Abstract
Human immune cells infected by HIV naturally contain high uracil content, and HIV reverse transcriptase (RT) does not distinguish between dUTP and dTTP. Many DNA viruses and retroviruses encode a dUTPase or uracil-DNA glycosylase (UNG) to counteract uracil incorporation. However, although HIV virions are thought to contain cellular UNG2, replication of HIV produced in cells lacking UNG activity does not appear to be impaired. Here we show that HIV reverse transcripts generated in primary human immune cells are heavily uracilated (>500 uracils per 10 kb HIV genome). We find that HIV DNA uracilation, rather than being dangerous, may promote the early phase of the viral life cycle. Shortly after reverse transcription, the ends of the HIV DNA are activated by the viral integrase (IN) in preparation for chromosomal insertion. However, the activated ends can attack the viral DNA itself in a suicidal side pathway, called autointegration. We find here that uracilation of target DNA inhibits the strand transfer of HIV DNA ends by IN, thereby inhibiting autointegration and facilitating chromosomal integration and viral replication. When uracilation is increased by incubating uracil-poor cells in the presence of increasing concentrations of dUTP or by infecting with virus that contains the cytosine deaminase APOBEC3G (A3G), the proportion of reverse transcripts that undergo suicidal autointegration decreases. Thus, HIV tolerates, or even benefits from, nonmutagenic uracil incorporation during reverse transcription in human immune cells.
After the HIV envelope fuses with a target cell plasma membrane to deliver the viral capsid into the cytoplasm, the viral genomic RNA is rapidly reverse transcribed into DNA. Soon thereafter within the HIV preintegration complex (PIC) in the host cell cytoplasm, HIV integrase (IN) activates the ends of the reverse transcripts by removing two nucleotides from the 3′ end of each strand through a process called 3′ processing to prepare for HIV DNA integration into chromosomal DNA. Three-prime processing makes the viral DNA vulnerable to autointegration, a suicidal side pathway in which the reactive ends attack viral DNA itself to produce deletion or inversion circles (1). Autointegration is a problem faced by retroviruses (2, 3) and mobile genetic elements, including bacteriophages and retrotransposons (4–6). Different retroviruses have evolved unique ways to control autointegration. Moloney murine leukemia virus employs the barrier-to-autointegration factor, which protects viral DNA through compaction to promote intermolecular integration (2, 7–9). We recently showed that the host SET complex, which contains three DNases (Ape1, Nm23-H1, and Trex1) and other DNA binding proteins, binds to the HIV PIC and protects it from autointegration (10). Knockdown of SET complex genes leads to increased HIV autointegration and correspondingly decreased chromosomal integration, resulting in ∼2- to 10-fold less HIV replication.
The SET complex is a base excision repair (BER) complex—Ape1 is the rate-limiting BER endonuclease and the two other nucleases in the complex are postulated to have BER proofreading functions (11, 12). An important function of BER is to recognize and remove misincorporated uracils in DNA. The main target cells of HIV infection, CD4 T cells and macrophages, naturally contain unusually high cellular uracil content (e.g., 8.4% of the total nucleotide pool or 1/4.4 dUTP/dTTP in macrophages, 12.2% of the total nucleotide pool or 1/4.2 dUTP/dTTP in peripheral blood T lymphocytes, compared with 0.2% of the total nucleotide pool or 1/100 dUTP/dTTP in HeLa and many other mammalian cells) (13–15). The HIV reverse transcriptase (RT) lacks proofreading activity and does not efficiently distinguish dUTP from dTTP. As such, HIV DNA generated in infected human immune cells likely contains uracils from misincorporation, although how much is unknown. Uracils in HIV DNA can also occur through cytosine deamination by APOBEC3 proteins that are highly expressed in immune cells. The HIV accessory protein Vif binds and inhibits A3G and APOBEC3F (16). To replicate in a uracil-rich cellular environment, many viruses, including some retroviruses, encode a dUTPase, which limits cellular uracil concentrations and/or a uracil-DNA glycosylase (UNG) to remove uracil from viral DNA. Replication and maintenance of latency of dUTPase-deficient herpes simplex virus (HSV-1) are severely impaired in the murine nervous system where dUTP levels are high and cellular dUTPase is low (17). Inactivation of the UNG gene of vaccinia virus also compromises its viability (18, 19). Retroviruses that replicate via DNA intermediates are also subjected to the same threat by uracil. Some retroviruses, including β-retroviruses (Mason-Pfizer monkey virus, murine mammary tumor virus) and nonprimate lentiviruses (feline immunodeficiency virus, equine infectious anemia virus), encode a dUTPase. The retrovirus-encoded dUTPase is dispensable for replication in dividing cells where the cellular uracil:thymidine ratio is low, but is essential for replication in nondividing cells, such as macrophages (20, 21). Surprisingly, primate lentiviruses, such as HIV, encode neither dUTPase nor UNG, but still manage to replicate efficiently in nondividing cells with a uracil-rich environment. Previous studies suggested that host UNG2 is incorporated in HIV particles through interaction with viral Vpr or IN and hypothesized a role in removing uracils from HIV DNA (22, 23). However, it is unclear whether HIV benefits from packaging UNG2, because viruses generated from human UNG2−/− cells are as competent as those from WT cells, and suppressing UNG activity does not significantly affect HIV infection in macrophages (24).
