Abstract
Self-cleavage at the N terminus of HIV-1 protease from the Gag-Pol precursor (autoprocessing) is crucial for stabilizing the protease dimer required for onset of mature-like catalytic activity, viral maturation, and propagation. Among nine clinical protease inhibitors (PIs), darunavir and saquinavir were the most effective in inhibiting wild-type HIV-1 group M precursor autoprocessing, with an IC50 value of 1–2 μM, 3–5 orders of magnitude higher than their binding affinities to the corresponding mature protease. Accordingly, both group M and N precursor–PI complexes exhibit Tms 17–21 °C lower than those of the corresponding mature protease–PI complexes suggestive of markedly reduced stabilities of the precursor dimer–PI ensembles. Autoprocessing of group N (natural variant) and three group M precursors bearing 11–20 mutations associated with multidrug resistance was either weakly responsive or fully unresponsive to inhibitors at concentrations up to a practical limit of approximately 150 μM PI. This observation parallels decreases of up to 8 × 103-fold (e.g., 5 pM to 40 nM) in the binding affinity of darunavir and saquinavir to mature multidrug resistant proteases relative to wild type, suggesting that inhibition of some of these mutant precursors will occur only in the high μM to mM range in extreme PI-resistance, which is an effect arising from coordinated multiple mutations. An extremely darunavir-resistant mutant precursor is more responsive to inhibition by saquinavir. These findings raise the questions whether clinical failure of PI therapy is related to lack of inhibition of autoprocessing and whether specific inhibitors can be designed with low-nM affinity to target autoprocessing.
Keywords: calorimetry, drug resistance, precursor processing, inhibitor binding, aspartic protease
The mature HIV-1 protease (PR) is synthesized as part of a large Gag-Pol polyprotein precursor (Fig. 1). It is responsible for its own release from the precursor (termed autoprocessing), thus promoting controlled proteolysis of the viral Gag and Gag-Pol polyproteins into the mature structural and functional proteins required for virus assembly, maturation and propagation (1, 2). PR is a dimeric aspartyl protease composed of two identical polypeptides of 99 amino acids each. The active site is formed along the dimer interface and each subunit contributes one of the two catalytic aspartic acid residues (Fig. 2) (1, 2). Because of its indispensable role in the viral life cycle, the mature PR dimer has been a successful target for structure-based design of drugs that bind specifically to the active site (3). Although these clinical PR inhibitors (PIs) curtail the progression of the disease, their long-term effectiveness has been limited due to rapidly evolving viral variants, proteases of which exhibit resistance with drastically lower affinity to the drugs than the wild-type PR (4). Selection of major drug resistance mutations (DRMs) is coordinated within an environment of natural variations in this highly polymorphic virus (Fig. 2 and Fig. S1C). Orders of magnitude decreases in binding affinity of PIs, however, may correspond to only subtle differences observed in PR–PI crystal structures (5).
Fig. 1.
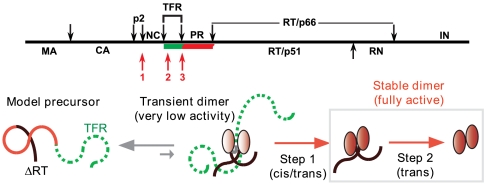
Schematic representation of the Gag-Pol polyprotein of HIV-1 (Top). Individual domains are MA, matrix; CA, capsid; NC, nucleocapsid; RT, reverse transcriptase; RN, RNase H; IN, integrase. Black arrows indicate specific sites of cleavage by PR. The green (TFR) and red (PR) bars denote the protease precursor mimetic used in the present studies. Autoprocessing occurs either stepwise in the order as listed (red arrows denoting sites 1, 2, and 3) at pH < 5 or directly at site 3 at pH < 5. Proposed mechanism for the processing of a model precursor comprising the TFR, PR, and truncated ΔRT domains (Bottom) (1, 2). Ovals indicate folded monomers in the transient precursor dimer, PR-ΔRT and mature PR (2, 6, 8). Transitioning ovals from light red to fully red depict appearance of catalytic activity and stable dimer formation of PR (6).
Fig. 2.
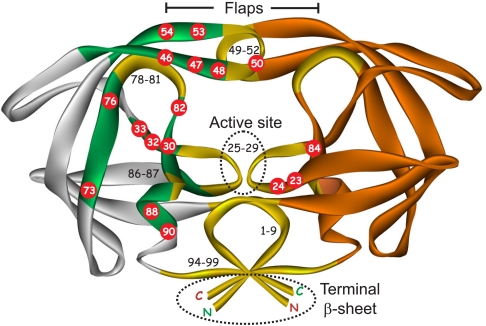
Ribbon representation of mature PRM structure showing the location of highly conserved regions under drug pressure (gold and black lettering) (25), regions of natural variability in PR among the four groups M, N, O, and P (gray), and naturally conserved regions where major DRMs are selected under drug pressure (green). Numbered red circles indicate the positions of major DRMs, as defined in the Stanford database and the red lettering in Fig. S1C. Dimer interfaces are shown in dotted ovals.
