Abstract
MicroRNAs and siRNAs interact with target sequences in mRNAs, inducing cleavage- and non-cleavage–based gene repression through the RNA-induced silencing complex (RISC) that consists of one of four mammalian Argonaute proteins, Ago1–Ago4. The process of how Dicer substrate small hairpin RNAs (shRNAs) are loaded into different mammalian Agos in vivo is not well established. Here we report that shRNAs are loaded into mammalian Agos in two stepwise processes, physical association and activation, with the latter being the rate-limiting step with noncleaving RISC. We establish that, although RNA duplexes processed from shRNAs bind to Agos in cells with similar affinity, the degree by which the complexes are activated (coupled with the removal of the passenger strand) correlates with the thermodynamic instability of RNA duplexes being loaded rather than the structure of the RNA, as was previously demonstrated in Drosophila. Interestingly, Ago loading of siRNAs is less sensitive to thermostability than that of their shRNA equivalents. These results may have important implications for the future design of RNAi-based therapeutics.
Keywords: RNA interference, gene silencing
MicroRNAs (miRNAs) and 21- to 23-nt siRNAs regulate gene expression in almost all eukaryotic organisms. Small RNA–mediated gene regulation is sequence-specific and believed to regulate at least one-half of all protein-encoding mRNAs (reviewed in refs. 1–4).
Argonaute proteins (Agos), which directly associate with small RNAs, are the core of all known RNA-induced silencing complexes (RISCs). These proteins are functionally specialized into distinct RNA silencing pathways. In Drosophila, Ago2-RISC is involved in target mRNA cleavage, and Ago1-RISC mediates translational repression (5, 6). The human genome encodes four Ago-like subfamily proteins (Ago1–Ago4) that have redundant functions as negative regulators of gene expression, but only Ago2 has cleavage activity (7–9).
In Drosophila, siRNAs and miRNAs are actively sorted into functionally distinct Ago-RISCs based on differences in structure rather than on their biogenesis (10, 11). Perfectly matched duplexes (siRNA-like) are preferentially incorporated into Ago2, and the loading is achieved by cleaving the passenger strand, whereas duplexes with central mismatched bulges (miRNA-like) are sorted to Ago1, and the unselected strand (asterisk strand) is eliminated through a “bypass” pathway, which is sensitive to the structure of the small RNA duplex and independent of the weak cleavage activity of Ago1 (12). A similar sorting mechanism exists in Caenorhabditis elegans, whereby small RNA duplexes with perfectly matched or bulged stems are channeled into RDE-1 and ALG-1, respectively (13). In plants, a different sorting mechanism has been reported. The composition and phosphorylation status of the 5′ nucleotide of small RNA duplexes influences their binding by the different Agos (14, 15). In mammalian cells, several studies have shown that miRNAs are similarly associated with four human Agos (7, 8, 16), suggesting the absence of a specific sorting mechanism.
Similar to what has been reported in flies (12, 17–20), mammalian Ago loading is a stepwise process (21, 22). First, small RNAs associate with Agos as a duplex with the help of chaperon proteins and consume ATP, presumably like their Drosophila homologs (23, 24). Then RISC is activated by converting the associated duplex RNA into a single strand (22). Although Ago2 can accomplish the activation step by cleavage of the passenger or asterisk strand RNA (21, 25), the mechanism of how the noncleaving Agos (Ago1/3/4) perform this step is not clear. Although previous reports (7, 8, 16) indicate that siRNAs and miRNAs associate with the four mammalian Agos indiscriminately, it is not clear whether an exogenously expressed small hairpin RNA (shRNA) would behave the same. More importantly, how the RISC assembly/activation process and RISC function are interrelated in vivo is not known.
Here, we investigated the loading mechanism of mammalian noncleaving Agos in Ago2 KO mouse embryonic fibroblast (MEF) cells (7) by using immunoprecipitation (IP) approaches to study the physical association between small RNAs and Agos, together with various functional tests based on well-designed reporter systems. We found that shRNAs with different structures associate with all four human Agos in a similar pattern. The conversion from inactive RISC (Ago associated with RNA duplex) to active RISC [Ago associated with single-stranded RNA (ssRNA)] is the rate-limiting step and is greatly influenced by the inherent thermodynamic stability of the RNA duplex being loaded. At the same time, the cleavage activation pathway specific to Ago2 is insensitive to duplex RNA thermostability.
Results
Small RNAs Processed from shRNAs with Different Stem Structures Associate Mammalian Agos in a Similar Pattern.
