most rapidly evolving category of proteins, that is, the “host defense ligands and receptors” (Murphy 1993). Class I members have been described in fishes (Sandra et al. 1995), birds (Burnside et al. 1991), and mammals, but class II only in mammals. As mainly shown using knockout mice, class II cytokine receptors (CRIIs) are active in various host defence mechanisms: antiviral (α/β or type I interferons) (Müller et al. 1994), antiparasitic (γ or type II interferon) (Huang et al. 1993), tolerance to bowel mucosal antigens [interleukin-10 (IL-10)] (Spencer et al. 1998), and blood coagulation [tissue factor (TF)] (Morrissey et al. 1987).
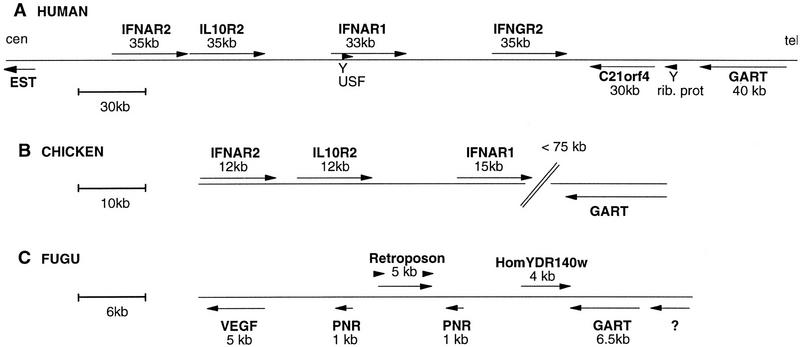
The two components of the receptors for α/β IFN (IFNAR1 and IFNAR2), for γ interferon (IFNGR1 and IFNGR2), and for IL-10 (IL10R1 and IL10R2) together with TF constitute the seven known CRII family members. With the exception of TF, which has virtually no intracellular domain, the genes coding the CRII have a similar intron/exon structure encoding the intracellular domain: They all have a single similarly placed phase 0 intron interrupting the coding region for the intracellular domain (Lutfalla et al. 1992, 1993, 1995; Raval et al. 1995; Rhee et al. 1996). They therefore have a close evolutionary relationship that is also reflected by the fact that four of them are in tandem on human chromosome 21 (Fig 1A): IFNAR2, IL10R2, IFNAR1, and IFNGR2. These four genes, together with the GART gene (Schild et al. 1990) that encodes three steps of purine synthesis, form a synteny group that is conserved in human and mouse (Cheng et al. 1993). The close proximity of some genes in this GART–CRII gene cluster (e.g., 500 bp between IFNAR2 and IL10R2) suggests that the large regions between the previously characterized genes could harbor other genes (Fig. 1A) (Lutfalla et al. 1995).
Figure 1.
Structure of the genomic loci downstream of the GART gene in human, chicken, and Fugu: Genes are indicated with their names, size, and orientation of transcription (arrows). Scale is indicated in each panel. (A) Human locus on chromosome 21. Arrowheads labeled Y represent processed pseudogenes. (rib. prot) Ribosomal protein S5. The orientation relative to the centromere (cen) and telomere (tel) is indicated. (EST) The presence of a spliced EST. Regions sequenced for this study: 5′ of IFNAR2 (accession no. AF039905), IL-10R2 to IFNAR1 (accession no. AF039904 ), IFNAR1 to IFNGR2 (accession no. AF039907), IFNGR2 to GART (accession no. AF039906). The accession number for C21orf4 cDNA is AF045606. (B) Chicken locus. The distance between IFNAR1 and GART has been measured by PFGE analysis (Fig. 5). Accession numbers are as follows: AF082664 (cDNA for cIFNAR1), AF082665 (cDNA for cIFNAR2), AF082666 (cDNA for cIL10R2), and AF082667 (CRII gene cluster). (C) Fugu locus (accession no. for the cosmid is AF083221): The question mark (?) represents a potential gene predicted with high confidence by both GRAIL (excellent exons) and GenScan (exon probability > 0.99).
Among the three biological systems in which receptors from the GART–CRII gene cluster are involved, the α/β IFN (type I IFN) is of special interest. In contrast to IL-10 and γ IFN (type II IFN) that are encoded by single genes, all species have several type I IFNs. Phylogenetic analysis of these type I IFNs based on nonsynonymous substitution has shown that β IFN diverged from α ∼250 Mya, that is, after the bird/mammal divergence (Hughes 1995). This is confirmed by the fact that the multiple bird type I IFNs can be neither classified as α nor as β (Sekellick et al. 1994; Sick et al. 1996). This major divergence between α and β IFNs early in mammalian evolution is reflected by the fact that they do not interact the same way with their shared receptor components (Lewerenz et al. 1998) and can induce distinct biological responses (Erlandsson et al. 1998). These observations therefore raise the question of the structure of the type I IFN receptor that the mammals inherited from their ancestors and how it managed to cope with such a diversification of its ligands.
Outside the mammalian family, the IFN system has been described and molecularly characterized only in birds with most of the efforts concentrated on chicken where the interferon system was first described by Isaacs and Lindenman (1957). The sequences of many different bird IFNs are now available (Sekellick et al. 1994; Digby and Lowenthal 1995), but the structure of their receptors still remains unknown. Chicken was therefore the first nonmammalian species of choice in which to look for the structure of the GART–CRII homologous locus. The pufferfish Fugu rubripes owing to its compact genome is the best suited fish species in which to study the structure of a gene cluster. We decided to use comparative genomic analysis to study how the type I IFN receptor genes developed during the evolution of the GART–CRII gene cluster in vertebrates. For this purpose, we first sequenced the gaps between the already known genes in the human GART–CRII gene cluster to have a complete description of its gene content. We then compared its large-scale structure with that of the chicken and of the pufferfish Fugu rubripes.
RESULTS
Complete Gene Repertoire of the Human GART–CRII Gene Cluster
One cosmid clone was obtained for each of the large intergenic regions of the GART–CRII gene cluster, plus two cosmids 5′ to IFNAR2. Cosmids Q18C2 and Q95D4 mapping 5′ to IFNAR2 were obtained from Dr. Soeda (Riken, Japan). Cosmid D66B10 spanning from IL10R2 to IFNAR1 was obtained from Dr. Soeda. Cosmid Q50G2 spanning from IFNAR1 to IFNGR2 was obtained by screening the chromosome 21-specific cosmid library LL21NCO2-Q with an IFNAR1 3′ UTR probe. Cosmid D16B8 (IFNGR2 to GART) was obtained from Dr. Soeda. Each cosmid was entirely sequenced. These new sequences, together with the sequences that we and others had determined for the previously described genes, cover a region of >350 kb spanning the entire GART–CRII gene cluster. Sequence analysis failed to identify any new gene within the CRII gene cluster.