A plausible pathway for the regulation of the protease emerges considering several in vivo and in vitro studies of precursor processing (Fig. 1) (1, 2). The PR that is flanked by the transframe region (TFR) and the reverse transcriptase (RT) at its N and C termini, respectively, in the Gag-Pol precursor presumably adopts a tertiary monomer fold spanning at least residues 10–90 (2). Prior to the cleavage at its N terminus (TFR/PR site), which precedes the C-terminal site (PR/RT), the dimer dissociation constant (Kd) of the protease is high and likely modulated by the TFR (Fig. 1) (2, 6). This high Kd could be essential to allow effective recruitment of polyproteins at the plasma membrane, prior to the onset of polyprotein processing (7). Dimer formation of the PR precursor occurs transiently to initiate N-terminal autoprocessing facilitated by interface interactions of at least the active site and the C-terminal residues and possibly stabilized further by the interaction of the N-terminal TFR/PR cleavage site sequence with the active-site and flap residues (Fig. 2) (2, 8). In the context of a Gag-Pol precursor, autoprocessing may proceed by stepwise cleavages from the farthest accessible site (i.e., p2/NC), followed by an internal TFR site, to the N terminus of PR (TFR/PR) (Fig. 1) (6, 9). However, appearance of full catalytic activity concomitant with stable dimer formation (Kd < 10 nM) (2) is consistent with a rate-limiting intramolecular (cis) cleavage at the TFR/PR junction (1, 6). Subsequent cleavages, at the C-terminal PR/RT site and other sites within the Gag-Pol, likely occur intermolecularly (trans) upon the release of a fully active PR (1, 2). Based on this pathway, we propose that perturbing the release of the active mature PR will adversely affect later steps in Gag and Gag-Pol maturation. Thus, the folding, transient dimerization and N-terminal autocatalytic cleavage steps present attractive targets for possible new antiviral agents.
In spite of a multitude of studies to improve the binding affinity of inhibitors to the mature PR and its DRMs, little is understood concerning inhibition of the protease prior to autoprocessing. To date four genetically distinct groups (M, N, O, and P) of HIV-1 have been identified, of which by far the predominant cause of infections worldwide is group M (10). The PR flanked at its N terminus by the TFR represents an ideal system to study its maturation without the complications that arise from using the entire Gag-Pol polyprotein containing multiple cleavage sites (Fig. 1). However, the depletion due to intrinsic autoprocessing of TFR-PR precursor during expression in Escherichia coli results in very low recovery of the protein after purification. Using these small quantities, details of the kinetics and pH dependent processing of the group M (TFR-PRM) and group N (TFR-PRN) precursors in vitro have been elucidated, as shown schematically in Fig. S1A (1, 2, 11).
Here we describe the use of PIs, added to the culture medium during the expression of PR precursors in E. coli, to block their autocatalytic conversion to the mature PR. This approach has led to (1) a simple, small-scale method to assess the efficacy of inhibition of autoprocessing of natural variants and DRM precursors and (2) the ability to accumulate sufficient quantities of these purified precursors to permit kinetics measurements with inhibitors in vitro and physical studies of the precursor-inhibitor complexes. Unique properties of the PR precursors as compared to the mature proteases are described, and implications of these findings are discussed.
Results and Discussion
The rationale for choosing the precursor constructs (TFR-PR, Fig. 1, green and red bars) for our analyses was based on their ability to undergo efficient autoprocessing when expressed in E. coli. It is also based on the assumption that under drug pressure, DRMs of the protease selected either after several passages in culture or after prolonged treatment of patients evolve to evade inhibition and propagate, and thus are not compromised in their catalytic activity for carrying out both autoprocessing and the various cleavages within the Gag and GagPol polyproteins required for viral maturation. Three DRMs were chosen out of a dozen constructs we screened for expression and autoprocessing activity.
Five precursor constructs and their corresponding mature proteases were used in this study (see Materials and Methods). Representative sequences of the mature proteases of groups M through P and three multi-DRMs are shown in Fig. S1C. The TFR is highly divergent in sequence within group M and among the various groups, ranging in length from 52–61 amino acids (Fig. S1B), whereas as expected, several regions within the mature PR domain are highly conserved (Fig. S1C). Fig. 2 shows the highly conserved and the variable regions in colors matching those shown for the sequences in Fig. S1C. It is noteworthy that most major DRMs (with the exception of I50 variants) are selected in regions between the naturally variable and the highly conserved regions, as shown in red circles (Fig. 2). In spite of the wide variation in the catalytic activity of mature PRM, PRN, PRM-11, PRM-P51, and PRM-20 (Table S1), all their precursor proteins undergo efficient processing with negligible residual precursor remaining after expression, consistent with efficient selection of DRMs (Figs. 3 and 4; 0 μM PI or control lanes). The DRMs PRM-11, PRM-P51, and PRM-20 exhibited 4–12 fold lower affinity for substrate (larger Km) than the wild-type PRM, which was partly compensated by the higher turnover rate of the enzyme-substrate complex (kcat) for PRM-20 but not for PRM-51. The natural variants TFR-PRP, which showed incomplete maturation upon expression in E. coli (Fig. 3D, lane 0), and TFR-PRO, which exhibited poor expression, were not pursued further.
Fig. 3.