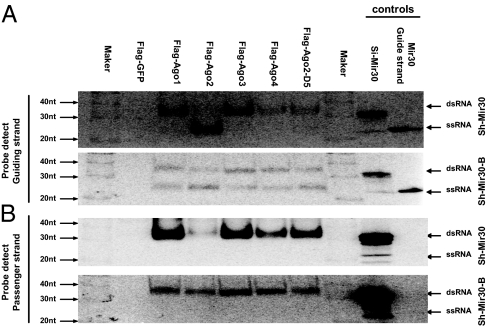
We investigated whether Ago loading in mammals was similar to what was observed in Drosophila, in which small RNAs were differentially sorted into various RISCs based on their structures (10, 11). To do this, we measured the association between mammalian Agos and small RNAs processed from shRNAs with different stem structures by IP. Two U6-driven shRNAs (sh-Mir30 and sh-Mir30-B) that generate the same guide-strand sequence with a different stem structure (Figs. 1 and 2C) were coexpressed with various Flag-tagged Agos or a control Flag-GFP expression cassette in Ago2 KO MEF cells. Western blotting experiments established that the Flag-tagged version of human Ago1, Ago2, and the two cleavage-deficient mutants, Ago2-D5 and Ago2-D6 (7), were expressed in nearly equivalent amounts in MEF cells (Fig. S1). Although the expression level of the original human Flag-Ago3 and Flag-Ago4 cDNA clones was low, they reached levels similar to the Flag-Ago1/2 clones after their codon sequences were optimized (Fig. S1). Quantification of Ago-containing sh-Mir30/sh-Mir30-B RNAs was determined 24 h posttransfection by anti-Flag IP and Northern blotting. The pull-down results were confirmed to be Ago-specific by the lack of signal in the GFP control (Fig. 1). Both the guide and passenger strands from either the bulged-stem or perfect-stem shRNAs were slightly enriched in Ago3/4 compared with Ago1/2(Fig. 1 A and B). Consistent with previous publications (7, 8, 16), endogenously expressed miRNA let-7 and mir-21 showed a similar affinity with all Agos (Fig. 1C) in the same experiments and were not enriched with the Ago3/4 pull-down as observed with the exogenously expressed shRNAs. The increase in enrichment for exogenously expressed small RNAs may imply that Ago3/4 can play a general role in the regulation of overly abundant small RNAs. Nonetheless, the association pattern of sh-Mir30 into different mammalian Agos was similar to that of sh-Mir30-B, indicating the stem structures of small duplex RNAs did not play a critical role in determining the association affinity between small RNAs and different Agos. In contrast to what was shown in Drosophila, small RNAs processed from shRNAs with different structures were not sorted into different Agos in mammalian cells.
Fig. 1.
Small RNAs processed from shRNAs with different stem structures associate mammalian Agos with similar pattern. (A and B) Perfect-stem sh-Mir30 (A) or bulged-stem shRNA-Mir30-B (B) coexpressed with either Flag-GFP (negative control) or various Flag-Agos in Ago2 KO MEF cells. At 24 h posttransfection, IP experiments with anti-Flag antibody were performed on cells lysates. RNA extracted from either 20% of input or IP were run on 10% polyacrylamide/7 M urea denaturing gels. Guide (bold and italic) and passenger-strand RNAs from each sample were identified by sequential Northern blotting. The intensity of bands was measured by phosphorimaging. The pull-down efficiency shown in the figure (in black) was calculated by dividing the IP signal by total input. The mean and SD of pull-down efficiency from three independent experiments are also provided (in italic). (C) Endogenous miRNAs let-7 and mir-21 were identified by using a similar IP/Northern blotting approach.
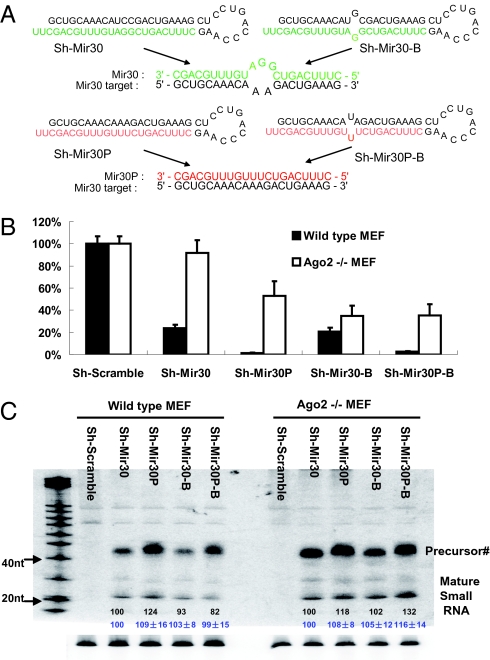
Fig. 2.