To investigate the impact of uracil misincorporation on HIV replication and to understand how HIV manages to replicate in the uracil-rich cellular environment in human immune cells, we developed a way to quantify uracil residues in HIV reverse transcripts during infection of its natural target cells. We find that HIV DNA reverse transcribed in primary human immune CD4 T cells and macrophages is heavily uracilated (>500 uracils per 10 kb genome, or more than one in five thymidines from each strand are replaced with uracils). These results suggest that virion-associated UNG2 does not effectively remove uracils from HIV DNA in infected human immune cells, supporting the notion that UNG2 may be dispensable for early stage HIV replication (24). We also found that uracilation of HIV DNA inhibits suicidal autointegration and increases viral infectivity using a single-round replication assay. Uracil misincorporation does not necessarily cause mutation per se because U:A pairs in chromosomal DNA are repaired faithfully by BER. Structural and enzymatic analysis of uracilated DNA and HIV IN suggests that uracil incorporation inhibits autointegration by inducing DNA distortions that block IN-mediated strand transfer, thereby favoring HIV DNA integration into chromosomal DNA in which uracil is efficiently removed. Thus, HIV tolerates, or may even benefit from, nonmutagenic uracil incorporation during reverse transcription in the uracil-rich environment of human immune cells.
Results
PCR Method to Estimate DNA Uracil Content.
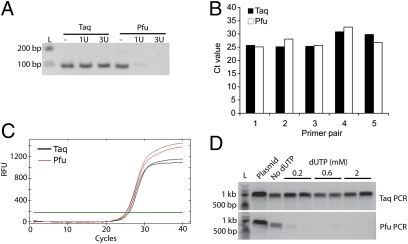
To estimate how many uracils are incorporated into HIV DNA in macrophages and CD4 T cells, we developed a PCR method. This method (“Taq/Pfu PCR”) is based on the fact that the archaebacterial DNA polymerase Pfu is strongly inhibited by uracil-containing DNA, whereas Taq DNA polymerase, which lacks proofreading like HIV RT, is not (25, 26). A similar type of assay has been used to quantify uracil in DNA (27). To assess the differential sensitivity of these polymerases to uracil, we tested the ability of both polymerases to amplify synthetic oligonucleotides (100 nucleotides in length of HIV gag sequence) containing zero, one, and three uracils as target templates using a pair of primers on the ends (Fig. 1A). Taq was similarly efficient at amplifying all three templates, whereas Pfu only generated the expected PCR product with the uracil-free DNA template, confirming the inability of Pfu to replicate across uracils. To verify that the difference in detection of Taq and Pfu PCR products by agarose gel electrophoresis was not due to differences in polymerase activity or in detecting their products, we used SYBR green dye to track DNA synthesis in real time in both reactions and found that Taq and Pfu yielded similar cycle threshold (Ct) values with five randomly chosen primer pairs in the HIV genome and plasmid HIV DNA as template (Fig. 1 B and C). We then prepared cytosolic DNA extracts from HeLa–CD4 cells, which have a low endogenous ratio of dUTP/dTTP (15), after overnight incubation with increasing amounts of dUTP in the culture medium and infection with a single-round vesicular stomatitis virus glycoprotein (VSV-G)–pseudotyped HIV bearing a luciferase reporter gene (HIV-Luc) (10) for 10 h. This time was chosen because reverse transcription is largely completed, whereas HIV DNA remains mostly in the cytosol and chromosomal integration has generally not yet occurred (10). Cytosolic DNA was used as template for Taq and Pfu amplification with HIV gag primers. Taq yielded comparable amounts of HIV products regardless of dUTP supplementation, whereas Pfu PCR only yielded the expected 1-kb PCR product when cells were grown in the absence of added dUTP (Fig. 1D). The absence of a Pfu PCR product suggests that at least one uracil was incorporated in each strand of the HIV reverse transcript within the region examined. We obtained similar results using two other pairs of HIV gag primers.
Fig. 1.
Taq/Pfu PCR method to assess DNA uracil content (A) DNA gel electrophoresis of Taq/Pfu PCR products using synthetic 100 nt DNA oligonucleotides, containing 0, 1, or 3 uracils as templates. Oligonucleotide sequences are provided in Table S1. L, DNA ladder (same below). (B and C) Real-time analysis of Taq/Pfu PCR. SYBR-green real-time PCR was carried out with DNA polymerase Taq or Pfu using five primer pairs randomly chosen from within the HIV genome and HIV-GFP plasmid DNA as a template. Ct value for each qPCR is shown in B. A representative qPCR trace plot (primer pair 1, in duplicate) is shown in C. (D) Taq/Pfu PCR assay using DNA isolated from infected HeLa–CD4 cells cultured with indicated amounts of dUTP supplementation. HIV-GFP plasmid was used as a control (“plasmid”). A and D show representative gel images from at least three independent experiments.
HIV DNA Synthesized in Infected Human Immune Cells Contains Many Uracils.