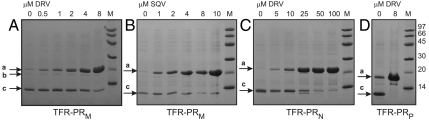
Inhibition of autoprocessing of protease precursors derived from natural variants, M (TFR-PRM), N (TFR-PRN), and P (TFR-PRP), by DRV and SQV in E. coli and analyses by SDS/PAGE. Letters a, b, and c correspond to the full-length precursor, intermediate precursor, and the mature protease, respectively, as shown in the schematic in Fig. S1A for TFR-PRM. M denotes standards in kDa.
Fig. 4.
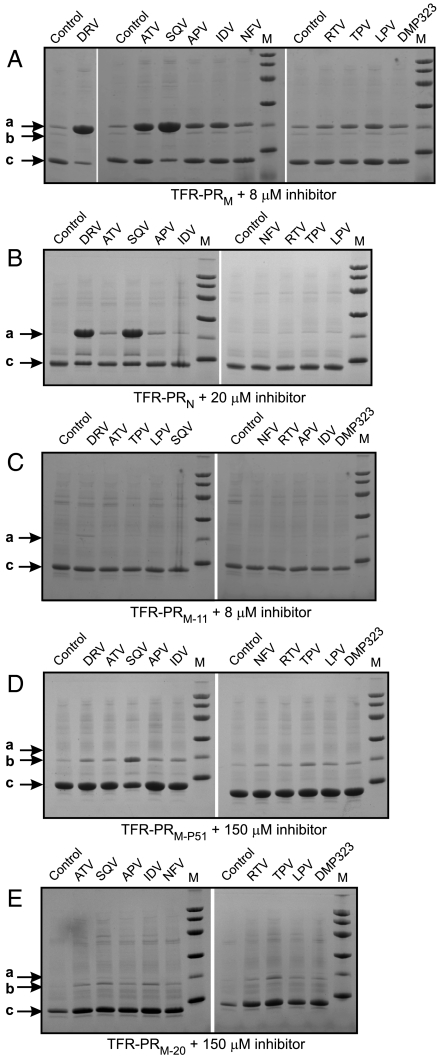
Evaluation of inhibition of autoprocessing of precursors derived from natural variants M and N (A and B) and three DRMs of group M (C–E) by nine clinical PIs and a symmetric inhibitor DMP323 (12). M denotes standards as indicated in Fig. 3. Abbreviations for clinical PIs are as listed in http://hivdb.stanford.edu/pages/drugSummaries.html.
Control experiments showed that PIs provided in the medium do not affect the rate of growth of E. coli BL21-DE3 either in the absence or presence of the plasmid encoding the wild-type TFR-PRM precursor (see Fig. S1C legend). Thus, expression of the various precursors was followed by the facile isolation of the proteins for SDS/PAGE. The optimized protocol to monitor autoprocessing and its inhibition is reproducible and semiquantitative, and it does not involve immunoblotting to visualize the proteins. Upon expression in E. coli the precursor undergoes maturation in a single step at the TFR/PR site to release the mature PR as illustrated in Fig. S1A for processing above pH 5 (upward arrows). Fig. 3 A and B shows the concentration dependence of inhibition of TFR-PRM autoprocessing in E. coli by two inhibitors. Very little conversion of the precursor to mature PRM is observed at the highest concentration of 8 μM DRV or 8–10 μM SQV provided in the medium. Autoprocessing of TFR-PRP is almost completely inhibited by 8 μM DRV (Fig. 3D). The dose-dependent inhibition of TFR-PRP was not assessed because complete processing was not observed even in the absence of inhibitor.
The effects of all nine clinical inhibitors of the mature PR in current use, and the symmetrical inhibitor DMP323 (12), on the processing of TFR-PRM were assessed at 8 μM inhibitor concentration under the same conditions (Fig. 4A). The strongest inhibition of TFR-PRM autoprocessing was observed with SQV and DRV, with slightly weaker inhibition by ATV of only TFR-PRM. These three inhibitors exhibit tight binding to mature PRM as indicated by the increase (ΔTm = Tm difference in the presence and absence of PI) of 19.3–22.4 °C in the thermal denaturation temperature upon inhibitor binding (13). However, SQV is a weaker binder than DRV to mature PRM as indicated by its approximately 40-fold greater inhibitor dissociation constant (400 pM as compared with ≤ 10 pM) (14, 15). Relative to TFR-PRM, processing of TFR-PRN (Figs. 3C and 4B) is somewhat less responsive to inhibition by DRV and SQV, requiring inhibitor concentrations of > 20 μM, and was not inhibited by ATV at this concentration. The weaker response of TFR-PRN relative to TFR-PRM (5–10 fold for DRV) follows a trend similar to that observed with the mature enzymes such that PRN exhibits a dissociation constant (KL) for DRV of 0.12 nM (11), approximately 10-fold larger than KL ≤ 0.010 nM for PRM (15).