The overall efficiency of Ago loading can be monitored by reporter assay in vivo. (A) Schematic illustration of various shRNAs and the interactions between target and guide-strand sequences. Notably, sh-Mir30 and sh-Mir30-B generate the same guide strand (green) that mismatches to the mir-30 targets, whereas sh-Mir30P and sh-Mir30P-B produce an identical guide strand (red) that match perfectly as a mir-30 target. (B) psiCHECK vector with four tandem mir30 target sites in the 3′ UTR were cotransfected with various shRNAs in either wild-type or Ago2 KO MEF cells. Dual-luciferase assays were performed at 24 h posttransfection. Firefly-luciferase activities were normalized with RL-luciferase, and the percentage of relative enzyme activity compared with the negative control (treated with sh-scramble) was plotted. Error bars represent the SD from 20 independent experiments, each performed in triplicate transfections. (C) Wild-type or Ago2 KO MEF cells were cotransfected with plasmids as described above. sh-RNAs expressed from U6-driven cassette were detected by Northern blotting. Notably, the size of the band labeled as Precursor# is shorter than expected. The quantitative values of mature strands are shown in black numbers together with the mean and SD from three independent experiments (in blue).
Interestingly, although both strands generated from the sh-Mir30 expression cassette were pulled down with Ago1/3/4 and Ago2-D5 [an Ago2 mutant deficient of slicer activity (7)], the guide, but not the passenger strand, was pulled down with Ago2 (Fig. 1A). Moreover, the association of both strands in Ago2-D5 established that the failure to detect the passenger strand in Ago2 was not because of inherent differences, such as strand bias, between Ago2 and Ago1/3/4 but likely a consequence of Ago2 cleavage of the passenger strand after or during loading (Fig. 1A). Notably, although the sh-Mir30-B passenger strand was detectable in the Ago2 pull-down, it was less abundant compared with what was found in the Ago1/3/4/D5 pull-downs (Fig. 1B). This observation indicates that Ago2 may be able to cleave the passenger strand during loading even if there is a bulge in the shRNA stem caused by the position of the mismatch in duplex stem, which may perturb but not eliminate cleavage of the passenger strand.
Overall Efficiency of Ago Loading Can Be Monitored by Reporter Assay in Vivo.
To quantify the amount of biologically active ssRNA-associated RISC, a direct product of the Ago loading process, we used a well-characterized reporter system in which four tandem mir-30 target sites were inserted in the 3′ UTR of the Renilla (RL)-luciferase reporter gene together with various shRNA-expressing plasmids. The plasmids expressing sh-Mir30 and sh-Mir30-B, as described previously, generated the same guide strands that were mismatched with the target and varied in their hairpin/stem structure (Fig. 2 A and C). In addition, we constructed sh-Mir30P and sh-Mir30P-B, which vary in their hairpin/stem structures but produce the same guide strands with perfect complementarity to the target (Fig. 2 A and C). Because each pair of shRNAs generates the same guide-strand sequence, the efficiency of shRNA-mediated repression on the reporter will be a direct measurement of the amount of active RISC from a particular shRNA.
In wild-type MEF cells, the bulged stem shRNAs (sh-Mir30-B and sh-Mir30P-B) resulted in a similar reduction in transgene expression compared with their perfectly matched stem shRNA (sh-Mir30 and sh-Mir30P) counterparts (Fig. 2B). However, in the absence of Ago2, and thus in the exclusive presence of noncleaving Agos (Ago2 KO MEF cells), introduction of a bulge in the shRNA stem substantially increased the repressive activity of sh-Mir30-B compared with sh-Mir30 while only moderately increasing the activity of sh-Mir30P-B compared with sh-Mir30P (Fig. 2B). The same result was found even when the Ago 2 KO cells were complemented with the noncleaving Ago2-D5 mutant (Fig. S2). Despite the fact that we demonstrated sh-Mir30 and sh-Mir30-B were similarly associated with noncleaving Agos (Fig. 1), the amounts of biologically active RISC resulting from these two shRNAs were dramatically different. Importantly, this variation did not result from differences in the levels of mature small RNAs generated from sh-Mir30 and sh-Mir30-B expression plasmids (Fig. 2C), suggesting that the improvement in RISC loading rather than shRNA loop processing/maturation is the reason for the higher amount of active noncleaving RISC (containing Ago1/3/4) from sh-Mir30-B.