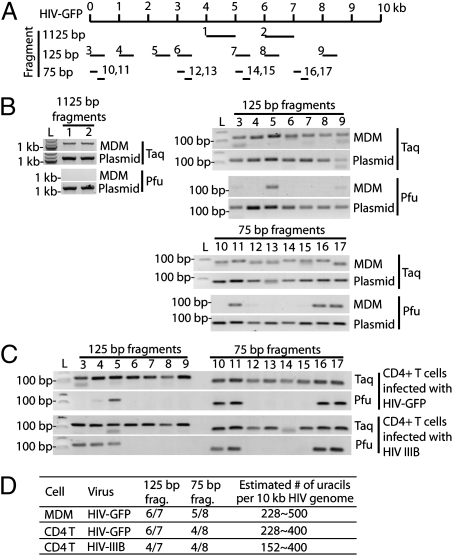
To estimate how many uracils are incorporated into HIV DNA in primary immune cells, we used the Taq/Pfu PCR assay to analyze cytosolic DNA from macrophages and CD4 T cells infected for 10 h with VSV-G pseudotyped or wild-type HIV (Fig. 2). Primer pairs were designed to amplify different size fragments from multiple locations throughout the HIV genome (Fig. 2A). Primer pairs amplifying longer regions are more likely to encounter a uracil (and thus block Pfu amplification) than primer pairs expanding shorter regions. The percentage of fragments of a given length that are “uracil positive” (amplified by Taq, but not Pfu), should provide a lower limit estimate of the number of total uracils incorporated into full length ∼10-kb HIV DNA. A “uracil positive” fragment should contain at least 2 uracils, one in each strand. Primer pairs (25 nucleotides long) were designed to examine regions of three sizes (1,075, 75, and 25 bp) to produce PCR fragments of 1,125, 125, and 75 bp, respectively. An array of Taq/Pfu PCR reactions was performed to determine which fragments are amplified by Taq, but not Pfu. In monocyte-derived macrophages (MDM) infected with VSV-G–pseudotyped HIV-GFP, Pfu amplified zero of two 1,075-bp regions, one of seven 75-bp regions, and three of eight 25-bp regions (Fig. 2B). HIV DNA contains 133 75-bp regions, of which 6 of 7 (86%) were Pfu PCR negative and are expected to contain at least two uracils. This suggests that there are at least 133 × 6/7 × 2 or 228 uracils per 10-kb HIV DNA. Performing a similar calculation on the basis of the 25-bp amplicons of which five of eight were Pfu PCR negative, we estimate that there are at least 400 × 5/8 × 2 or 500 uracils per 10-kb HIV DNA. Smaller fragment size and more sampling should provide better resolution and a more accurate estimate. Pfu PCR negative fragments are not more AT rich than the other fragments in our panel. We repeated this uracil mapping experiment using CD4 T cells infected with VSV-G–pseudotyped HIV-GFP or wild-type HIVIIIB (Fig. 2 C and D). Analysis of the 75-bp regions suggested a lower limit of 228 and 152 uracils, respectively, and analysis of the 25-bp regions suggested ≥400 uracils per HIV reverse transcript for both viruses. Thus, HIV reverse transcripts are heavily uracilated to a similar extent during infection of both macrophages and T cells.
Fig. 2.
HIV DNA is heavily uracilated in infected human immune cells. (A) Schematic showing the locations of primer pairs (numbered) in the HIV-GFP genome used for uracil mapping. The numbers in this diagram correspond to labels for the gel images in B and C. (B) Taq/Pfu PCR using DNA isolated from HIV-GFP–infected MDM or plasmid DNA (as a control). (C) Taq/Pfu PCR using DNA isolated from CD4 T cells infected with HIV-GFP or HIVIIIB virus. (D) A table summarizing the findings from B and C. All primer pairs yielded expected PCR products in Taq-PCR or PCR reactions using plasmid as a template. Primer pairs that failed to yield a PCR product in Pfu-PCR were scored as uracil positive for that fragment. See text for details on how the estimated number of uracils per 10 kb HIV genome was calculated.
Because our assay uses DNA extracted from multiple infected cells rather than a single infected cell and the lack of a Pfu signal requires that all of the HIV DNA copies in the sample contain uracil in at least one position on each strand, it is likely that the actual number of uracils in each HIV DNA may be higher than our lower limit estimate of 15–50 per kb. Because HIV DNA is AT rich (∼60%), at least 15–50 of 600 AT residues or ∼2.5–8.3% of thymidines were replaced by uracils during reverse transcription. This number should be compared with estimates of the ratio of dUTP/dTTP in macrophages and lymphocytes of 20–25% (14), which could be considered an upper limit of the uracil-to-thymidine ratio of HIV DNA synthesized in those cells. In contrast, in human chromosomal DNA it is estimated that 70–200 uracils are generated by cytosine deamination per day (<0.0001 per kb) (28), and Ung−/− mouse embryo fibroblasts have ∼2,000 uracils per genome (0.001 per kb) (29). In fact Taq and Pfu similarly amplified HIV chromosomal DNA in infected T cells. Therefore, HIV cytosolic DNA is heavily uracilated in human immune cells relative to chromosomal DNA.
HIV Reverse Transcripts Generated in Uracil-Rich Human Macrophages Are Less Susceptible to Autointegration than Transcripts Generated in Uracil-Poor HeLa Cells.