Expression of precursor constructs corresponding to three drug-resistant PRM mutants derived from clinical isolates or by selection by PI exposure in cell culture (Fig. S1) was assessed in E. coli with the panel of 10 inhibitors essentially as described for the wild-type precursor. None of the mutant precursors were inhibited by any of the PIs at 8 μM concentration, as shown for TFR-PRM-11 (Fig. 4C). The highly drug-resistant mutant, TFR-PRM-20 was not inhibited by any PI even at high concentrations (Fig. 4E). In the presence of 150 μM of each inhibitor, with the exception of TPV (tested at 75 μM because of poor solubility), relative band intensities indicated that ≥90% of the protein was converted to mature PRM-20. No significant accumulation of the full-length precursor was detected with any of the inhibitors. TFR-PRM-P51 was also unresponsive to most of the PIs and only partially inhibited by SQV at 150 μM concentration (Fig. 4D). The intermediate molecular weight protein (b) shown in Fig. 4 D and E that migrates slightly faster than expected for the full-length TFR corresponds to the product of the F8/L9 cleavage within the TFR (cf. Fig. S1A). A comparable site was shown to be less responsive to inhibition than the TFR/PR cleavage that produces the mature, active PRN (11). Similarly the p2/NC cleavage, which was reported to occur preceding the internal TFR cleavage under specific conditions, could be far less responsive to inhibition (2, 9). The unresponsiveness of these sites to inhibition by PIs could be explained by the entropic advantage of these cleavages, which presumably occur via intramolecular processes (2).
The dependence of inhibition of processing on DRV concentrations up to 100 μM was systematically examined for wild-type TFR-PRM, TFR-PRN, and three DRMs (Fig. 5, data quantified from gels shown in Fig. 3C and Fig. S2) under identical conditions. For TFR-PRM, half maximal inhibition in the E. coli system, as indicated by precursor accumulation, is observed at approximately 1–2 μM (Fig. 3A), and virtually complete inhibition occurs in the presence of 10 μM DRV. Similar inhibition by DRV observed on the autoprocessing of a chimeric precursor in mammalian cells monitored by immunoblotting (16) confirms that inhibition of the native TFR-PR constructs in E. coli provides a rapid and reliable prediction of the effect of PIs on autoprocessing. This effect is in the same range as the maximum concentrations (6–10 μM) of DRV in plasma (17) or of SQV in cells (18) achieved on administration of PIs to human subjects. By contrast, the DRM TFR-PRM-11 required between 10 and 25 μM DRV for half maximal inhibition, and TFR-PRM-20 and TFR-PRM-P51 were not significantly inhibited by DRV at the highest concentrations tested, consistent with the single point assays shown using 150 μM DRV in Fig. 4 D and E. The relative susceptibilities of these mutants to DRV inhibition are consistent with known PI-resistance profiles for specific major DRMs according to the Stanford PI-resistance database and the International AIDS Society–USA panel for antiretroviral drug resistance (4). TFR-PRM-20 contains four major mutations (V32I, I47V, 154L, and I84V) plus an accessory (minor) mutation (L33F) associated with DRV resistance and treatment failure (19). Interestingly, TFR-PRM-P51 obtained by selection in cell cultures by DRV bears mutations at four of these sites (V32I, L33F, I54M, and I84V). TFR-PRM-11, from a patient failing IDV therapy, contains three major DRMs, two of which (G73S and I84V) are associated with both DRV and IDV resistance. All three mutants also bear other DRMs as defined in (4). Reduced DRV response appears to require three or more DRV-resistance mutations accompanied by a high background of other DRMs (19).
Fig. 5.
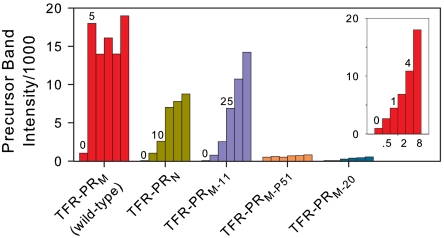
Comparison of dose-response profiles for inhibition of autoprocessing of natural variants and DRMs by DRV. Integrated intensities of the precursor bands (Fig. 3C for TFR-PRN and Fig. S2) at 0, 5, 10, 25, 50, and 100 μM DRV are shown for each precursor. Intensity data for TFR-PRM were corrected for a twofold lower concentration applied to the gel relative to the other samples. The inset shows data from Fig. 3A for TFR-PRM. DRV concentrations (μM) are indicated above or below the bars.
We assessed if any one of a few chosen major DRMs (shown as red circles in Fig. 2) is critical to drug resistance in the ensemble of mutations. Revertants of TFR-PRM-20 were constructed and analyzed for the inhibition of autoprocessing by DRV. The results demonstrate that any single (N30D, I32V, V84I) or double (N30D/I32V, V47I/L54I, D88N/M90L) revertant mutation in TFR-PRM-20 did not restore inhibition of autoprocessing by 100 μM DRV or SQV (Fig. S3). I32V and D88N/M90L were mildly affected in their processing, and N30D/I32V more significantly so, even in the absence of inhibitor (Fig. S3), but no discernible inhibition was observed in the presence of the inhibitor. This result clearly supports the clinical observations that combinations of major DRMs selected in conjunction with mutations in the variable regions produce a coordinated effect of extreme drug resistance as observed in PRM-P51 and PRM-20, but collectively these mutations preserve the autoprocessing activity crucial for viral maturation and propagation.