Activation/Unwinding Is the Rate-Limiting Step of Noncleaving Agos Loading.
As demonstrated above, less functional RISC programmed with sh-Mir30 was generated than that with sh-Mir30-B. The discrepancy between the association data that was measured by IP (Fig. 1) and the functional data that was monitored by reporter assay (Fig. 2) strongly suggested that the activation/unwinding process, and not the association step, was the rate-limiting process for mammalian Ago loading. We reasoned that, although both shRNAs equally associated with the Agos, a large proportion of the poorly loaded sh-Mir30 RNA would exist as a duplex, whereas more of the sh-Mir30-B RNA would be present as a single-stranded guide RNA in noncleaving Agos. To directly test this idea, we analyzed the same Ago-IP samples with a native gel providing a means to distinguish between cellular Ago-associated ssRNAs and duplexed RNAs (Fig. 3). The single-band signal found in each IP sample detected in the denaturing gel (Fig. 1 A and B) was resolved into two bands in the native gel (Fig. 3). Using siRNA duplex and single-stranded guide-strand RNAs as controls, we confirmed that the faster-running band represented the single-stranded small RNA in activated RISC, whereas the slower-running band corresponded to the small RNA as a duplex in likely inactive RISC (Fig. 3). Just as our model predicted, the poorly loaded sh-Mir30 RNAs associated with noncleaving Agos (Ago1/3/4) were almost exclusively found as passenger/guide-strand duplexes, whereas the less stable sh-Mir30-B associated with noncleaving Agos was more prominently found as single-stranded guide-strand RNA (Fig. 3A). In contrast to the differences in the sh-Mir30 and sh-Mir30-B guide strands, virtually all of the passenger strands from both shRNAs found associated with the noncleaving Agos were present as duplex RNA forms (Fig. 3B). These results were consistent with the idea that the guide and passenger strands of sh-Mir30-B become incorporated into the noncleaving Agos as duplexes but that only the guide and not the passenger strand becomes incorporated into the active RISC. The nonincorporated passenger strands were likely degraded rapidly, resulting in the observed higher steady-state concentration of Ago-associated guide strands.
Fig. 3.
Activation/unwinding is the rate-limiting step of noncleaving Ago loading. IP samples from Fig. 1 were separated on polyacrylamide native gels. Then, 20 fmol synthetic RNAs (si-Mir30 duplex and single-stranded guide-strand RNA of Mir-30) were also loaded on the gel as controls. Guide (A) and passenger-strand (B) RNAs were identified by sequential Northern blotting. Notably, the si-Mir30 control ran as a mixture of dsRNAs and trace amount of ssRNAs. The marker - RNA Decade (Ambion).
To further support the idea that the passenger strands were indeed truly double-stranded and not in an active RISC, we set up a reporter assay to determine the amount of active RISC containing a single-stranded passenger strand. To do so, we constructed a psiCHECK-based reporter system in which the four original target sequences were replaced by four custom target sequences specific to the passenger strands. As predicted, no noticeable gene repression was observed when various shRNAs were coexpressed with this reporter (Fig. S3). The lack of passenger strand–mediated gene repression confirmed the high prevalence of passenger strand as nonfunctional duplex RNAs in noncleaving Agos.
The native gel results, combined with our earlier observations, strongly support the stepwise loading model of noncleaving Agos. More important, although association of the duplex is independent of the sequence/structure, our results indicate that activation is the rate-limiting step of RISC function.
Noncleaving Ago Loading Efficiency Is Determined by Thermodynamic Stability Rather Than by Stem Structure of shRNAs.
Next, we investigated whether the stem structure of the small RNAs being loaded was a critical factor in the rate-limiting step of RISC activation, explaining the observed difference in RISC function between sh-Mir30 (perfectly matched stem) and sh-Mir-30-B (bulged stem) (Fig. 2B). To extend this result to a second shRNA sequence, we replaced the mir-30 target sites with the Drosophila bantam miRNA target sequences and designed two pairs of shRNAs based on bantam sequences (26) (Fig. 4A). We carried out similar cotransfection studies using coexpressed shRNAs with either perfect or bulged stems in wild-type and Ago2 KO MEF cells (Fig. 4A). In contrast to the mir-30 results, either marginal (sh-Bantam vs. sh-Bantam-B) or no (sh-BantamP vs. sh-BantamP-B) difference in the repression efficiency in the MEF KO cells was observed when a bulge was introduced into the stem of shRNAs (Fig. 4B).