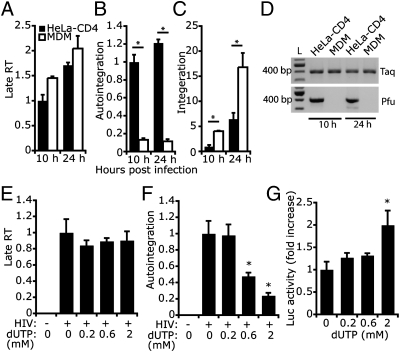
We next examined how uracil incorporation affects the early steps of the HIV life cycle. We first compared the relative numbers of HIV late reverse transcripts, autointegrants, and integrated DNA copies in human MDM (which have dUTP/dTTP ∼1/4) (14) to HeLa–CD4 cells (dUTP/dTTP ∼1/100) (15) after infection with an equal amount of VSV-G–pseudotyped HIV. Reverse transcription, autointegration, and chromosomal integration were measured 10 and 24 h postinfection (hpi) (Fig. 3 A–C). HIV infection in both cells generated similar amounts of late RT products, suggesting that HIV reverse transcription is not affected by high cellular uracil content in MDMs. In contrast, autointegration was severely reduced in MDMs (11–13% of that in HeLa–CD4 cells), and chromosomal integration increased in MDMs by three- to four-fold. Taq/Pfu PCR analysis (of a 400-bp region) of HIV cytosolic DNA from infected cells confirmed that HIV DNA is more uracilated in MDMs (Fig. 3D). Thus, the high cellular uracil content in MDMs does not appear to impede HIV replication. On the contrary, the uracil environment may be beneficial for HIV to complete the early stages of the life cycle, by inhibiting suicidal autointegration and promoting productive chromosomal integration.
Fig. 3.
Uracilation of HIV DNA inhibits autointegration. (A–D) HIV RT products produced in infected macrophages are less susceptible to autointegration than those produced in HeLa–CD4 cells. HeLa–CD4 and monocyte-drived macrophages (MDM) were infected with an equal amount of HIV-GFP. Late RT (A), autointegration (B), and integrated DNA (C) were measured 10 h and 24 h postinfection. Late RT was normalized to human mitochondrial DNA; autointegration was normalized to late RT; integrated DNA was normalized to human β-globin DNA. *P < 0.05, Student's t test. Error bars indicate SD of three independent experiments. Differential uracil incorporation in HIV DNA was verified by Taq/Pfu PCR (D). (E–G) Culturing HIV-infected HeLa–CD4 cells with increasing amounts of dUTP reduces autointegration and promotes infection. dUTP was added to the medium the night before infection with VSV-G–pseudotyped HIV-Luc virus at a multiplicity of infection of 1. HIV reverse transcription (late RT, E) and autointegration (F) were measured 10 h postinfection, and Luciferase activity was measured 24 h postinfection (G). Values were normalized as above. *P < 0.05, Student's t test. Error bars indicate SD of three independent experiments.
Uracil Incorporation in HIV DNA Inhibits Autointegration.
To further investigate whether uracil incorporation promotes early steps of the HIV life cycle, we added increasing amounts of dUTP to the culture medium of HeLa–CD4 cells and then infected them 1 d later with VSV-G–pseudotyped HIV-Luc and measured HIV reverse transcription and autointegration 10 hpi and Luc activity 24 hpi (Fig. 3 E–G). Addition of dUTP did not affect cell growth or viability during the 2-d period of the assay. dUTP treatment also did not affect HIV DNA synthesis in HeLa–CD4 cells (Fig. 3E), consistent with the fact that HIV RT does not distinguish between dUTP and dTTP. In contrast, HIV autointegration was inhibited in a dose-dependent manner by adding dUTP (Fig. 3F). Luc activity increased twofold in cells treated with the highest dose of dUTP, suggesting that dUTP treatment increased HIV replication, perhaps by reducing suicidal autointegration.
C-to-U Hypermutation by APOBEC3G Inhibits HIV Autointegration.
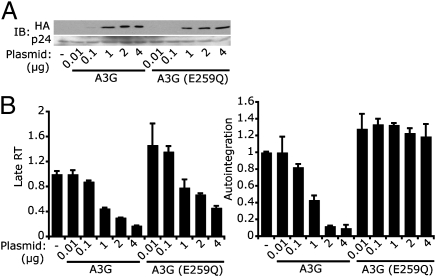
We next examined the effect on HIV autointegration of introducing uracils into HIV DNA by an alternative method, cytosine deamination by A3G (30). A3G is an intrinsic antiviral host protein counteracted by HIV Vif (16). We generated HIV-LucΔvif viruses containing no A3G or increasing amounts of wild-type A3G or A3G-E259Q (an enzymatically defective mutant) (31) by transfecting virus-producing 293T cells with increasing amounts of A3G expression plasmids (Fig. 4A). Viral stocks were normalized on the basis of HIV capsid (p24) level measured by ELISA and were used at equal capsid protein levels to infect HeLa–CD4 cells (which do not express A3G). Incorporation of A3G and A3G-E259Q viruses reduced HIV late RT, consistent with a previous report that A3G blocks HIV reverse transcription independently of its deaminase activity (Fig. 4B and ref. 32). To assess the effect of increased uracilation of HIV reverse transcripts, we normalized the number of autointegration events to the number of reverse transcripts generated under each condition. Incorporation of enzymatically dead A3G into virions did not affect the amount of autointegration per reverse transcript, whereas wild-type A3G inhibited autointegration. Thus, uracilation of HIV DNA by A3G also inhibited HIV autointegration. However, we did not observe a corresponding increase in overall HIV-Luc activity, perhaps because A3G inhibits multiple steps of the HIV life cycle (32, 33), offsetting its effect of preventing autointegration.