Autoprocessing is dependent on dimer formation of the precursor which was suggested to occur transiently in its initial phase from studies employing enzyme kinetics and NMR (1, 2, 8). The apparent Kd for TFR-PRM has been estimated from kinetic experiments to be at least 130-fold larger than for the mature PR (6). As it is impossible to accumulate TFR-PRM-P51 and TFR-PRM-20 precursors for isolation and comparison with the wild-type TFR-PRM, Kds were evaluated only for the corresponding mature proteases as described (11). Mature proteases PRN, PRM-11, PRM-P51, and PRM-20 all exhibited larger values of the Kd relative to PRM (Table S1). Although larger Kd values for the mature enzymes might be expected to correlate with a reduced tendency to form the transient dimer of the precursors, required for initiating autoprocessing and inhibitor binding, the magnitudes of Kd did not appear to correlate with the IC50 for inhibition of autoprocessing of the corresponding precursors.
The protocol described here permits the isolation and detailed study of PR precursors that were previously extremely difficult to obtain due to their intrinsic autoprocessing during expression. A major goal of the present work has been to compare their responses to PIs with those of the corresponding mature enzymes, and to assess the effect of DRMs on these responses. In order to use the data obtained in E. coli as a semiquantitative measure of precursor binding/inhibition by the PIs it was necessary to establish a relationship between their IC50 values in E. coli and those determined under defined, similar conditions in vitro. The lowest protein concentration at which in vitro autoprocessing reactions can be reliably measured by SDS/PAGE on Tris-Tricine gels and detection by Coomassie staining is approximately 1 μM dimer (approximately 500 ng/lane) because of detectability limits and also because the high Kd of the precursor may compromise the rate of the autoprocessing reaction even in the absence of inhibitor. Assessment of the dose-response profile for DRV with wild-type TFR-PRM indicated an IC50 ≤ 1 μM for DRV, such that stoichiometric titration of the protein occurs even at the lowest practical concentration of enzyme and inhibitor, and thus a reliable estimate of the IC50 could not be obtained under our conditions. Consequently, binding of a weaker inhibitor, SQV, to the mutant TFR-PRM-11, which was expected to have an affinity in the conveniently measurable range, was chosen for a comparative dose-response study in vitro. There is reasonable agreement between the IC50 values measured in vitro and in vivo for inhibition of autoprocessing of TFR-PRM-11 by SQV (Fig. S4). Near neutral pH, the IC50 in vitro is approximately 25–50 μM. A two- to fourfold higher IC50 value observed in E. coli could be due to different intracellular conditions or because the precursor is being continuously produced rather than present at a fixed initial concentration. Similarly, while inhibition of autoprocessing of TFR-PRN by SQV in E. coli occurs in the range of 5–10 μM, inhibition in vitro was observed at 1–2 μM (Fig. S4). Thus we conclude that IC50 values for inhibitors measured in the E. coli system follow the same trend and are slightly higher than the values estimated in vitro.
Table 1 summarizes the data for DRV and SQV binding to mature PRM, the natural variant PRN and three DRMs, as measured by ITC from literature values or this work, and their comparison with observed IC50 values for the corresponding precursors. For the mature enzymes, inhibitor dissociation constants (KL) for DRV are in the sub-nM to low nM range whereas the IC50 values for the precursors are 103–105 fold larger, suggesting a drastic difference in the affinities of DRV for the mature and precursor proteins. A significant contribution to this difference may result from overcoming the less-favorable monomer-dimer equilibrium for the precursor in order to form the ternary dimer-PI complex. For example, DRV, which was specifically designed for exceptionally tight binding to the mature dimeric protease, has a KL for PRM that is 1–2 × 105 fold smaller (15, 20) than its IC50 for inhibition of the precursor. For PRM-11 and its precursor the difference is similar (2 × 105). The differences are significantly smaller for SQV inhibition, with the largest difference being approximately 5 × 103-fold for PRM and its precursor, largely resulting from poorer inhibition of the mature protease by SQV relative to DRV that is not reflected in its inhibition of the precursor. The next largest difference (approximately 2.7 × 103) for inhibition mediated by SQV is between TFR-PRM-P51 and PRM-P51. An estimate for the inhibition of TFR-PRM-20 and TFR-PRM-P51 autoprocessing is hard to achieve because of the practical limitation of DRV and SQV concentrations (approximately 150 μM) in the medium. For DRV inhibition, only lower limits were obtained for the IC50, which may be expected to be in the high μM or mM range based on the difference in the KL values observed for mature PRM compared to PRM-20 or PRM-P51, showing increases of approximately 8,000-fold (see Table 1). It is noteworthy that the TFR-PRM-P51, an extreme DRV DRM, is significantly sensitive to inhibition of autoprocessing by SQV (Fig. 4D, SQV lane). Thus, understanding the atomic details of interactions of precursor-PI complexes may help to develop and/or identify compounds that are specific for inhibiting autoprocessing with higher affinity.
Table 1.