Between IFNGR2 and GART, we identified one gene, C21orf4 (Fig. 1A). This seven-exon gene that matches ESTs in humans and mice encodes a small (160-amino-acid) protein with four potential transmembrane domains. As shown in Figure 2, Caenorhabditis elegans, Schizosaccharomyces pombe, and Saccharomyces cervisiae have genes encoding proteins with significant similarities to C21orf4. Transmembrane topology of each of these four proteins was studied independently using “Tmpred.” As shown in Figure 2, the predicted topology for these proteins is one of four transmembrane domains with both the amino and carboxyl termini cytoplasmic. The better conserved regions correspond to the transmembrane domains. Whereas the yeast genes are devoid of introns, the human ORF is interrupted by five introns, and the C. elegans ORF is interrupted by two, one being at the same position in both species: a phase I intron interrupting the G codon proximal to TM3 in the TM2–TM3 loop. This confirms that these genes are homologous.
Figure 2.
Sequence alignment of the human C21orf4 with its related proteins in C. elegans and in yeast. Predicted transmembrane domains are indicated TM1–TM4. YOL129w is from S. cerevisiae, SPC8D2_2 is from S. pombe, and G_YK3109 is from C. elegans. Identical (*), similar (:), and related (.) residues are indicated.
At the IFNAR2 extremity of the cluster lies a potential gene with three spliced ESTs (i.e., corresponding to spliced mRNAs and not to contaminating genomic DNA) from cDNA libraries from different tissues (GenBank accession nos. AA400173, AA868122, and AA934973). There is no evidence for a coding phase. This potential gene is therefore the centromeric border of the CRII gene cluster.We now have a complete description of the human CRII gene cluster. The recent release in GenBank of genomic sequences extending almost 600 kb centromeric and >100 kb telomeric to the sequences we have determined (GenBank accession nos. AP000037 to AP000042, AP000046, and AP000047) has allowed us to confirm that no other CRII genes lie in this 1-Mb region of human chromosome 21.
Large-Scale Structure of the Chicken GART–CRII Gene Cluster
Probe hIL10R2 was hybridized on a zooblot containing murine, chicken, and fish DNA. As shown in Figure 3, it gives clearly specific signals on murine and chicken DNA but not on fish DNA. A chicken genomic fragment hybridizing to that probe was cloned and sequenced from a chicken λ genomic library and was found to harbor potential exons encoding a homolog of hIL10R2. Four other λ genomic clones that together cover 46 kb were cloned by chromosomal walking and were sequenced. The analysis of the sequence reveals the presence of potential exons encoding homologs of the human proteins IFNAR1, IFNAR2, and IL10R2. Oligonucleotides were designed for these exons; internal RT–PCR, 3′ and 5′ RACE were used to clone and sequence the corresponding cDNAs from neonate chicken liver mRNAs. Three expressed genes were identified using these cDNAs; their respective chromosomal positions (Fig. 1B) and the similarity of their encoded proteins to the corresponding human proteins (Fig 4) allow us to ascribe them as chicken IFNAR2 (cIFNAR2), cIL10R2, and cIFNAR1 with amino acid identities of 28%, 42%, and 36%, respectively.
Figure 3.
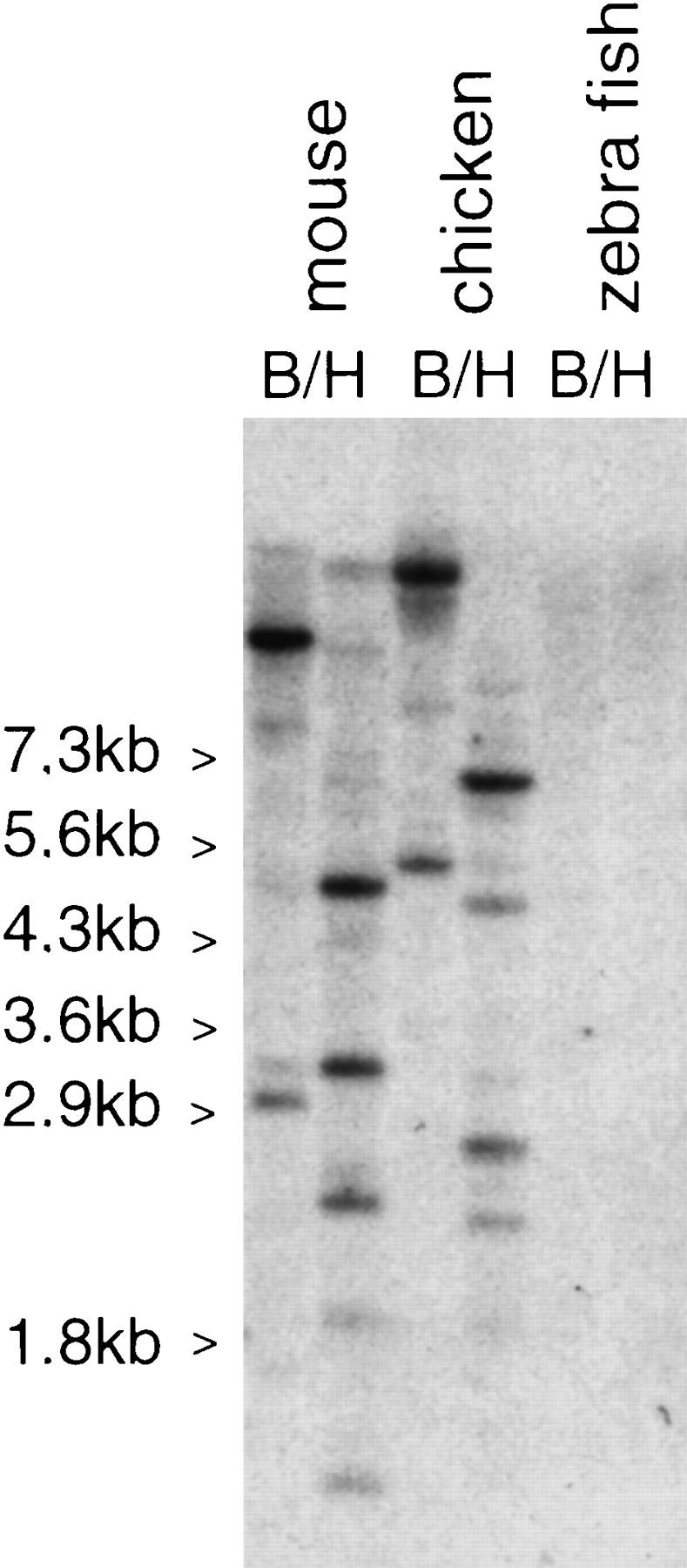
Zoo blot hybridized at low stringency with a human IL10R2 probe. Murine, chicken, and zebrafish DNAs were digested either with BamHI or with HindIII, blotted to a nylon membrane, and hybridzed with a human IL10R2 probe. Sizes are indicated.