Fig. 4.
APOBEC3G-mediated cytosine deamination inhibits HIV autointegration. HeLa–CD4 cells were infected with VSV-G–pseudotyped HIV-LucΔvif viruses containing increasing amounts of wild-type A3G or deaminase-defective A3G (E259Q). The amount of A3G or A3G (E259Q) plasmid used for producing viruses in 293T cells is indicated on the Bottom. (A) Immunoblot (IB) analysis showing the amount of A3G and A3G (E259Q) in viruses. Both A3G and A3G (E259Q) are HA-tagged. p24 (HIV capsid) was used as a loading control. (B) HIV late RT and autointegration were measured 10 h postinfection and normalized as in Fig. 3.
Uracil Incorporation Alters HIV DNA Structure.
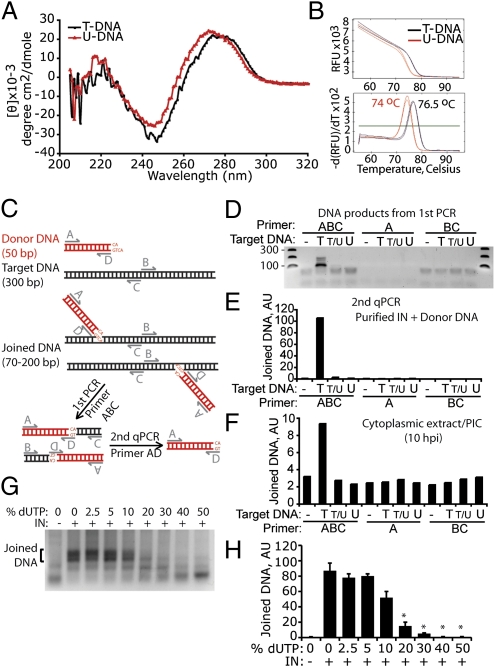
To understand why HIV DNA uracilation might inhibit autointegration, we first asked whether replacing thymidines with uracils (i.e., removing the 5-methyl group of thymidine) distorts HIV DNA structure. The 5-methyl group of thymidine protrudes into the major groove of DNA and is important for base stacking and structural stability of the DNA. Removing the 5-methyl group increases the minor groove size and affects DNA–protein interactions (34–36). The structure of HIV gag DNA (50-bp sequence) synthesized with either thymidines (T-DNA) or uracils in place of thymidine (U-DNA) was compared by circular dichroism (CD) spectrometry and thermal denaturation (Fig. 5). U-DNA displayed a significant blueshift in the 245- to 300-nm range of the CD spectrum compared with T-DNA (Fig. 5A). A similar blueshift was also observed when Bromo-dU was used to replace thymidine and has been attributed to turn structure distortion (37). U-DNA also showed reduced thermostability compared with T-DNA by melt curve analysis, in which the release of the SYBR green DNA dye was measured as a function of temperature (Fig. 5B, Upper). U-DNA consistently displayed a melting temperature 2° to 3° lower than T-DNA (Fig. 5B, Lower), indicating that uracil-containing DNA has reduced thermostability. Taken together, we conclude that uracilation significantly alters the structure and stability of DNA.
Fig. 5.
Uracil inhibits IN-mediated integration. (A and B) Uracil distorts HIV DNA as assessed by circular dichroism (A) and melt curve (B) analysis of a synthetic 50 bp (HIV gag sequence, 48% GC) dsDNA, containing thymidines (T-DNA) or uracils in place of thymidines (U-DNA). See Materials and Methods for sequences. Melt curve plot was generated by measuring the dissociation of SYBR green dye from dsDNA as a function of temperature (B, Upper). A derivative plot (B, Lower) is used to calculate the melting temperature (labels on peaks). Results in A and B are representative of three experiments. (C–F) Target DNA uracilation inhibits IN-mediated integration. (C) Diagram of half-site integration assay catalyzed by recombinant HIV IN. The donor DNA (preprocessed to have the 5′-AC dinucleotide overhang) is in red and the target DNA is in black. (D) DNA gel electrophoresis of PCR amplified integration products. Target DNA used in the in vitro integration assay contained either all thymidine (T), an equal mixture of thymidine and uracil (T/U), or all uracil (U). Primers used for amplification are shown on Top. Expected donor-target joining product length ranges from 70 to 200 bp. (E) qPCR analysis (with primers A and D) of PCR products obtained in D. AU, arbitrary unit (same below). (F) In vitro integration assay was carried out as in E using HIV PICs isolated from infected HeLa–CD4 cells to replace recombinant IN and donor DNA. Results in D–F are representative of two experiments. (G and H) Ten to 20% of uracil in target DNA inhibits integration. Dose–response experiment using varying levels of uracil in target DNA (as indicated) in an in vitro integration assay as in D. DNA gel electrophoresis of PCR-amplified integration products is shown in G, and quantification of the joined DNA, analyzed by qPCR as in E for two independent experiments, is shown in H. % dUTP, percentage of dUTP of total dUTP plus dTTP. Data represent mean ± SD. *P < 0.01, Student's t test (compared with 0% dUTP plus IN).