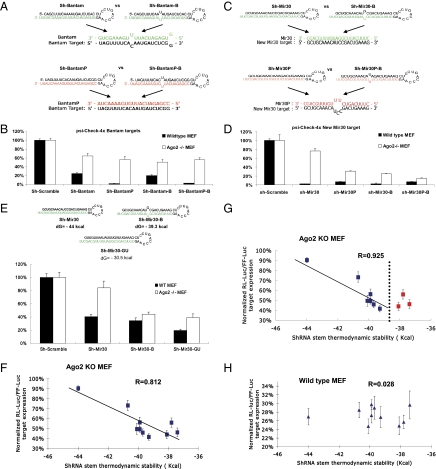
Fig. 4.
RISC function of noncleaving Agos is determined by thermodynamic stability rather than by stem structure of shRNAs. (A) Design of various shRNAs and their associations with targets based on Drosophila miRNA bantam sequence. (B) Wild-type or Ago2 KO MEF cells were cotransfected with the psiCHECK reporter with four tandem bantam target sites in the 3′ UTR and a plasmid expressing various shRNAs. Dual-luciferase assays were performed at 24 h posttransfection, and the results were plotted as described previously. Error bars represent the SD from three independent experiments, each performed in triplicate. (C) Schematic illustration of various shRNAs and interactions between target and guide-strand sequences. Different from Mir30 targets presented in Fig. 2, sh-Mir30 and sh-Mir30-B generate the same guide strand (green), which was a perfect match to the new-mir-30 targets, whereas sh-Mir30P and sh-Mir30P-B produced an identical guide strand (red), which was a mismatch to the new-mir-30 target. (D) psiCHECK vector with four tandem new-mir30 target sites in the 3′ UTR were cotransfected with various shRNA in either wild-type or Ago2 KO MEF cells. Dual-luciferase assays were performed at 24 h posttransfection, and results are plotted as described previously. Error bars represent the SD from three independent experiments, each performed in triplicate transfections. (E) psiCHECK-4xMir30 reporter constructs were cotransfected with the newly designed sh-Mir30-GU, which shares the same guide strand as sh-Mir30 and sh-Mir30-B (green), in either WT or Ago2 KO MEF cells. Results were plotted as described above. Error bars represent the SD from three independent experiments, each performed in triplicate. (F) psiCHECK-4xMir40 reporter constructs were cotransfected with sh-Mir30-3 to sh-Mir30-19 (Fig. S4) individually in Ago2 KO MEF cells. The thermodynamic stability of each shRNA was predicted by mfold (37, 38). The whole sequence, including the loop, was taken into the calculation as an ssRNA. The normalized luciferase reporter level (sh-scramble–treated sample was set as 100%) vs. the shRNA stem thermodynamic stability was plotted. A strong correlation between shRNA-mediated repression efficiency and stem thermodynamic stability was observed (R = 0.812). The more stable the stem, the less repression observed for a particular shRNA via the noncleaving Agos in Ago2 KO MEF cells. More detailed information can be found in Fig. S5 and Table S1. (G) A higher correlation (R = 0.925) was observed when shRNAs with a thermostability lower than a potential threshold (dotted line) were excluded (red squares). (H) The same experiments were performed in wild-type MEF cells. No correlation (R = 0.028) was observed. The results were plotted as described above. Each data point in F and H was the average of 12 trials (four independent experiments, each performed in triplicate) ± SD. The folding energies of shRNAs were also calculated without the stem loop sequences (only mature small RNA duplexes sequences after Dicer processing); the R values were 0.887 and 0.117, respectively.
These results led us to question the role of the stem structure as the major determinant factor in the noncleaving Ago loading process. Another interesting observation was the striking difference in noncleaving Ago-mediated transgene knockdown between sh-Mir30 and sh-Mir30P, even though they contained the same perfect stem structure (Fig. 2). However, because sh-Mir30 and sh-Mir30P have a different hybridization between their respective guide strand and the same target sequence, we designed studies to eliminate this variable as a possible explanation for differential knockdown activity by noncleaving Agos. Specifically, we replaced the original target sequences within the target 3′ UTR with four custom tandem sequences designed to provide a perfect match with the guide strand of sh-Mir30/sh-Mir30-B but mismatched with the guide strands of sh-Mir30P/sh-Mir30P-B, respectively (Fig. 4C). Importantly, the repression mediated by sh-Mir30P was still higher compared with that directed by sh-Mir30 (Fig. 4D), strongly suggesting that the reduced functionality of sh-Mir30 was a result of the intrinsic duplex sequence rather than of the structure of the stem.