Figure 4.
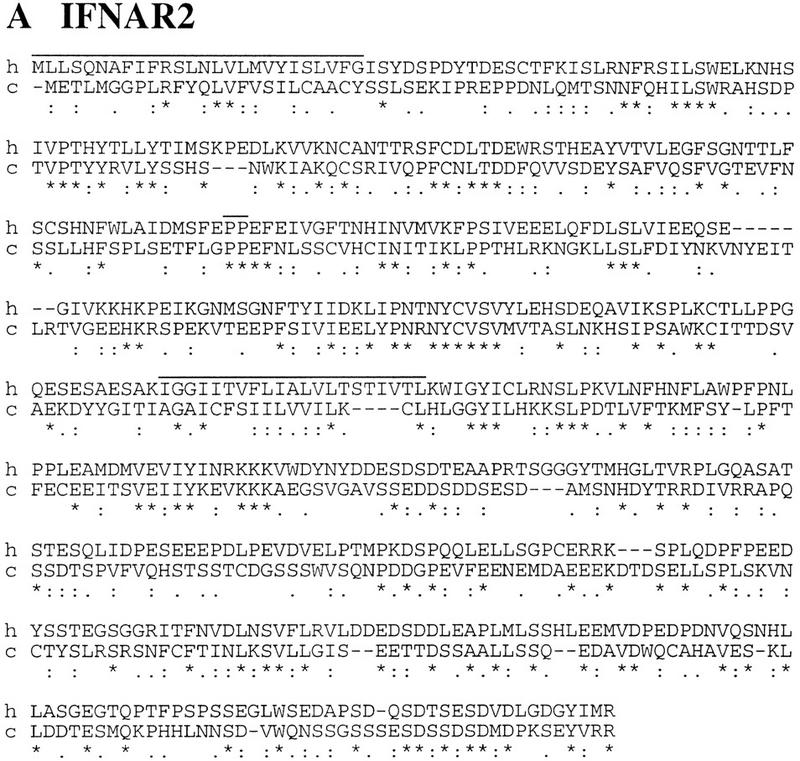


Sequence alignment of human IFNAR2 (A), IL10R2 (B), and IFNAR1 (C) to their chicken counterparts. Long overlines indicate leader peptides and transmembrane domains; short overlines represent links between SD100 domains. In the IFNAR1 alignment, (###) indicating the link between the two D200s highlights the duplication of the external domain. Identical (*), similar (:), and related (.) residues are indicated. Respective percents of identity for IFNAR2, IL10R2, and IFNAR1 are 28%, 42%, and 36%.
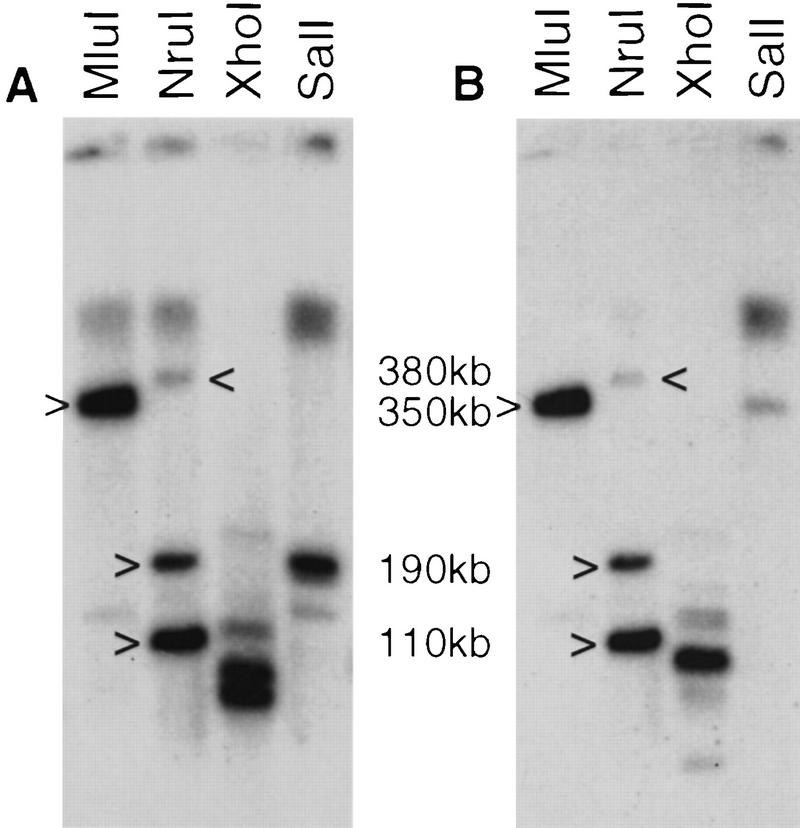
Figure 5 presents the physical linkage between this CRII gene cluster and the chicken GART gene. The same MluI and NruI chicken genomic fragments hybridize to both a cIFNAR1 and a chicken GART probe. The common MluI band is 350 kb, and the smallest common NruI fragment is 110 kb. As the NruI site in the CpG island upstream of the 15-kb IFNAR1 gene is unmethylated and therefore cut to completion and as the chicken GART gene spans >20 kb (not shown), this leaves a distance of <75 kb between IFNAR1 and GART. Figure 1B presents the schematic structure of the GART–CRII gene cluster in chicken. The structure of the locus is the same as in humans; sizes and distances are roughly three times smaller, which is in agreement with the overall difference in genome size between mammals and birds (Tiersch and Wachtel 1991). The intron–exon structure of these first described nonmammalian CR genes is the same as that of the mammals. Each SD100 is bordered by phase 1 introns; SD100A and SD100B, respectively, have phase 2 and phase 1 introns. In particular, similar to the mammalian IFNAR1, the cIFNAR1 gene codes for a double external domain.
Figure 5.