Uracil-DNA Inhibits Integration by HIV IN.
We next examined whether uracil-induced DNA structural changes in target DNA might inhibit HIV IN strand transfer. We used an in vitro integration assay in which recombinant HIV IN catalyzes half-site integration (joining of only one strand of dsDNA) of a donor and a target DNA (38). This assay is often used to characterize IN strand transfer activity and assess host factors, such as LEDGF, that influence integration (39). In the assay, the donor DNA (50 bp, HIV U5 sequence) contains a recessed CA dinucleotide end that mimics the processed viral DNA end. The target DNA contains non-HIV sequence (300 bp, AT = 53%; Materials and Methods), which allowed us to perform the assay with either recombinant IN and synthesized donor DNA or with HIV PIC isolated from infected cells. Target DNA samples that contain all T, half T/half U, or all U were used to determine the effect on integration of uracil in the target strand. U5 DNA integration products were detected by a semiquantitative nested PCR assay [similar to the Alu-PCR method used for quantifying integrated HIV DNA (ref. 40 and Fig. 5C)]. In the first PCR, a successfully integrated product containing both a donor and a target DNA molecule can be amplified only by using all three primers (ABC), but not by A or BC alone. PCR products from the first PCR step are heterogeneous in length (Fig. 5D), representing integration events that occurred at multiple sites along the target DNA. All DNA products from the first PCR contain the donor DNA sequence, which was then amplified by “nested” qPCR (with primers A and D). Recombinant HIV IN robustly catalyzed half-site integration into target DNA that contained no uracil, as previously described (Fig. 5 D and E and refs. 38, 39). In contrast, half T and half U (T/U) or all U in the target DNA completely inhibited integration, suggesting that IN strongly disfavors uracil-containing target DNA. To confirm these results, cytoplasmic extract from infected HeLa–CD4 cells (10 hpi) was used as a source of native HIV IN and donor DNA, which was incubated with the same target DNA oligonucleotides as above, containing varying T/U ratios. Although PICs support efficient concerted integration of endogenous U3 and U5 donor DNA ends (41), our assay monitored U5 end joining only. Integration occurred with T-DNA, but not with T/U- or U-DNA (Fig. 5F). Although HIV PIC IN formed only a fraction of integration products compared with the recombinant enzyme, these results clearly demonstrate that uracilation of target DNA inhibits HIV integration in vitro.
To determine how much uracil in target DNA is required to inhibit integration, we performed a dose–response in vitro integration experiment using varying levels of dU in the target DNA (Fig. 5 G and H). Integration was inhibited by only 10% dU (of total dT + dU) in target DNA; 20% dU almost completely blocked integration. Because at least 8%, and possibly as much as 25%, of thymidines in HIV DNA in macrophages and CD4 T cells are replaced by uracil, our data suggest that HIV uracilation contributes to blocking autointegration in a physiologically relevant way.
Discussion
In this study, we provide direct evidence that HIV DNA generated in infected primary human immune cells is heavily uracilated and that uracilation protects HIV DNA from the suicidal pathway of autointegration, facilitating chromosomal integration. Our results suggest that at least 8% of thymidines in HIV DNA in macrophages and CD4 T cells are replaced by uracils. A more accurate estimate of the degree of uracilation could be obtained by refining the Taq/Pfu PCR assay to amplify smaller segments than the smallest region we examined (25 bp) using fewer infected cells. In the immune cells that HIV infects, uracil:thymidine ratios approach 25%, and because HIV RT uses either base equivalently, freshly reverse transcribed HIV DNA likely contains about 20–25% uracils in place of thymidine. Several studies have suggested that UNG2, the major mammalian UNG, is packaged into HIV virions. However, it is unknown whether UNG2 is contained within the reverse transcription complex and retained as it matures into the PIC or whether it acts on nascent HIV reverse transcripts. Our results do not exclude the possibility that viral-associated UNG2 is active. However, much of the incorporated uracil in HIV DNA is retained in cytosolic HIV DNA. Virions produced in cells lacking UNG2 activity have no replication defect (24), indicating that HIV tolerates a high degree of uracilation. Because BER is highly efficient within the nucleus, it is likely that incorporated uracils in integrated chromosomal HIV DNA are efficiently removed and faithfully replaced by thymidines once integration has occurred.
HIV IN favors integration (for both integration and autointegration) (10, 42, 43) into target DNAs at sites that loosely match an 11-bp consensus sequence containing approximately four to seven thymidines. Thus, many available autointegration sites would contain at least one nearby uracil. We showed here that uracilation of target DNA inhibits IN strand transfer. Thus, uracil-rich HIV cytosolic DNA is a poor target for (auto)integration compared with uracil-poor chromosomal DNA. Lentiviral IN may have evolved to enhance chromosomal integration in uracil-rich cells. Structural changes caused by uracilation could have a detrimental effect on IN targeting. Indeed, the recently solved structure of the prototype foamy virus (PFV) IN multimer in complex with DNA oligomers revealed that the major groove is severely widened and minor groove compressed at the site of integration (44, 45). Replacing thymidines with uracils, which involves removal of the 5-methyl group of thymidines, expands the minor groove, thereby potentially creating structural hindrance for IN access to inhibit strand transfer. 5-methyl groups of thymidines lying within the core of the target DNA integration site directly contact PFV IN in the cocrystal structure (44). Thus, replacing these thymidines with uracils could affect IN binding.