Because the thermodynamic stability of sh-Mir30 predicted by mfold (dG = −44 Kcal/mol) was significantly greater than that of sh-Mir30P (dG = −38.7 Kcal/mol), we elected to determine whether shRNAs with less stable stems would be more effectively loaded by noncleaving Agos. To do so, we designed a custom shRNA (sh-Mir30-GU) that shared the same guide strand as sh-Mir30/sh-Mir30B by replacing all of the C residues with U residues in the passenger strand (Fig. 4E). The G:U pairs between the guide and passenger strands allow sh-Mir30-GU to share the same perfectly matched stem structure as sh-Mir30 RNA but contain an even lower thermodynamic stability than sh-Mir30-B RNA. The increased repression mediated by the sh-Mir30-GU in Ago2 KO MEF cells (Fig. 4E) further established that the thermodynamic stability, and not the structure of the stem per se, was the determining factor sensed by noncleaving Agos during the loading process.
Finally, we designed a number of different shRNAs that shared the same guide-strand sequences but contained single-base bulges at various positions within the passenger strand (Fig. S4). Cotransfection of the reporter and the shRNAs in Ago2 KO MEF cells showed a strong correlation (R = 0.812) between the repression efficiency and shRNA thermodynamic stability (Fig. 4F). Interestingly, shRNAs with folding energy greater than −38.5 kCal/mol had an even higher correlation coefficient (R = 0.925) (Fig. 4G), suggesting that there may be a specific folding energy threshold at which point the Ago loading process will not be further enhanced. In addition, the repression results obtained from the other three pairs of shRNA (sh-Mir30P vs. sh-Mir30P-B, sh-Bantam vs. sh-Bantam-B, and sh-BantamP vs. sh-BantamP-B) were consistent with the comparative thermodynamic results when the threshold value (−38.5 kCal/mol) was taken into consideration (Table S1). Our experiments established that it is the thermodynamic stability rather than the bulge structure itself that correlates with noncleaving Ago loading, presumably at the activation/unwinding step. Of note, no correlation (R = 0.028) was observed when parallel experiments were performed in wild-type MEF cells (Fig. 4H), indicating that Ago2’s loading process was insensitive to the thermodynamic stability.
Loading of siRNAs Is Less Sensitive to Thermodynamic Stability Than That of shRNAs.
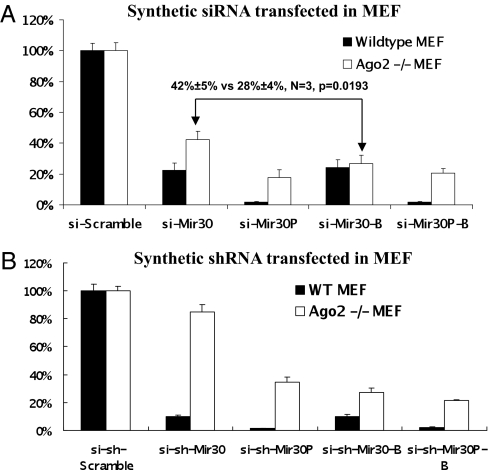
To ask whether synthetic siRNA duplexes were loaded into RISC in a manner similar to their shRNA counterparts, we designed siRNA duplexes based on the mir-30 shRNA sequences, with one extra mismatch at the 5′ end of the guide strand to favor RISC incorporation (Fig. S6). When cotransfection studies were carried out in Ago2 MEF cells, an overall similar pattern of reporter gene knockdown was observed when comparing these siRNA duplexes to their corresponding shRNA equivalents. Interestingly, si-Mir30 and si-Mir30-B, which differ in their thermodynamic stability just as their shRNA counterparts do, resulted in a much lesser difference in target repression in Ago2 KO MEF cells (Fig. 5A). To rule out that this discrepancy was a result of differences in the intracellular biogenesis of small RNAs (chemically synthesized vs. expressed), synthetic shRNAs (si-shRNAs) were designed and transfected into cells with the same concentration of siRNAs counterparts. Thermostable synthetic shRNA (si-sh-Mir30) paralleled the repressive activity of the expressed sh-Mir-30, but not si-Mir30, strongly suggesting that the differences in transgene expression repression observed between the siRNA and shRNAs were attributable to differences in siRNA/shRNA processing/maturation (Fig. 5B). This observation further implies that siRNAs may bypass the RISC loading complex (RLC) and join RISC directly, as opposed to shRNAs that undergo an obligatory Dicer processing step before Ago loading.