Physical linkage between the CRII gene cluster and the GART gene in chicken. DNAs from chicken embryonic fibroblasts was digested with MluI, NruI, XhoI, and SalI, electrophoresed, and blotted to a membrane. The same membrane was hybridized sequentially with a chicken IFNAR1 3′ probe (A) and, after stripping, with a chicken GART probe (B). Common bands are highlighted. Multiple NruI hybridizing fragments using probes devoid of NruI sites are attributable to partial methylation of the NruI sites.
The Fugu GART Locus
Despite evidence for an antiviral host defence system, no molecular confirmation has been obtained, and hence, the presence of interferon in fish is still controversial (Wilson et al. 1983; Tamai et al. 1993; Uzé et al. 1995). The absence of specific hybridization of mammalian CRII probes on fish DNA prompted us to use the genomic approach to investigate the possible presence of CRII genes. The well-conserved GART gene was used as an entry point to the locus. A Fugu GART probe was obtained by screening a size-selected Fugu plasmid library with human and chicken GART probes. The Fugu GART probe was used to screen a Fugu cosmid library at high stringency. A set of overlapping cosmid clones was obtained, and the cosmid clone extending the farthest 3′ of the GART gene was entirely sequenced (cosmid F177L18). Identifying a unique cosmid contig indicates that the GART gene is single copy in Fugu. This was confirmed by Southern blot analysis, as exemplified in Figure 6. The Fugu GART probe is a HindIII fragment with an internal BamHI site. It hybridizes to a single HindIII fragment and to two BamHI fragments of the same size as those predicted by the sequence of the cosmid, further establishing the colinearity between the cosmid and the Fugu genome. The results of the analysis of the sequence of the cosmid are shown in Figure 1C: No CR gene is present on that cosmid. The GART gene has the same structure as in mouse and human, with 20 coding exons and similarly placed introns, but it spans only 6.5 kb compared with 28 kb in mouse (Kan and Moran 1995) and 38 kb in human (GenBank accession no. AP000046). The encoded protein is 67% identical to that of both mouse and human. 5′ of GART lies a gene predicted by both GRAIL and GenScan programs. The predicted polypeptide, however, has no homologies to any known gene; it does not resemble the SON3 gene that is present 5′ to GART in human and mouse (Cheng et al. 1993). 3′ to GART is the Fugu homolog of the yeast YDR140w gene whose function is unknown (38% amino acid identities). The stop codons of these two genes are separated by only 122 bp. The cosmid also harbors duplicated copies of a Fugu homolog of the intronless human putative neurotransmitter receptor gene (PNR) (Zeng et al. 1998) that border the insertion of a retroposon. The sequence similarity between the two PNR copies (95% amino acid identities) extends 5′ of the coding regions but not 3′ suggesting that both copies are functional. This PNR duplication is probably the result of the recent insertion of the retroposon. At the extremity of the cosmid lie potential exons of a Fugu vascular endothelial growth factor (VEGF) gene. Sequence analysis indicates that it belongs to subgroup VEGF-C/VEGF-D that has up to now only been described in mammals (Yamada et al. 1997). Because exons corresponding to regions of difference between VEGF-C and VEGF-D have not been identified on the cosmid, more precise assignment to either subtype C or D is not possible.
Figure 6.
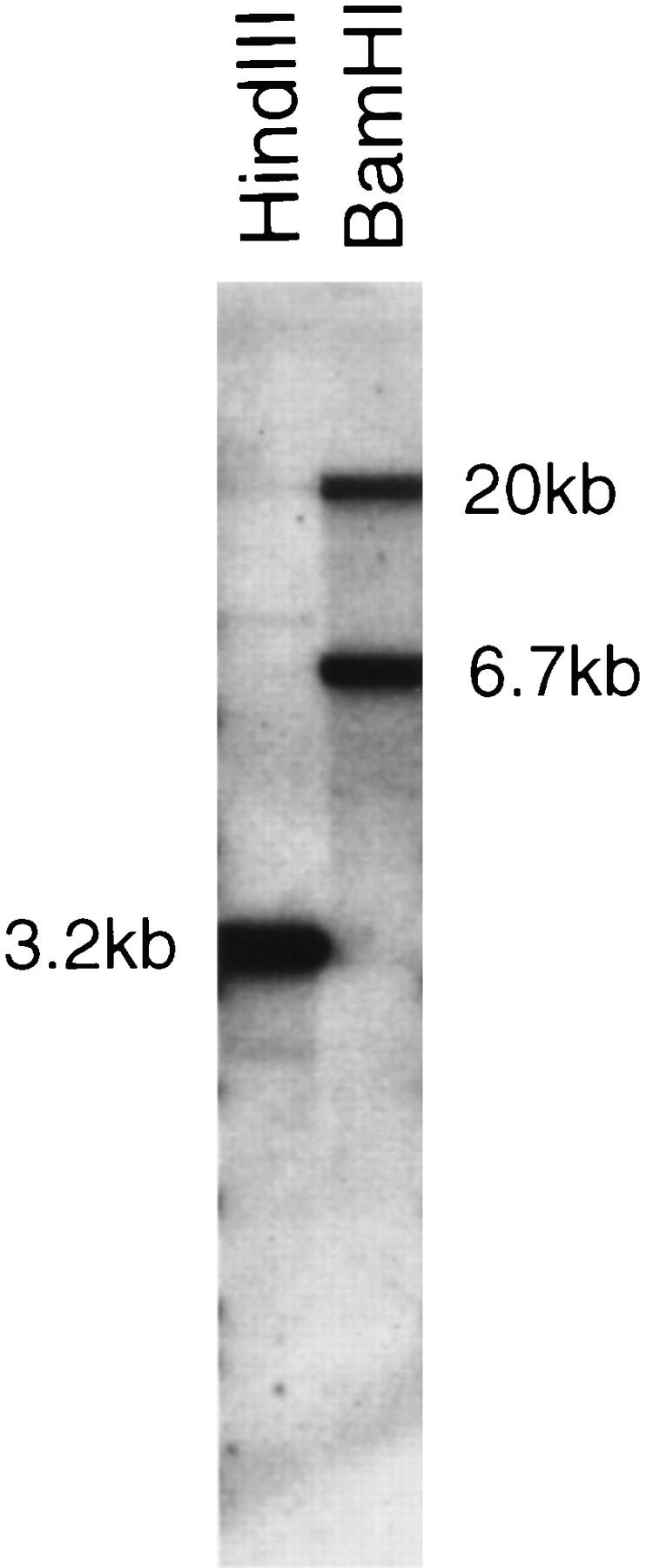
A unique GART locus in Fugu. Fugu DNA was digested with HindIII and BamHI, electrophoresed, and transfered to a nylon membrane. The membrane was hybridized with the Fugu GART probe.