In addition to a role in protecting HIV from autointegration, uracilation of HIV reverse transcripts might influence other events in the early phase of HIV replication. We recently found that HIV DNA can be recognized by an unknown innate immune DNA cytosolic sensor in macrophages and T cells to trigger production of type I interferons. Cytosolic HIV DNA uracilation might interfere with nucleic acid detection in the cytosol that could potentially activate innate immunity (46).
We previously showed that the host SET complex inhibits HIV autointegration by binding to HIV DNA soon after reverse transcription in the cytosol (10). Here we found that intrinsic uracilation of HIV DNA in natural target cells also reduces autointegration. Both processes (SET complex binding and HIV DNA uracilation) can independently inhibit autointegration. For example, the SET complex inhibits HIV autointegration in HeLa–CD4 cells, where HIV DNA is not uracilated (10), and uracilated target DNA blocks IN-mediated strand transfer in vitro in the absence of the SET complex (Fig. 5D). However, it is likely that these two mechanisms also act in concert. The SET complex has BER activity (11, 12). Because one BER function is to repair misincorporated uracils, the SET complex may recognize and bind HIV DNA via its uracils. In fact, HMGB2, a component of the SET complex, preferentially binds to distorted DNA structures. We envision a model in which HIV DNA is uracilated during reverse transcription, which attracts the SET complex to bind. SET complex binding to these uracilated regions of HIV DNA in the cytosol then shields them from autointegration. It may be that the SET complex does not repair HIV DNA uracils in the cytosol, where the abundance of other downstream BER pathway enzymes may be limiting. Because the SET complex shuttles between the cytosol and the nucleus (47), it may traffic with HIV DNA into the nucleus for integration and then mark the spots of uracil incorporation for BER within the nucleus. Further studies are needed to investigate these possibilities.
Materials and Methods
Human CD4+ T cells and macrophages were isolated from PBMC as in ref. 46. HIVIIIB, HIV-Luc, and HIV-GFP were generated as in ref. 46. HIV DNA products were analyzed as in ref. 10. In vitro integration assay was performed as in ref. 38. Taq/Pfu PCR details and other additional information are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Peter Cherepanov (Imperial College London) for insightful discussions, Dana Gabuzda (Dana-Farber Cancer Institute) for HIV-GFP plasmid, Ned Landau (New York University) for A3G plasmid, Stephen Blacklow (Harvard Medical School) for help with the CD measurement, and members of the J.L. laboratory for helpful discussions. This work was supported by National Institutes of Health Grants AI45587 (to J.L.), T32 HL066987-10 (to N.Y.), and AI052014 (to A.E.) and an Endowed Scholars Award from University of Texas Southwestern (to N.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102943108/-/DCSupplemental.
References
- 1.Li Y, et al. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: Identification of replication-competent and -defective viral genomes. J Virol. 1991;65:3973–3985. doi: 10.1128/jvi.65.8.3973-3985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee MS, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki Y, Craigie R. Regulatory mechanisms by which barrier-to-autointegration factor blocks autointegration and stimulates intermolecular integration of Moloney murine leukemia virus preintegration complexes. J Virol. 2002;76:12376–12380. doi: 10.1128/JVI.76.23.12376-12380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garfinkel DJ, et al. Retrotransposon suicide: Formation of Ty1 circles and autointegration via a central DNA flap. J Virol. 2006;80:11920–11934. doi: 10.1128/JVI.01483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benjamin HW, Kleckner N. Intramolecular transposition by Tn10. Cell. 1989;59:373–383. doi: 10.1016/0092-8674(89)90298-5. [DOI] [PubMed] [Google Scholar]
- 6.Maxwell A, Craigie R, Mizuuchi K. B protein of bacteriophage mu is an ATPase that preferentially stimulates intermolecular DNA strand transfer. Proc Natl Acad Sci USA. 1987;84:699–703. doi: 10.1073/pnas.84.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee MS, Craigie R. Protection of retroviral DNA from autointegration: Involvement of a cellular factor. Proc Natl Acad Sci USA. 1994;91:9823–9827. doi: 10.1073/pnas.91.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl Acad Sci USA. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skoko D, et al. Barrier-to-autointegration factor (BAF) condenses DNA by looping. Proc Natl Acad Sci USA. 2009;106:16610–16615. doi: 10.1073/pnas.0909077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan N, Cherepanov P, Daigle JE, Engelman A, Lieberman J. The SET complex acts as a barrier to autointegration of HIV-1. PLoS Pathog. 2009;5:e1000327. doi: 10.1371/journal.ppat.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Postel EH, Abramczyk BM, Levit MN, Kyin S. Catalysis of DNA cleavage and nucleoside triphosphate synthesis by NM23-H2/NDP kinase share an active site that implies a DNA repair function. Proc Natl Acad Sci USA. 2000;97:14194–14199. doi: 10.1073/pnas.97.26.14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Höss M, et al. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J. 1999;18:3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 14.Aquaro S, et al. Macrophages and HIV infection: Therapeutical approaches toward this strategic virus reservoir. Antiviral Res. 2002;55:209–225. doi: 10.1016/s0166-3542(02)00052-9. [DOI] [PubMed] [Google Scholar]