Comparison of inhibition of mature wild-type and DRM proteases (KL) and their precursors (IC50) by DRV and SQV based on dose-response experiments and ITC
| Construct | DRV |
SQV |
||
| Mature protease |
Precursor |
Mature protease |
Precursor |
|
| KL, nM | ∼IC50, nM | KL, nM | ∼IC50, nM | |
| PRM/TFR-PRM | 0.005-0.01* | 1,000 | 0.4 ± 0.01† | 2,000 |
| PRN/TFR-PRN | 0.12‡ | 10,000 | nd | 10,000§ |
| PRM-11/TFR-PRM-11 | 1.6 ± 0.7 | 30,000 | 2,330 ± 264† | 100,000 |
| PRM-P51/TFR-PRM-P51 | 37 ± 5 | > 150,000 | 54 ± 7 | 150,000§ |
| PRM-20 and TFR-PRM-20 | 41 ± 1 | > 250,000¶ | 930 ± 93 | > 150,000 |
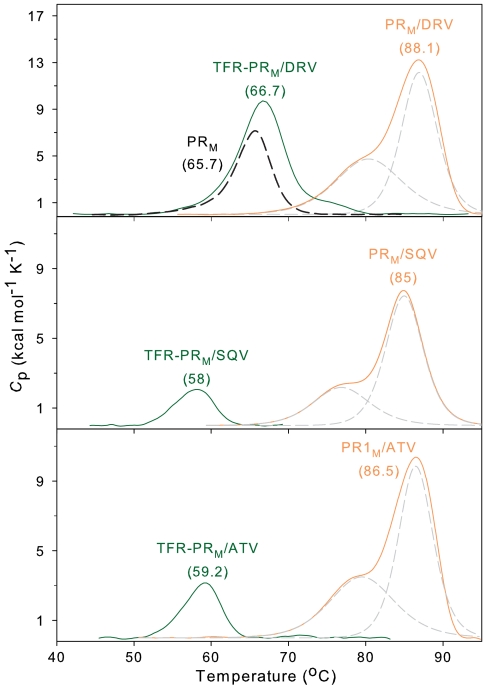
DSC experiments in the absence of an inhibitor are not possible with precursors because significant processing of the folded precursor occurs rapidly within 10 min (6, 21). However, when folded in the presence of active-site-directed inhibitors, TFR-PR precursors are expected to form a ternary complex based on NMR observations of inactive TFR-PRD25N precursor (2) as well as mass determination of the active TFR-PRN precursor in the presence of DRV by size exclusion chromatography coupled with multiangle light scattering/refractive index measurements (11). An inactive precursor construct with an active-site mutation that allows DSC in the absence of PI is not a valid model to evaluate inhibitor binding because the D25N mutation drastically reduces the binding affinity of mature PRM to DRV by approximately 106-fold (5). The large differences in Tm (ΔTm) between uninhibited mature proteases and their complexes with inhibitors (Fig. 6 and Fig. S5 and Table S2) provide an estimate of the relative strength of inhibitor binding (5, 13) to the mature enzymes. Importantly, observed Tms for precursor–DRV complexes are drastically lower than for the corresponding mature PR–DRV complexes, as shown in Fig. 6 for TFR-PRM and PRM. The differences in Tm (mature protease minus precursor as their dimer/PI complexes), Tm.diff, reflect the much weaker binding interaction and dimer stabilization of the precursors by inhibitors as compared to the mature proteases. Similar effects of DRV were observed for PRN (Fig. S5). For TFR-PRM/SQV the Tm is lower and the Tm.diff is larger than that for TFR-PRM/DRV, indicating that TFR-PRM/SQV may be less stable than TFR-PRM/DRV. Consistent with this observation, substantial processing of TFR-PRM occurred in vitro in the presence of 1 μM SQV, whereas 1 μM DRV completely blocked processing. SQV is a better inhibitor of TFR-PRM-P51 autoprocessing than DRV with an IC50 of ∼150 μM (see Table 1 and Fig. 4D). Binding of these two inhibitors to mature PRM-P51 is similar, as shown by KL values that differ by a factor of < 2 (Table 1) and a difference in ΔTm of < 1 °C for its DRV and SQV complexes (Table S2). In contrast, the complex of DRV with mature PRM is significantly tighter than for SQV as shown by its 30- to 40-fold smaller KL (14, 20) and its 3 °C larger ΔTm (13).
Fig. 6.
Comparison of the thermal stabilities of the precursor (TFR-PRM) and the mature protease (PRM) complexed to PIs and PRM (dashed black) in the absence of PI. Data for PRM (Top) are from (5, 13) for comparison. DSC scans were carried out using approximately 14 μM PRM and 8–10 μM TFR-PRM (as dimers) in the presence of approximately twofold molar excess of DRV, SQV, or ATV. Tm values (°C) are shown in parentheses. For TFR-PRM bound to DRV, SQV, and ATV, values of Tm.diff (Tm of mature PR-PI complex minus Tm of TFR-PRPI complex) are 20.2, 27.0, and 27.3 °C, respectively. Dashed gray lines depict the deconvolution of the two apparent transitions discussed in ref. 5.
Concluding Remarks
Autoprocessing to release the fully functional mature protease is indispensable for polyprotein processing, HIV maturation and propagation. Although studies of interactions of PIs with drug-resistant mature HIV-1 proteases have been widespread, to date little has been described about interactions of inhibitors with the corresponding precursors. Development of a simple protocol has now permitted rapid assessment of the inhibition of maturation of both natural precursor variants and DRMs by available PIs, paving the way for potential applications in screening of candidate drugs that target maturation of specific drug-resistant TFR-PR mutants.