DISCUSSION
Comparative Analysis of the Human Chromosome 21 GART–CRII Gene Cluster with its Chicken and Fugu Homologs
Somatic cell genetics had shown that human chromosome 21 harbors genes involved in the IFN type I and type II responsiveness (Slate et al. 1981). This was confirmed by the cloning of the human IFNAR1 gene coding one chain of the IFN-α/β-receptor and its mapping to human chromosome 21q22.1 in the vicinity of the GART gene (Lutfalla et al. 1992). However, it rapidly appeared that the responsiveness toward human IFNs of heterospecific cells expressing hIFNAR1 did not mimic the interspecific hybrids harboring this region of human chromosome 21 (Uzé et al. 1990, 1992). This observation was the starting point for the positional cloning of the two neighbors of the hIFNAR1 gene: the hIFNGR2 gene that is the second component of the functional IFN γ-receptor (Soh et al. 1994) and the initially named CRFB4 orphan receptor gene that turned out to be the IL10R2 gene encoding the second component of the functional IL-10 receptor (Lutfalla et al. 1993; Kotenko et al. 1997; Spencer et al. 1998). The cloning of a new cDNA encoding a transmembrane protein capable of binding type I IFN then allowed the cloning of the IFNAR2 gene next to the IL10R2 gene (Novick et al. 1994; Domanski et al. 1995; Lutfalla et al. 1995). This recurrent discovery of CRII genes in this region of human chromosome 21 led us to complete the sequencing of the whole locus. After analysis of the sequence of the gaps between the already identified genes in the GART–CRII gene cluster, we were unable to identify a new CR gene. We have considered as the centromeric border of the cluster an EST-based gene mapping 50 kb 3′ to IFNAR1. At the telomeric end of the cluster, between IFNGR2 and GART, we have identified a new gene, C21orf4 of unknown function. From its small size and its four predicted transmembrane domains, the encoded protein is structurally related to the peripheral myelin protein PMP22 that is encoded by a gene whose alterations are associated with various hereditary peripheral neuropathies in both mice and humans (Suter and Snipes 1995). The presence in C. elegans of a homologous gene offers an excellent animal system to test for its involvement in the nervous system.
To understand how the GART–CRII gene cluster has evolved, we have investigated its structure in two nonmammalian species: chicken and Fugu. Owing to the respective diversity of the type I IFN systems in birds and mammals, we have concentrated our efforts on the identification of the chicken type I IFN receptor components. We show that the gene content is similar to that of its human counterpart. The order of the genes is the same, with the IL10R2 gene between IFNAR2 and IFNAR1 (providing the first evidence for an IL-10 system in birds) and the GART gene lying downstream of the IFNAR1 gene. The relative spacing of the genes is conserved, and all the genes harbor the same introns at the same position as their human counterparts (see GenBank accession no. AF0822667). Furthermore, for these three receptors, the primary protein structure is sufficiently conserved to state that both the locus and the structure of the type I IFN receptor were fixed in an ancestor of the birds and mammals. Molecular and paleontological data agree to propose that synapsids (mammals’ ancestors) and sauropsids (birds’ and reptiles’ ancestors) diverged early in amniote evolution a little more than 300 million years ago in the Carboniferous period (Benton 1990; Kumar and Hedges 1998). Thus, the similarity of the genomic organization and of the encoded proteins that we describe in birds and mammals suggests that the functions corresponding to these genes were fixed before the divergence of the two species, that is, in an ancestor of the amniotes. This is expected for the GART gene (involved in de novo purine biosynthesis) but was unsuspected either for IL-10, which has never been described in birds, or for the type I IFN system whose ligands have diversified so much during the evolution of birds and mammals.
Sequencing the gap between the chicken IFNAR1 gene and the chicken GART gene would probably be a good strategy to allow access to the chicken IFNGR2 gene.
Contrary to the frequent conservation of synteny between Fugu and mammals, the well-conserved Fugu GART gene is in a different genetic environment. The human homolog of YDR140w is not yet mapped and the human PNR gene maps to 6q23 (Zeng et al. 1998), whereas the human VEGF-C and VEGF-D map respectively to 4q34 (Paavonen et al. 1996) and Xq22 (Yamada et al. 1997), and therefore, none of theses genes are physically linked in the human genome. An obvious explanation would be that this region only harbors genes that have been translocated during the evolution of the fish lineage or during the evolution of the tetrapods before the amniote situation was fixed. Alternatively, the fishes and the tetrapods would have kept a different tetralog for the GART functions after the tetraploidization of the vertebrate genome early in their evolution, and the evolution of the VEFG-C/VEFG-D subgroup plus the insertion of the retroposon would have erased the ancestral paralogy. We still have no idea whether fishes have CRIIs.
Evolution of the IFN System
The rapidly evolving (Murphy 1993) CR family offers interesting examples of redundancy (several ligands on the same receptor), two of which are paradigmal: One is the growth hormone/prolactin (GH/PRL) family; the other is the type I IFN family.
As reviewed by Goffin et al. (1996), GH and PRL diverged from a common ancestor early in the vertebrate lineage, with GH being linked to growth and morphogenesis and PRL to osmoregulation. Their receptors also diverged from a common ancestor, and the general rule is that GH and PRL do not crossreact with each other’s receptor. Early in mammalian evolution, the PRL signal transduction pathway was selected to play a key role in the mammalian specific reproduction. In most mammalian species, the PRL gene has been duplicated, one copy keeping a pituitary-specific expression but new copies [called the placental lactogens (PL)] acquiring new tissue-specific expression (trophoblast, placenta, etc.). In primates, owing to the unique (among GHs) property of GH to bind PRLR, it is the GH gene that has been duplicated to generate hPL that interact with the PRLR but lost most of their affinity to the GHR. Available data suggest that despite differences in modes of interaction for a given receptor, the same protein complexes are formed whichever the ligand and the same signal is transduced (Goffin et al. 1996).