- 15.Mahagaokar S, Orengo A, Rao PN. The turnover of deoxyuridine triphosphate during the HeLa cell cycle. Exp Cell Res. 1980;125:86–94. doi: 10.1016/0014-4827(80)90192-5. [DOI] [PubMed] [Google Scholar]
- 16.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: An innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 17.Pyles RB, Thompson RL. Evidence that the herpes simplex virus type 1 uracil DNA glycosylase is required for efficient viral replication and latency in the murine nervous system. J Virol. 1994;68:4963–4972. doi: 10.1128/jvi.68.8.4963-4972.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellison KS, Peng W, McFadden G. Mutations in active-site residues of the uracil-DNA glycosylase encoded by vaccinia virus are incompatible with virus viability. J Virol. 1996;70:7965–7973. doi: 10.1128/jvi.70.11.7965-7973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millns AK, Carpenter MS, DeLange AM. The vaccinia virus-encoded uracil DNA glycosylase has an essential role in viral DNA replication. Virology. 1994;198:504–513. doi: 10.1006/viro.1994.1061. [DOI] [PubMed] [Google Scholar]
- 20.Lichtenstein DL, et al. Replication in vitro and in vivo of an equine infectious anemia virus mutant deficient in dUTPase activity. J Virol. 1995;69:2881–2888. doi: 10.1128/jvi.69.5.2881-2888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagaman PC, et al. Molecular cloning and characterization of deoxyuridine triphosphatase from feline immunodeficiency virus (FIV) Virology. 1993;196:451–457. doi: 10.1006/viro.1993.1501. [DOI] [PubMed] [Google Scholar]
- 22.Selig L, et al. Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J Virol. 1997;71:4842–4846. doi: 10.1128/jvi.71.6.4842-4846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willetts KE, et al. DNA repair enzyme uracil DNA glycosylase is specifically incorporated into human immunodeficiency virus type 1 viral particles through a Vpr-independent mechanism. J Virol. 1999;73:1682–1688. doi: 10.1128/jvi.73.2.1682-1688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaiser SM, Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J Virol. 2006;80:875–882. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lasken RS, Schuster DM, Rashtchian A. Archaebacterial DNA polymerases tightly bind uracil-containing DNA. J Biol Chem. 1996;271:17692–17696. doi: 10.1074/jbc.271.30.17692. [DOI] [PubMed] [Google Scholar]
- 26.Hogrefe HH, Hansen CJ, Scott BR, Nielson KB. Archaeal dUTPase enhances PCR amplifications with archaeal DNA polymerases by preventing dUTP incorporation. Proc Natl Acad Sci USA. 2002;99:596–601. doi: 10.1073/pnas.012372799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horváth A, Vértessy BG. A one-step method for quantitative determination of uracil in DNA by real-time PCR. Nucleic Acids Res. 2010;38:e196. doi: 10.1093/nar/gkq815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 29.Nilsen H, et al. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- 30.Harris RS, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 31.Newman EN, et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 32.Bishop KN, Verma M, Kim E-Y, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mbisa JL, et al. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J Virol. 2007;81:7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 35.Bailly C, Crow S, Minnock A, Waring MJ. Demethylation of thymine residues affects DNA cleavage by endonucleases but not sequence recognition by drugs. J Mol Biol. 1999;291:561–573. doi: 10.1006/jmbi.1999.2979. [DOI] [PubMed] [Google Scholar]
- 36.Marathe A, Bansal M. The 5-methyl group in thymine dynamically influences the structure of A-tracts in DNA at the local and global level. J Phys Chem B. 2010;114:5534–5546. doi: 10.1021/jp911055x. [DOI] [PubMed] [Google Scholar]
- 37.Augenlicht L, Nicolini C, Baserga R. Circular dichroism and thermal denaturation studies of chromatin and DNA from BrdU-treated mouse fibroblasts. Biochem Biophys Res Commun. 1974;59:920–926. doi: 10.1016/s0006-291x(74)80067-7. [DOI] [PubMed] [Google Scholar]
- 38.Dar MJ, et al. Biochemical and virological analysis of the 18-residue C-terminal tail of HIV-1 integrase. Retrovirology. 2009;6:94. doi: 10.1186/1742-4690-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hare S, et al. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 2009;5:e1000515. doi: 10.1371/journal.ppat.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol. 2002;76:10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen H, Engelman A. Asymmetric processing of human immunodeficiency virus type 1 cDNA in vivo: Implications for functional end coupling during the chemical steps of DNA transposition. Mol Cell Biol. 2001;21:6758–6767. doi: 10.1128/MCB.21.20.6758-6767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holman AG, Coffin JM. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc Natl Acad Sci USA. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shun MC, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chowdhury D, Lieberman J. Death by a thousand cuts: Granzyme pathways of programmed cell death. Annu Rev Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.