Extremely tight binding to the wild-type mature PR has been achieved with pM inhibitors such as DRV and SQV. These highly stable complexes exhibit Tms of 85–88 °C. In contrast, these PIs form complexes with TFR-PR precursors with much lower thermal stability (Tms of 58–67 °C), and their IC50 values for inhibition of autoprocessing are 103–105 times larger than their binding constants to the mature proteases. In particular, autoprocessing of two of the DRM precursors examined was not inhibited by any PI in current clinical use, even at high micromolar concentrations. We suggest that inability of PIs to inhibit autoprocessing of these mutant precursors may contribute significantly to treatment failure in the clinic. Thus, possible structure-based design of PIs with stronger affinity to the active site of the precursor or as dimerization inhibitors of the autoprocessing reaction merits consideration as an alternative new approach to overcome drug resistance. Despite significant research efforts (2), dissociative inhibitors of mature PR have no clinical applications to date, in part because of the requirement for extremely tight binding to compete with the strong interaction (low Kd) between the monomer units of the mature enzyme, in contrast to the transient dimers formed by the precursors, which are likely to exhibit significantly higher Kds (2, 6, 8). Thus, design of inhibitors that interfere with precursor dimerization may provide a more practical approach.
Materials and Methods
Protein Designations.
In the present study, the precursor and mature PR derived from Group M (subtype B-HXB2) are designated as TFR-PRM and PRM (6), respectively, and similarly, from HIV-1 group N as TFR-PRN and PRN (11). All DRMs were derived from group M, and their precursors bear the B-HXB2 TFR sequence flanking the N terminus of the PR as shown in Fig. S1B. Two DRMs are designated according to the number of mutations in the PR domain as PRM-11 (14) and PRM-20 (22) for the mature proteases and as TFR-PRM-11 and TFR-PRM-20 for their precursors. A DRV resistant mutant (23) is designated as PRM-P51 and TFR-PRM-P51 accordingly.
Expression Vectors.
Genes encoding the seven precursor constructs, four natural variants, and three DRMs were synthesized and cloned in pET11a vector between Nde1 and BamH1 sites. Site-directed mutagenesis of TFR-PRM-20 was carried out using the Quik-Change mutagenesis kit (Agilent Technologies). Expressed constructs were verified by both DNA sequencing and mass spectrometry.
Small-Scale Cultures for Screening Inhibition of Autoprocessing During Expression in E. coli.
Small-scale cultures (0.5–1 ml) of E. coli bearing the appropriate construct were grown in the absence or presence of added PIs. Protein expression was induced at an optical density of 0.7 for a period of 2 h with 2 mM isopropyl thiogalactoside, following which the cells were harvested and lysed by sonication. The expressed protein was partially purified and analyzed by SDS/PAGE on 20% homogeneous PhastGels (GE Healthcare). Compositions of the media were kept constant.
Preparation of Purified Mature Proteases and Precursor Proteins.
To allow isolation of sufficient quantities of purified precursor proteins for DSC and kinetic studies of the autoprocessing reactions in vitro, cultures were scaled up to a total volume of 100 ml in the presence of PI. Mature proteases derived from cultures grown in the absence of PI, and protease precursors were both purified from inclusion bodies as described (11).
Characterization by Kinetics and Calorimetry.
The kinetic parameters, kcat and Km, and the Kds, for the mature proteases were determined in 50 mM sodium acetate, pH 5, 0.25 M NaCl using a spectrophotometric assay as described (11). Values are listed in Table S1.
For DSC, precursors were folded by addition of 5 mM sodium acetate buffer, pH 5.3 with an approximately twofold molar excess of inhibitor in the quench buffer. Mature proteases were folded similarly or by the quench protocol as described previously (final pH 5.0) (24) in the absence or presence of the same molar excess of inhibitor. Thermal denaturation scans using a MicroCal VP-DSC microcalorimeter (GE Healthcare) and data handling were as described previously (5, 13). Values of the Tm (Table S2) were determined from the maxima of the transitions.
Inhibitor binding to mature PR mutants in 50 mM sodium acetate buffer, pH 5, at 28 °C was assessed using a MicroCal high-precision iTC200 titration calorimeter (GE Healthcare).
Supplementary Material
Acknowledgments.
This research was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (NIH), and the Intramural AIDS-Targeted Program of the Office of the Director, NIH, and Grant GM062920 from the NIH. We are grateful to Ad Bax for reading and conveying critical comments. PIs used in this study were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH, and DMP323 from DuPont Pharmaceuticals.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102278108/-/DCSupplemental.