The main other example of redundancy in the CR family is the type I IFN system. Of the four groups of type I IFN that have been described in mammals (α, β, Ω, and τ) (Roberts et al. 1997), only two are present in all mammalian species: α and β; this discussion will therefore only deal with them. The number of β IFN genes in mammals varies from one (human and mice) to at least five in cattle (Wilson et al. 1983). In constrast, there are multiple genes for α IFN genes in all mammalian species so far examined. The current theory for this diversification is that the different α IFN genes duplicated independently from their progenitor gene after the major eutherian orders diverged but that the β and α IFN genes arose from a duplication event occuring early in the evolution of mammals (250 Mya) after the bird/mammal divergence (Hughes 1995). Consistent with this fact, the numerous bird IFNs can neither be classified as β nor as α (Sekellick et al. 1994; Sick et al. 1996). In contrast to what has been described with the GH/PRL family, the α and β IFNs do not interact the same way with their shared receptor components; sites exist that are subtype specific (Lewerenz et al. 1998). The question was therefore opened of whether the structural organization of the receptor that allows these two kinds of interactions had been selected after the α/β IFN divergence or whether it pre-existed. The presence of two receptor components and their identical domain organization establish that the main characteristics of the type I IFN receptor were fixed before the duplication/divergence of its ligands. IFNAR1 has two D200 in its extracellular domain and a short (100 amino acids) intracellular domain, whereas IFNAR2 has a single D200 and a long intracellular domain. The fact that IFNAR1 (with its duplicated ligand-binding domain) and IFNAR2 can offer different binding sites to their ligands (Lewerenz et al. 1998) has probably been instrumental in this observed diversification of the type I IFNs. The appearance of new viruses during the expansion of these animal families has likely been the environmental pressure that has selected this unique diversification of a ligand family in the context of a fixed receptor structure.
METHODS
Human Cosmids
The screening of the chromosome 21-specific cosmid library LL21NCO2-Q was as described previously (Lutfalla et al. 1995). The IFNAR1 3′ UTR probe corresponds to positions 31964–32906 of GenBank accession number X60459.
Cloning of the Chicken CRII gene cluster
The previously described CRF2-4 probe (Lutfalla et al. 1993) was used as a IL10R2 probe to screen the λ FIXII chicken genomic library from Stratagene (La Jolla, CA) using 30% formamide (Uzé et al. 1992). One positive clone (ph78) was isolated that harbors the cIL10R2 gene. A terminal probe from ph78 (postions 11864–12366 of GenBank accession no. AF082667), 5′ to IL10R2, was used to clone phage ph51 (harboring cIFNAR2) and one 3′ to IL10R2 (positions 25256–25965 of AF082667) was used to clone phage ph2. A terminal probe from ph2 (positions 32304–33154 of GenBank accession no. AF082667) was used to clone phage ph3 that harbors cIFNAR1. The cIFNAR1 3′ probe corresponds to positions 1312–2425 of GenBank accession number AF082664.
Cloning of the Fugu GART Locus
DNA was prepared from a slice of Fugu purchased from a Japanese market. The size of the HindIII fragment hybridizing (30% formamide) with both chicken and human GART cDNA probes was determined using Southern blots. Corresponding DNA fragments were size-selected and cloned in Bluescript. Clones hybridizing with the GART probes were isolated and sequenced. Their sequence corresponds to position 31751–34938 of sequence AF083221 and shows evidence for the presence of a GART gene. It was used as a Fugu GART probe to screen the “UK HGMP Resource Centre” Fugu genomic cosmid library. The obtained contig of cosmids was oriented with respect to the GART gene using 3′ and 5′ GART probes. Identity of the sequence of the Fugu GART probe we cloned to the cosmid sequence confirmed that the slice of fish we got from the market was from Fugu. The chicken GART probe corresponds to the total sequence of GenBank accession number X54200. The human GART probe spans positions 481–3036 of GenBank accession number X54199.
Sequence
Clones were sonicated and shotgun-cloned in blunt-ended Bluescript vector. DNA from the subclones was prepared manually in 96-well plates. Sequencing was performed using the Dye-Terminator dichlororhodamine technology (Perkin-Elmer) and analyzed on an ABI377 sequencer. Individual sequences were assembled using the “acembly” package (mieg@crbm.cnrs-mop.fr). Four levels of sequence analysis were performed: first, sequence comparisons (BLAST and FAST) either as DNA or as six-phase translation versus DNA or protein data libraries; second, a search for spliced ESTs; third, exon/gene predicitions using GENSCAN and GRAIL; and fourth, by looking in six-phase translations for conserved protein motifs. The integration of these analyses was done manually. “Prediction of Transmembrane Regions and Orientation” (TMpred) has been run on the ISREC web server at: http://www.isrec.isb-sib.ch/software/TMPRED_form.html. Sequence alignments were done using Clustalw.
Pulsed-Field Gel Analysis, RNA Isolation, RT–PCR, 5′ and 3′ RACE
Pulsed-field gel electrophoresis (PFGE) was performed (250 V, 20 hr, 15-sec pulse) using the TAFE system (Beckman) as described previously (Lutfalla et al. 1993). RNA isolations using the RNA-B solution (Bioprobe systems) were as described previously (Uzé et al. 1992). RT–PCR on oligo(dT) reverse transcriptions was as described previously (Uzé et al. 1992) with internal oligonucleotides: for IFNAR2, PC6 (CTAACAACTTTCAGCACATTTT) and PC7R (GTGGTTCTCAGTTATTCATCTT) with annealing at 55°C; for IL10R2, PC2 (CCTGCTGCTGTGCGTGTCTG) and PC2R (AGTCTGGTTGGCTCTTTCTTTG) with annealing at 60°C; for IFNAR1, PC4 (TGTGGAACTACACTGGAGATGG) and PC4R (ACTGTCGCTATTGTCTGTTTTG) with annealing at 55°C. RACE (5′) was as described previously (Uzé et al. 1992) with the following oligonucleotides: for IFNAR2, priming with PC11 (GAGAGCGGTGAAAAATGGAGTA) and PCR between BamC10 and PC10 (TGTTTCGCAATCTTCCAGTTAC) with annealing at 60°C; for IFNAR1, priming with PC12 (TACTTCTGACCTTTTCTACATTGG) and PCR between BamC10 and PC13 (CTTCCCTTCTTTCAGCCCTTATGC) or PC15 (GTGACTGACATTCTGGCAACC) with annealing at 60°C. RACE (3′) for IFNAR1 was as follow: Reverse transcriptions were initiated using the oligonucleotide SpU1T18 (ACCTCCCAGTTCAGCATTACT18), and PCR was between an internal oligonucleotide PC9 (CAAAGTGGCAGAAGGTATCAGG) and SpU1T3 (ACCTCCCAGTTCAGCATTACT3) with annealing at 55°C. All PCR was performed using Taq polymerase (Boehringer Mannheim) with the buffer supplied by the furnisher (2 mm MgCl2 final concentration) for 30 cycles. Products were purified using spin columns, and were both directly sequenced from extremities and subcloned for complete sequencing and further analysis.