Fig. 5.
The loading of siRNAs is less sensitive to thermodynamic stability than that of shRNAs. (A) Wild-type or Ago2 KO MEF cells were cotransfected with psiCHECK reporter with four tandem mir30 target sites in the 3′ UTR and various synthetic siRNA duplexes. Dual-luciferase assays were performed at 24 h posttransfection, and the results are plotted as described previously. Error bars represent the SD from three independent experiments, each performed in triplicate. (B) The same experiment was performed with synthetic shRNAs, the sequences of which were exactly as illustrated in Fig. 2A.
Discussion
Our results provide genetic and cellular confirmation of a previously reported model (reviewed in ref. 27) that suggests that mammalian Ago loading is divided into two distinct processes, physical association and activation. We demonstrated that vector-expressed shRNAs, similar to endogenous miRNAs, were not sorted into Ago-specific RISCs in mammals but rather become physically associated with all Agos with similar affinity. This process is followed by a “maturation” step during which the passenger strand is eliminated. We established that activation and not RNA association is the rate-limiting step in Ago loading. Importantly, the efficiency of the cleavage-independent activation process (unwinding) is substantially enhanced by the thermodynamic instability of the duplex for noncleaving Agos. Alternatively, cleavage-dependent activation (cleavage of passenger/star strand) is evoked when Ago2-RISC is occupied by a completely or near-completely base-paired stem (Fig. 1A). This Ago2-specific activation pathway is apparently robust and insensitive to duplex thermodynamic stability. Although unstable shRNAs were more effectively loaded and in turn resulted in greater repression by noncleaving Agos, this effect was easily masked in wild-type MEF cells where Ago2 was present (Figs. 2B and 4H).
This model is further strengthened by a recent study (22) using a different biochemical approach. Ago complexes isolated from cells mixed with radio-labeled siRNAs underwent conversion from a duplex to ssRNA state confirming the activation/unwinding step. However, unlike their cell-free studies, we did not observe a dependence on structure as the determining factor in either the association or activation step. Instead, we show that in vivo the activation step of Ago loading is the rate-limiting step, which is dependent on the thermostability of the RNA duplexes.
A recent report using a cell-free reconstitution system indicated that mammalian Ago1 has weak cleavage activity (28), which was not detected by our biochemical approach possibly because of the sensitivity of our assay. Nonetheless, such weak activity would provide Ago1 an advantage in loading/activating perfect-stem RNA duplexes compared with Ago3/4, consistent with the observation that Ago1/2 can mediate better repression on mismatched target sequences compared with Ago3/4 only when perfect-stem shRNAs were used (9).
In mammals, the RLC contains Dicer, the transactivation response RNA(TAR) binding protein TRBP, and individual Agos (29). The mechanistic detail of how the RLC facilitates Ago loading is still unknown. It is generally believed that the miRNA duplexes are released from Dicer after processing and handed off to RISC to initiate the Ago loading process. In support of this idea, it is of note that the mature RNA sequences, but not the precursors, were enriched in the Ago pull-down experiments (Fig. 1), indicating that the association between Agos and precursor RNAs within the RLC were weak and indirect in relation to the association between Agos and mature small RNAs. However, the detection of Ago2-cleaved precursor miRNA (ac-pre-miRNA), an intermediate product with an intact loop and cleaved passenger strand (30), indicates that the processing of the hairpin loop and elimination of the passenger strand are highly coupled processes, suggesting a possible role of Dicer in Ago loading.
The majority of mammalian miRNAs contain an extensive bulge structure in their stems, making the thermodynamic stability lower than the threshold that might be sensed by the Ago loading process. As a result, most of the endogenous miRNAs can be effectively loaded into mammalian Agos by the cleavage-independent unwinding pathway. Given the fact that Ago2 is more active in the miRNA repression pathway, it is intriguing to ask why miRNAs are being effectively loaded into Ago1/3/4. Perhaps subtle differences in the thermodynamic stability provide Ago-miRNA hierarchies, especially if Agos are limiting.