References
- 1.Louis JM, Weber IT, Tozser J, Clore GM, Gronenborn AM. HIV-1 protease: Maturation, enzyme specificity, and drug resistance. Adv Pharmacol. 2000;49:111–146. doi: 10.1016/s1054-3589(00)49025-3. [DOI] [PubMed] [Google Scholar]
- 2.Louis JM, Ishima R, Torchia DA, Weber IT. HIV-1 Protease: Structure, dynamics, and inhibition. Adv Pharmacol. 2007;55:261–298. doi: 10.1016/S1054-3589(07)55008-8. [DOI] [PubMed] [Google Scholar]
- 3.Mitsuya H, Maeda K, Das D, Ghosh AK. Development of protease inhibitors and the fight with drug-resistant HIV-1 variants. Adv Pharmacol. 2008;56:169–197. doi: 10.1016/S1054-3589(07)56006-0. [DOI] [PubMed] [Google Scholar]
- 4.Johnson VA, et al. Update of the drug resistance mutations in HIV-1: December 2009. Top HIV Med. 2009;17:138–145. [PubMed] [Google Scholar]
- 5.Sayer JM, Liu F, Ishima R, Weber IT, Louis JM. Effect of the active-site D25N mutation on the structure, stability and ligand binding of the mature HIV-1 protease. J Biol Chem. 2008;283:13459–13470. doi: 10.1074/jbc.M708506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louis JM, Clore GM, Gronenborn AM. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat Struct Biol. 1999;6:868–875. doi: 10.1038/12327. [DOI] [PubMed] [Google Scholar]
- 7.Haraguchi H, et al. Intracellular localization of human immunodeficiency virus type 1 Gag and GagPol products and virus particle release: Relationship with the Gag-to-GagPol ratio. Microbiol Immunol. 2010;54:734–746. doi: 10.1111/j.1348-0421.2010.00276.x. [DOI] [PubMed] [Google Scholar]
- 8.Tang C, Louis JM, Aniana A, Suh JY, Clore GM. Visualizing transient events in amino-terminal autoprocessing of HIV-1 protease. Nature. 2008;455:693–696. doi: 10.1038/nature07342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pettit SC, Everitt LE, Choudhury S, Dunn BM, Kaplan AH. Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism. J Virol. 2004;78:8477–8485. doi: 10.1128/JVI.78.16.8477-8485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roques P, et al. Phylogenetic characteristics of three new HIV-1 N strains and implications for the origin of group N. AIDS. 2004;18:1371–1381. doi: 10.1097/01.aids.0000125990.86904.28. [DOI] [PubMed] [Google Scholar]
- 11.Sayer JM, Agniswamy J, Weber IT, Louis JM. Autocatalytic maturation, physical/chemical properties, and crystal structure of group N HIV-1 protease: Relevance to drug resistance. Protein Sci. 2010;19:2055–2072. doi: 10.1002/pro.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamazaki T, et al. Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy. Protein Sci. 1996;5:495–506. doi: 10.1002/pro.5560050311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sayer JM, Louis JM. Interactions of different inhibitors with active-site aspartyl residues of HIV-1 protease and possible relevance to pepsin. Proteins. 2009;75:556–568. doi: 10.1002/prot.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muzammil S, Ross P, Freire E. A major role for a set of non-active site mutations in the development of HIV-1 protease drug resistance. Biochemistry. 2003;42:631–638. doi: 10.1021/bi027019u. [DOI] [PubMed] [Google Scholar]
- 15.King NM, et al. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J Virol. 2004;78:12012–12021. doi: 10.1128/JVI.78.21.12012-12021.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L, Chen C. Autoprocessing of human immunodeficiency virus type 1 protease miniprecursor fusions in mammalian cells. AIDS Res Ther. 2010;7:27. doi: 10.1186/1742-6405-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scholler-Gyure M, et al. Pharmacokinetics of darunavir/ritonavir and TMC125 alone and coadministered in HIV-negative volunteers. Antivir Ther. 2007;12:789–796. [PubMed] [Google Scholar]
- 18.Ford J, et al. Intracellular and plasma pharmacokinetics of saquinavir-ritonavir, administered at 1,600/100 milligrams once daily in human immunodeficiency virus-infected patients. Antimicrob Agents Chemother. 2004;48:2388–2393. doi: 10.1128/AAC.48.7.2388-2393.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKeage K, Perry CM, Keam SJ. Darunavir: A review of its use in the management of HIV infection in adults. Drugs. 2009;69:477–503. doi: 10.2165/00003495-200969040-00007. [DOI] [PubMed] [Google Scholar]
- 20.Brower ET, Bacha UM, Kawasaki Y, Freire E. Inhibition of HIV-2 protease by HIV-1 protease inhibitors in clinical use. Chem Biol Drug Des. 2008;71:298–305. doi: 10.1111/j.1747-0285.2008.00647.x. [DOI] [PubMed] [Google Scholar]
- 21.Louis JM, Wondrak EM, Kimmel AR, Wingfield PT, Nashed NT. Proteolytic processing of HIV-1 protease precursor, kinetics and mechanism. J Biol Chem. 1999;274:23437–23442. doi: 10.1074/jbc.274.33.23437. [DOI] [PubMed] [Google Scholar]
- 22.Dierynck I, et al. Binding kinetics of darunavir to human immunodeficiency virus type 1 protease explain the potent antiviral activity and high genetic barrier. J Virol. 2007;81:13845–13851. doi: 10.1128/JVI.01184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koh Y, et al. In vitro selection of highly darunavir-resistant and replication-competent HIV-1 variants by using a mixture of clinical HIV-1 isolates resistant to multiple conventional protease inhibitors. J Virol. 2010;84:11961–11969. doi: 10.1128/JVI.00967-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishima R, Torchia DA, Louis JM. Mutational and structural studies aimed at characterizing the monomer of HIV-1 protease and its precursor. J Biol Chem. 2007;282:17190–17199. doi: 10.1074/jbc.M701304200. [DOI] [PubMed] [Google Scholar]
- 25.Ceccherini-Silberstein F, et al. Identification of the minimal conserved structure of HIV-1 protease in the presence and absence of drug pressure. AIDS. 2004;18:F11–F19. doi: 10.1097/01.aids.0000131394.76221.02. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.