Acknowledgments
We thank M. Lewerenz, Dr. C. Bonnerot, and Dr. E. Mogensen for their help and advice throughout this work. We are indebted to Dr. E. Soeda for the gift of human cosmids and to Dr. G. Elgar, Dr. S. Takeshita, and Dr. M.L. Yaspo for their help in starting the Fugu work. We thank Professor P. Staeheli and Dr. U. Schultz for their help in starting the chicken work, Professor G. Gillet for the gift of chicken embryonic fibroblasts and Dr. Y. Kohara for access to C. elegans ESTs. This work was supported in part by grants from the Association pour la Recherche sur le Cancer, the Ligue Nationale contre le Cancer, European Commission (BIOMEDII), and the National Institutes of Health (grants CA78213 and HD17449).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lutfalla@infobiogen.fr; FAX +33 467 04 02 31.
REFERENCES
- Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MJ. Phylogeny of the major tetrapod groups: Morphological data and divergence dates. J Mol Evol. 1990;30:409–424. doi: 10.1007/BF02101113. [DOI] [PubMed] [Google Scholar]
- Burnside J, Liou SS, Cogburn LA. Molecular cloning of the chicken Growth Hormone Receptor complementary deoxyribonucleic acid: Mutation of the gene in sex-linked dwarf chickens. Endocrinology. 1991;128:3183–3192. doi: 10.1210/endo-128-6-3183. [DOI] [PubMed] [Google Scholar]
- Cheng S, Lutfalla G, Uzé G, Chumakov IM, Gardiner K. GART, SON, IFNAR, and CRF2-4 genes cluster on human chromosome 21 and mouse chromosome 16. Mamm Genome. 1993;4:338–342. doi: 10.1007/BF00357094. [DOI] [PubMed] [Google Scholar]
- Digby MR, Lowenthal JW. Cloning and expression of the chicken interferon-gamma gene. J Interferon Cytokine Res. 1995;15:939–945. doi: 10.1089/jir.1995.15.939. [DOI] [PubMed] [Google Scholar]
- Domanski P, Witte M, Kellum M, Rubinstein M, Hackett R, Pitha P, Colamonici O. Cloning and expression of the long form of the beta subunit of the interferon alpha/beta receptor that is required for signaling. J Biol Chem. 1995;270:21606–21611. doi: 10.1074/jbc.270.37.21606. [DOI] [PubMed] [Google Scholar]
- Erlandsson L, Blumenthal R, Eloranta ML, Engel H, Alm G, Weiss S, Leanderson T. Interferon beta is required for interferon alpha production in mouse fibroblasts. Curr Biol. 1998;8:223–226. doi: 10.1016/s0960-9822(98)70086-7. [DOI] [PubMed] [Google Scholar]
- Goffin V, Shiverick KT, Kelly PA, Martial JA. Sequence-function relationships within the expanding family of prolactin, growth hormone, placental lactogen and related proteins in mammals. Endocr Rev. 1996;17:385–410. doi: 10.1210/edrv-17-4-385. [DOI] [PubMed] [Google Scholar]
- Gorman DM, Itoh N, Jenkins NA, Gilbert DJ, Copeland NG, Miyajima A. Chromosomal localization and organization of the murine genes encoding the beta subunits (AIC2A and AIC2B) of the interleukin 3, granulocyte/macrophage colony-stimulating factor, and interleukin 5 receptors. J Biol Chem. 1992;267:15842–15848. [PubMed] [Google Scholar]
- Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel RM, Aguet M. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- Hughes AL. The evolution of the type I interferon gene family in mammals. J Mol Evol. 1995;41:539–548. doi: 10.1007/BF00175811. [DOI] [PubMed] [Google Scholar]
- Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B. 1957;147:258–267. [PubMed] [Google Scholar]
- Kan JL, Moran RG. Analysis of a mouse gene encoding three steps of purine synthesis reveals use of an intronic polyadenylation signal without alternative exon usage. J Biol Chem. 1995;270:1823–1832. doi: 10.1074/jbc.270.4.1823. [DOI] [PubMed] [Google Scholar]
- Kotenko SV, Krause CD, Izotova LS, Pollack BP, Wu W, Pestka S. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO J. 1997;16:5894–5903. doi: 10.1093/emboj/16.19.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer E, Baker E, D’Andrea RJ, Slim R, Phillips H, Moretti PAB, Lopez AF, Petit C, Vadas MA, Sutherland GR, Goodal GJ. A cytokine receptor gene cluster in the X-Y pseudoautosomal region? Blood. 1993;82:22–28. [PubMed] [Google Scholar]
- Kumar S, Hedges B. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Lewerenz M, Mogensen KE, Uzé G. Shared receptor components but distinct complexes for alpha and beta interferons. J Mol Biol. 1998;282:585–599. doi: 10.1006/jmbi.1998.2026. [DOI] [PubMed] [Google Scholar]
- Lutfalla G, Gardiner K, Proudhon D, Vielh E, Uzé G. The structure of the human interferon alpha/beta receptor gene. J Biol Chem. 1992;267:2802–2809. [PubMed] [Google Scholar]
- Lutfalla G, Gardiner K, Uzé G. A new member of the cytokine receptor gene family maps on chromosome 21 at less than 35 kb from IFNAR. Genomics. 1993;16:366–373. doi: 10.1006/geno.1993.1199. [DOI] [PubMed] [Google Scholar]
- Lutfalla G, Holland SJ, Cinato E, Monneron D, Reboul J, Rogers NC, Smith JM, Stark GR, Gardiner K, Mogensen KE, Kerr IM, Uzé G. Mutant U5A are complemented by an interferon alpha beta receptor subunit generated by alternative processing of a new member of a cytokine receptor gene cluster. EMBO J. 1995;14:5100–5108. doi: 10.1002/j.1460-2075.1995.tb00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey JH, Fakhrai H, Edgington TS. Molecular cloning of the cDNA for tissue factor, the cellular receptor for the initiation of the coagulation cascade. Cell. 1987;50:129–135. doi: 10.1016/0092-8674(87)90669-6. [DOI] [PubMed] [Google Scholar]
- Müller U, Steinhoff U, Reis LFL, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- Murphy PM. Molecular mimicry and the generation of host defense protein diversity. Cell. 1993;72:823–826. doi: 10.1016/0092-8674(93)90571-7. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Kosugi H, Miyajima A, Arai K, Yokota T. Structure of the gene encoding the alpha subunit of the human granulocyte-macrophage colony stimulating factor receptor. J Biol Chem. 1994;269:10905–10912. [PubMed] [Google Scholar]