A helicase activity was predicted to be responsible for the separation of two strands during Ago loading in cleavage-independent unwinding. Consistent with this idea, multiple helicases, including p68, p72, RNA helicase A, RCK/p54, TNRC6B, Gemin3/4, and Mov10, have been identified as factors associated with RLC and may potentially function in RISC loading (31–35). However, a recent in vitro reconstitution assay indicated the lack of helicase requirement during RISC activation (22). Our results favor the latter scenario because it is unlikely that an enzymatic unwinding process would be sensitive to the thermostability of target duplexes. In fact, we showed that the unwinding process of Dicer substrate shRNAs was more sensitive to thermostability than that of their siRNA counterparts. One intriguing model to explain these results is that there are two pathways involved in the Ago-mediated unwinding of an RNA duplex. In Dicer-dependent loading (shRNAs and presumable miRNA precursors), no or only a weak inherent helicase activity of Dicer itself (36) would predominate making this process depend more on duplex RNA thermostability. In contrast, Dicer-independent loading of siRNAs may bypass the RLC and join RISC as ssRNA with the help of endogenous helicases. These two pathways are not mutually exclusive, and the process used may also rely on which Ago the small RNA is being loaded onto. This result would also explain why perfectly complementary siRNAs cannot be unwound by recombinant noncleaving Agos themselves in vitro (22) yet remain highly functional in vivo (Fig. 5B).
Whereas our report provides mechanistic insights into how small duplex RNAs can modulate gene expression in mammals, the study provides strong support that the thermodynamic stability of shRNA/siRNAs will be a factor to consider in future therapeutic applications. Moreover, our findings that siRNAs and shRNAs with virtually the same duplex sequences can be differentially loaded into RISC might explain why there is not always concordance between siRNA and shRNAs designed to target the same sequence. Together, our results will also provide insights into how to design potent si/shRNAs with minimal off-target effects for biological knockdown of important genes and/or treatment of diseases.
Materials and Methods
Agos Co-IP of Small RNAs.
In brief, Ago2 KO MEF cells were cotransfected with plasmids expressing Flag-tagged Agos and various shRNAs. At 36 h posttransfection, immunoprecipitations from cell extracts were performed using anti-Flag M2 agarose beads. Small RNAs associated with Agos were analyzed by either native or denaturing gels and Northern blotting.
See SI Methods for additional details.
Supplementary Material
Acknowledgments
We thank Dirk Haussecker for critically reading the manuscript, Greg Hannon for the MEF Ago 2 KO cells, and Carl Novina for discussions. This work was supported by grants from the National Institutes of Health's National Institute of Allergy and Infectious Diseases (to M.A.K.) and National Institute of Diabetes and Digestive and Kidney Diseases (to M.A.K.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018023108/-/DCSupplemental.
References
- 1.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 5.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 6.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 8.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Förstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct Argonaute complexes after production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- 13.Steiner FA, et al. Structural features of small RNA precursors determine Argonaute loading in Caenorhabditis elegans. Nat Struct Mol Biol. 2007;14:927–933. doi: 10.1038/nsmb1308. [DOI] [PubMed] [Google Scholar]
- 14.Mi S, et al. Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montgomery TA, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 16.Azuma-Mukai A, et al. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci USA. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 18.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 20.Kim K, Lee YS, Carthew RW. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA. 2007;13:22–29. doi: 10.1261/rna.283207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoda M, et al. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyoshi T, Takeuchi A, Siomi H, Siomi MC. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol. 2010;17:1024–1026. doi: 10.1038/nsmb.1875. [DOI] [PubMed] [Google Scholar]
- 24.Iwasaki S, et al. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell. 2010;39:292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 25.Shin C. Cleavage of the star strand facilitates assembly of some microRNAs into Ago2-containing silencing complexes in mammals. Mol Cells. 2008;26:308–313. [PubMed] [Google Scholar]
- 26.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 27.Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35:368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Wang B, et al. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol. 2009;16:1259–1266. doi: 10.1038/nsmb.1712. [DOI] [PubMed] [Google Scholar]
- 29.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diederichs S, Haber DA. Dual role for Argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 31.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 32.Meister G, et al. Identification of novel Argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 33.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salzman DW, Shubert-Coleman J, Furneaux H. P68 RNA helicase unwinds the human let-7 microRNA precursor duplex and is required for let-7-directed silencing of gene expression. J Biol Chem. 2007;282:32773–32779. doi: 10.1074/jbc.M705054200. [DOI] [PubMed] [Google Scholar]
- 35.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 36.Soifer HS, et al. A role for the Dicer helicase domain in the processing of thermodynamically unstable hairpin RNAs. Nucleic Acids Res. 2008;36:6511–6522. doi: 10.1093/nar/gkn687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.