- Novick D, Cohen B, Rubinstein M. The human interferon alpha/beta receptor: Characterization and molecular cloning. Cell. 1994;77:391–400. doi: 10.1016/0092-8674(94)90154-6. [DOI] [PubMed] [Google Scholar]
- Paavonen K, Horelli-Kuitunen N, Chilov D, Kukk E, Pennanen S, Kallioniemi OP, Pajusola K, Olofsson B, Eriksson U, Joukov V, Palotie A, Alitalo K. Novel human vascular endothelial growth factor genes VEGF-B and VEGF-C localize to chromosomes 11q13 and 4q34, respectively. Circulation. 1996;93:1079–1082. doi: 10.1161/01.cir.93.6.1079. [DOI] [PubMed] [Google Scholar]
- Raval P, Obici S, Russell SW, Murphy WJ. Characterisation of the 5′ flanking region and gene encoding the mouse interferon-gamma receptor. Gene. 1995;154:219–223. doi: 10.1016/0378-1119(94)00869-t. [DOI] [PubMed] [Google Scholar]
- Rhee S, Ebensperger C, Dembic Z, Pestka S. The structure of the gene for the second chain of the human IFN-gamma receptor. J Biol Chem. 1996;271:28947–28953. doi: 10.1074/jbc.271.46.28947. [DOI] [PubMed] [Google Scholar]
- Roberts RM, Liu L, Alexandro A. New and atypical families of type I interferons in mammals: Comparative functions, structures and evolutionary relationships. Prog Nucleic Acid Res Mol Biol. 1997;56:287–325. doi: 10.1016/s0079-6603(08)61008-9. [DOI] [PubMed] [Google Scholar]
- Sandra O, Sohm F, de Luze A, Prunet P, Edery M, Kelly PA. Expression cloning of a cDNA encoding a fish prolactin receptor. Proc Natl Acad Sci. 1995;92:6037–6041. doi: 10.1073/pnas.92.13.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D, Brake AJ, Kiefer MC, Young D, Bar PJ. Cloning of three human multifunctional de nove purine biosynthetic genes by functional complementation of yeast mutations. Proc Natl Acad Sci. 1990;87:2916–2920. doi: 10.1073/pnas.87.8.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekellick MJ, Ferrandino AF, Hopkins DA, Marcus PI. Chicken interferon gene: Cloning, expression and analysis. J Interferon Res. 1994;14:71–79. doi: 10.1089/jir.1994.14.71. [DOI] [PubMed] [Google Scholar]
- Sick C, Schultz U, Staeheli P. A family of genes coding two serologically distinct chicken interferons. J Biol Chem. 1996;271:7635–7639. doi: 10.1074/jbc.271.13.7635. [DOI] [PubMed] [Google Scholar]
- Slate DL, Ruddle FH, Tan YH. Interferon 3. London, UK and New York, NY: Academic Press; 1981. Genetic control of the interferon system; pp. 65–76. [Google Scholar]
- Soh J, Donnelly RJ, Kotenko S, Mariano TM, Cook JR, Wang N, Emanuel SL, Schwartz B, Miki T, Pestka S. Identification and sequence of an accessory factor required for activation of the human interferon gamma receptor. Cell. 1994;76:793–802. doi: 10.1016/0092-8674(94)90354-9. [DOI] [PubMed] [Google Scholar]
- Spencer SD, Di Marco F, Hooley J, Pitts-Meek S, Bauuer M, Ryan AM, Sordat B, Gibbs VC, Aguet M. The orphan receptor CRF2-4 is an essential subunit of the interleukin 10 receptor. J Exp Med. 1998;187:571–578. doi: 10.1084/jem.187.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter U, Snipes GJ. Biology and genetics of hereditary motor and sensory neuropathies. Annu Rev Neurosci. 1995;18:45–75. doi: 10.1146/annurev.ne.18.030195.000401. [DOI] [PubMed] [Google Scholar]
- Tamai T, Shirahata S, Noguchi T, Sat N, Kimura S, Murakami H. Cloning and expression of flatfish (Paralichthys olivaceous) interferon cDNA. Biochim Biophys Acta. 1993;1174:182–186. doi: 10.1016/0167-4781(93)90112-q. [DOI] [PubMed] [Google Scholar]
- Thoreau E, Petridou B, Kelly PA, Djiane J, Mornon J-P. Structural symmetry of the extracellular domain of the cytokine/growth hormone/prolactin receptor family and interferon receptors revealed by hydrophobic cluster analysis. FEBS Lett. 1991;282:16–31. doi: 10.1016/0014-5793(91)80437-8. [DOI] [PubMed] [Google Scholar]
- Tiersch TR, Wachtel SS. On the evolution of the genome size of birds. J Hered. 1991;82:363–368. doi: 10.1093/oxfordjournals.jhered.a111105. [DOI] [PubMed] [Google Scholar]
- Uzé G, Lutfalla G, Gresser I. Genetic transfer of a functional human interferon alpha receptor into mouse cells: Cloning and expression of its cDNA. Cell. 1990;60:225–234. doi: 10.1016/0092-8674(90)90738-z. [DOI] [PubMed] [Google Scholar]
- Uzé G, Lutfalla G, Bandu M-T, Proudhon D, Mogensen KE. Behavior of a cloned murine interferon alpha beta receptor expressed in homospecific or heterospecific background. Proc Natl Acad Sci. 1992;89:4774–4778. doi: 10.1073/pnas.89.10.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzé G, Lutfalla G, Mogensen KE. alpha and beta interferons and their receptor and their friends and relations. J Interferon Cytokine Res. 1995;15:3–26. doi: 10.1089/jir.1995.15.3. [DOI] [PubMed] [Google Scholar]
- Wilson V, Jeffreys AJ, Barrie PA, Boseley PG, Slocombe PM, Easton A, Burke DC. A comparison of vertebrate gene families detected by hybridization with human IFN-β mRNA. J Mol Biol. 1983;166:457–475. doi: 10.1016/s0022-2836(83)80281-2. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Nezu J, Shimane M, Hirata Y. Molecular cloning of a novel vascular endothelial growth factor, VEGF-D. Genomics. 1997;42:483–488. doi: 10.1006/geno.1997.4774. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Fan P, Rand E, Kyaw H, Su K, Madike V, Carter KC, Li Y. Cloning of a putative human neurotransmitter receptor expressed in skeletal muscle and brain. Biochem Biophys Res Commun. 1998;242:575–578. doi: 10.1006/bbrc.1997.7591. [DOI] [PubMed] [Google Scholar]